Molecular and Structural Basis of the Proteasome α Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer
Abstract
:1. Introduction
2. Results and Discussion
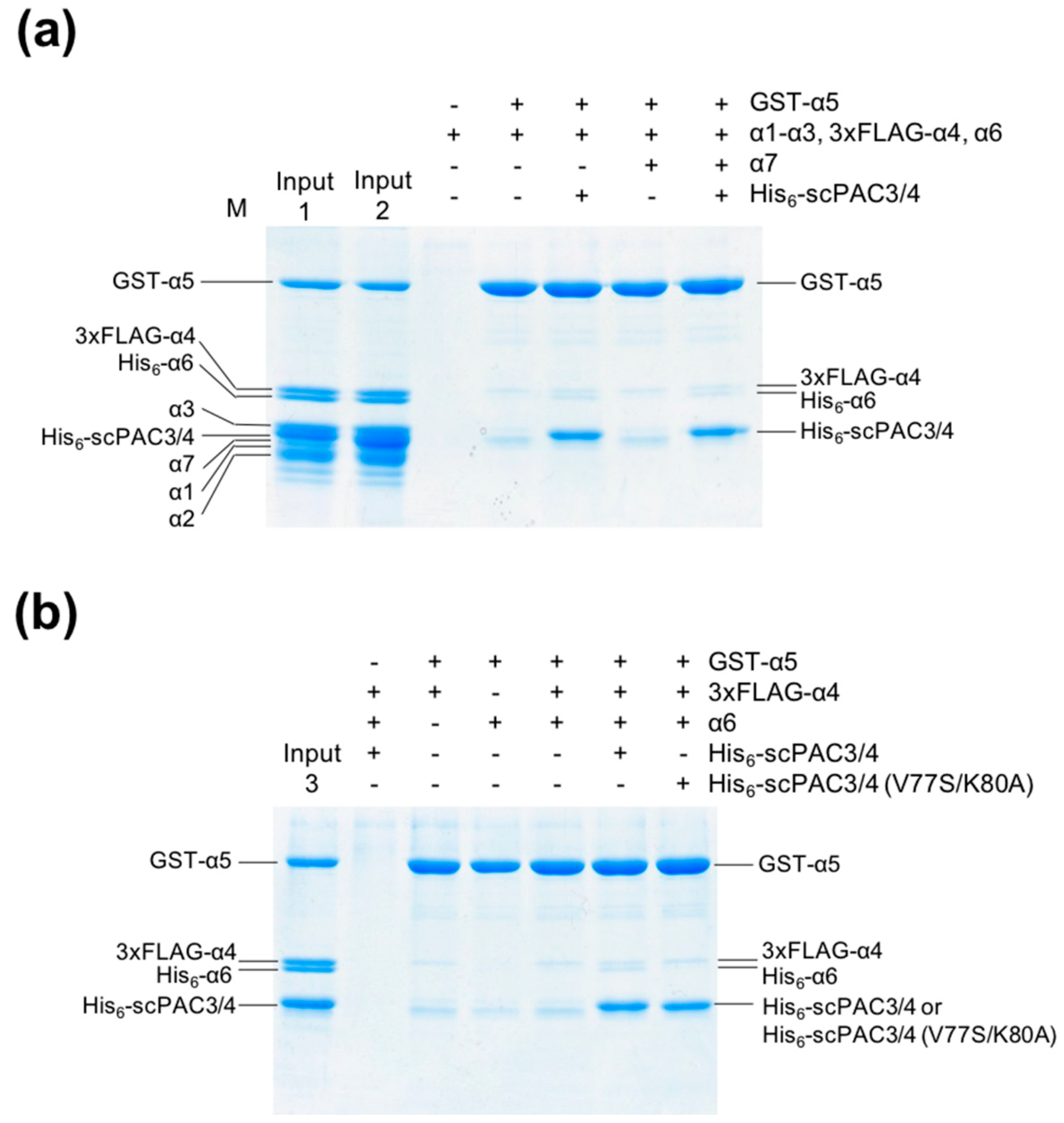
2.1. The PAC3-PAC4 Heterodimer Interacts Primarily with α5
2.2. The PAC3-PAC4 Heterodimer Acts as Molecular Matchmaker in α4-α5-α6 Assembly
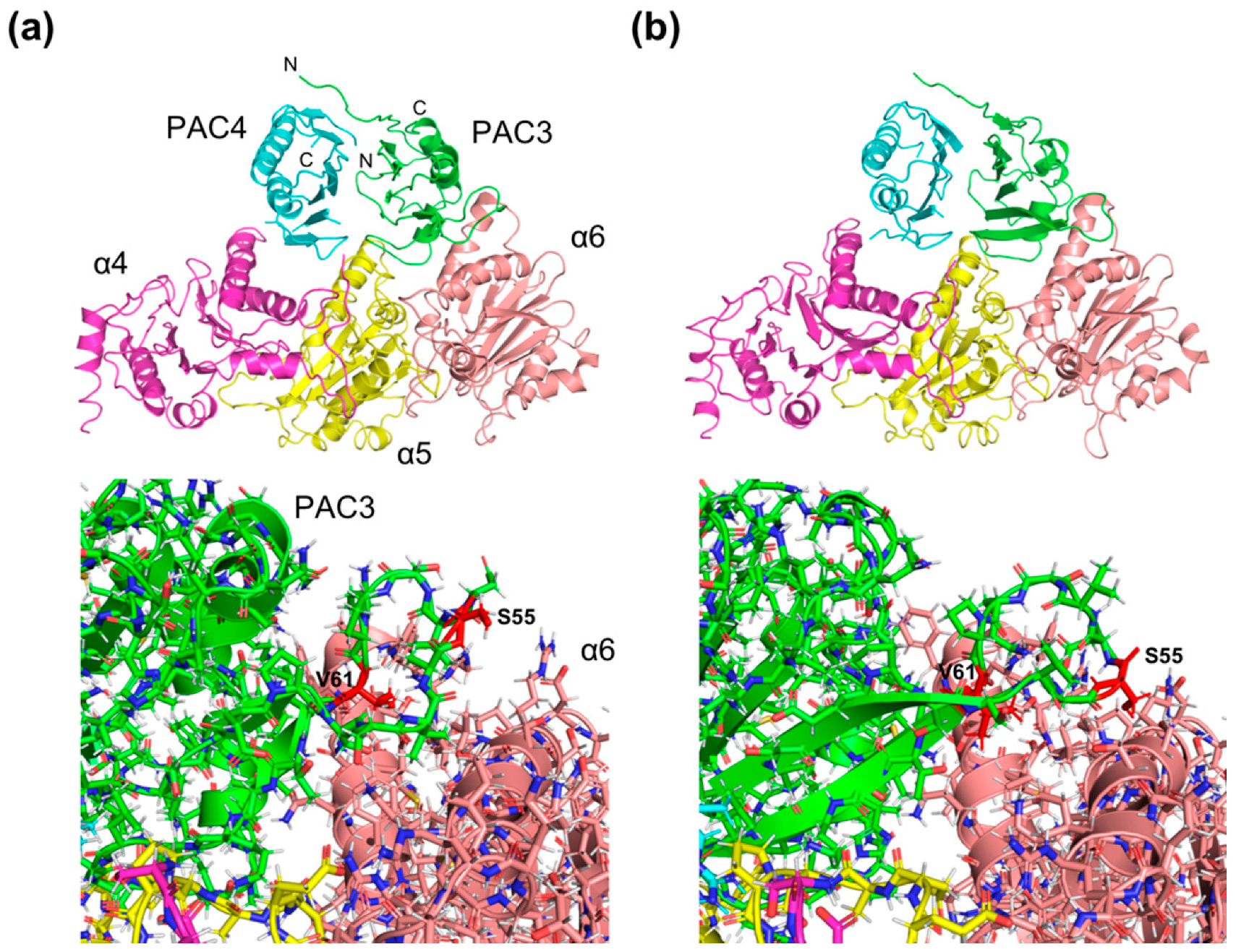
2.3. Structural Insights into the Mechanisms Underlying PAC3/PAC4-Dependent α4-α5-α6 Assembly
3. Methods
3.1. Sample Preparation
3.2. Pull-Down Experiments
3.3. Crystallization, X-ray Data Collection, and Structure Determination
3.4. Computer-Aided Model Building
3.5. NMR Spectroscopy
3.6. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CBB | Coomassie Brilliant Blue |
| CP | Core particle |
| GST | Glutathione S-transferase |
| HSQC | Heteronuclear single-quantum correlation |
| NMR | Nuclear magnetic resonance |
| NOE | Nuclear Overhauser effect |
| PAC | Proteasome-assembling chaperone |
| RP | Regulatory particle |
| SC | Single-chain |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
References
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef] [Green Version]
- Baumeister, W.; Walz, J.; Zuhl, F.; Seemuller, E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell 1998, 92, 367–380. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murata, S.; Yashiroda, H.; Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009, 10, 104–115. [Google Scholar] [CrossRef]
- Kish-Trier, E.; Hill, C.P. Structural biology of the proteasome. Annu. Rev. Biophys. 2013, 42, 29–49. [Google Scholar] [CrossRef]
- Tomko, R.J., Jr.; Hochstrasser, M. Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 2013, 82, 415–445. [Google Scholar] [CrossRef]
- Kato, K.; Satoh, T. Structural insights on the dynamics of proteasome formation. Biophys. Rev. 2018, 10, 597–604. [Google Scholar] [CrossRef]
- Hirano, Y.; Hayashi, H.; Iemura, S.; Hendil, K.B.; Niwa, S.; Kishimoto, T.; Kasahara, M.; Natsume, T.; Tanaka, K.; Murata, S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol. Cell 2006, 24, 977–984. [Google Scholar] [CrossRef]
- Le Tallec, B.; Barrault, M.B.; Courbeyrette, R.; Guerois, R.; Marsolier-Kergoat, M.C.; Peyroche, A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell 2007, 27, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Almond, J.B.; Cohen, G.M. The proteasome: A novel target for cancer chemotherapy. Leukemia 2002, 16, 433–443. [Google Scholar] [CrossRef]
- Doi, T.; Yoshida, M.; Ohsawa, K.; Shin-ya, K.; Takagi, M.; Uekusa, Y.; Yamaguchi, T.; Kato, K.; Hirokawa, T.; Natsume, T. Total synthesis and characterization of thielocin B1 as a protein–protein interaction inhibitor of PAC3 homodimer. Chem. Sci. 2014, 5, 1860–1868. [Google Scholar] [CrossRef]
- Zhang, X.; Schulz, R.; Edmunds, S.; Kruger, E.; Markert, E.; Gaedcke, J.; Cormet-Boyaka, E.; Ghadimi, M.; Beissbarth, T.; Levine, A.J.; et al. MicroRNA-101 Suppresses Tumor Cell Proliferation by Acting as an Endogenous Proteasome Inhibitor via Targeting the Proteasome Assembly Factor POMP. Mol. Cell 2015, 59, 243–257. [Google Scholar] [CrossRef]
- Chen, D.; Frezza, M.; Schmitt, S.; Kanwar, J.; Dou, Q.P. Bortezomib as the first proteasome inhibitor anticancer drug: Current status and future perspectives. Curr. Cancer Drug Targets 2011, 11, 239–253. [Google Scholar] [CrossRef]
- Wu, W.; Sahara, K.; Hirayama, S.; Zhao, X.; Watanabe, A.; Hamazaki, J.; Yashiroda, H.; Murata, S. PAC1-PAC2 proteasome assembly chaperone retains the core α4-α7 assembly intermediates in the cytoplasm. Genes Cells 2018, 23, 839–848. [Google Scholar] [CrossRef]
- Yashiroda, H.; Mizushima, T.; Okamoto, K.; Kameyama, T.; Hayashi, H.; Kishimoto, T.; Niwa, S.; Kasahara, M.; Kurimoto, E.; Sakata, E.; et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat. Struct. Mol. Biol. 2008, 15, 228–236. [Google Scholar] [CrossRef]
- Takagi, K.; Saeki, Y.; Yashiroda, H.; Yagi, H.; Kaiho, A.; Murata, S.; Yamane, T.; Tanaka, K.; Mizushima, T.; Kato, K. Pba3-Pba4 heterodimer acts as a molecular matchmaker in proteasome α-ring formation. Biochem. Biophys. Res. Commun. 2014, 450, 1110–1114. [Google Scholar] [CrossRef]
- Kurimoto, E.; Satoh, T.; Ito, Y.; Ishihara, E.; Okamoto, K.; Yagi-Utsumi, M.; Tanaka, K.; Kato, K. Crystal structure of human proteasome assembly chaperone PAC4 involved in proteasome formation. Protein Sci. 2017, 26, 1080–1085. [Google Scholar] [CrossRef]
- Ishii, K.; Noda, M.; Yagi, H.; Thammaporn, R.; Seetaha, S.; Satoh, T.; Kato, K.; Uchiyama, S. Disassembly of the self-assembled, double-ring structure of proteasome α7 homo-tetradecamer by alpha6. Sci. Rep. 2015, 5, 18167. [Google Scholar] [CrossRef]
- Kozai, T.; Sekiguchi, T.; Satoh, T.; Yagi, H.; Kato, K.; Uchihashi, T. Two-step process for disassembly mechanism of proteasome α7 homo-tetradecamer by α6 revealed by high-speed atomic force microscopy. Sci. Rep. 2017, 7, 15373. [Google Scholar] [CrossRef] [Green Version]
- Schrader, J.; Henneberg, F.; Mata, R.A.; Tittmann, K.; Schneider, T.R.; Stark, H.; Bourenkov, G.; Chari, A. The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science 2016, 353, 594–598. [Google Scholar] [CrossRef]
- Sugiyama, M.; Hamada, K.; Kato, K.; Kurimoto, E.; Okamoto, K.; Morimoto, Y.; Ikeda, S.; Naito, S.; Furusaka, M.; et al. SANS simulation of aggregated protein in aqueous solution. Nucl. Instrum. Methods Phys. Res. A 2009, 600, 272–274. [Google Scholar] [CrossRef]
- Sugiyama, M.; Kurimoto, E.; Yagi, H.; Mori, K.; Fukunaga, T.; Hirai, M.; Zaccai, G.; Kato, K. Kinetic asymmetry of subunit exchange of homooligomeric protein as revealed by deuteration-assisted small-angle neutron scattering. Biophys. J. 2011, 101, 2037–2042. [Google Scholar] [CrossRef]
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P.R. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 282–292. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Langer, G.; Cohen, S.X.; Lamzin, V.S.; Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008, 3, 1171–1179. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Spassov, V.Z.; Yan, L. A fast and accurate computational approach to protein ionization. Protein Sci. 2008, 17, 1955–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Uekusa, Y.; Mimura, S.; Sasakawa, H.; Kurimoto, E.; Sakata, E.; Olivier, S.; Yagi, H.; Tokunaga, F.; Iwai, K.; Kato, K. Backbone and side chain 1H, 13C, and 15N assignments of the ubiquitin-like domain of human HOIL-1L, an essential component of linear ubiquitin chain assembly complex. Biomol. NMR. Assign. 2012, 6, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Koeller, D.G. Sparky, Version 3.0; University of California: San Francisco, CA, USA, 1993.
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Satoh, T.; Yagi-Utsumi, M.; Okamoto, K.; Kurimoto, E.; Tanaka, K.; Kato, K. Molecular and Structural Basis of the Proteasome α Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer. Int. J. Mol. Sci. 2019, 20, 2231. https://doi.org/10.3390/ijms20092231
Satoh T, Yagi-Utsumi M, Okamoto K, Kurimoto E, Tanaka K, Kato K. Molecular and Structural Basis of the Proteasome α Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer. International Journal of Molecular Sciences. 2019; 20(9):2231. https://doi.org/10.3390/ijms20092231
Chicago/Turabian StyleSatoh, Tadashi, Maho Yagi-Utsumi, Kenta Okamoto, Eiji Kurimoto, Keiji Tanaka, and Koichi Kato. 2019. "Molecular and Structural Basis of the Proteasome α Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer" International Journal of Molecular Sciences 20, no. 9: 2231. https://doi.org/10.3390/ijms20092231
APA StyleSatoh, T., Yagi-Utsumi, M., Okamoto, K., Kurimoto, E., Tanaka, K., & Kato, K. (2019). Molecular and Structural Basis of the Proteasome α Subunit Assembly Mechanism Mediated by the Proteasome-Assembling Chaperone PAC3-PAC4 Heterodimer. International Journal of Molecular Sciences, 20(9), 2231. https://doi.org/10.3390/ijms20092231






