Biochemical Characterization of a New β-Agarase from Cellulophaga algicola
Abstract
:1. Introduction
2. Results
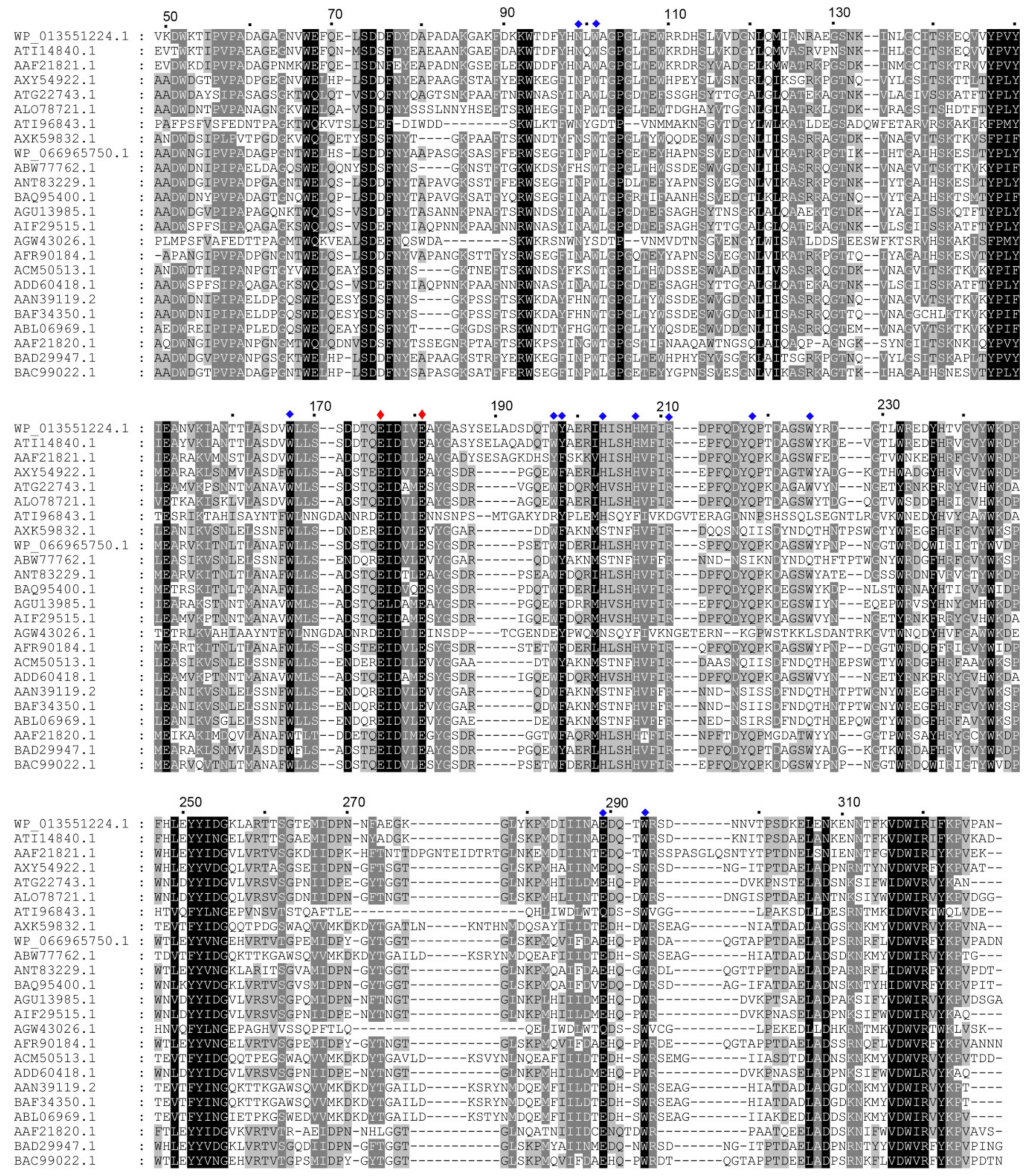
2.1. Amino Acid Sequence Analysis of CaAga1
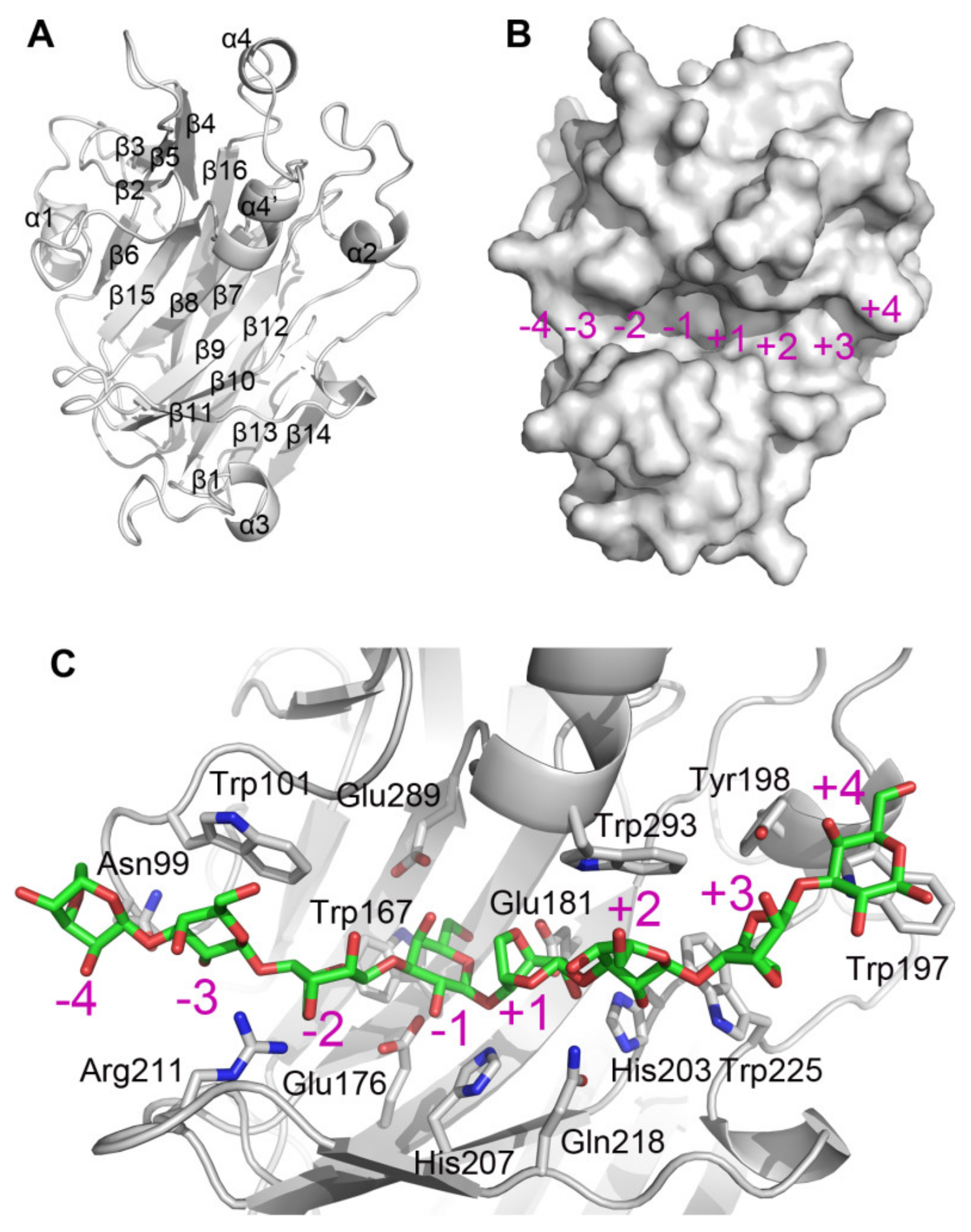
2.2. Structural Characteristic of CaAga1
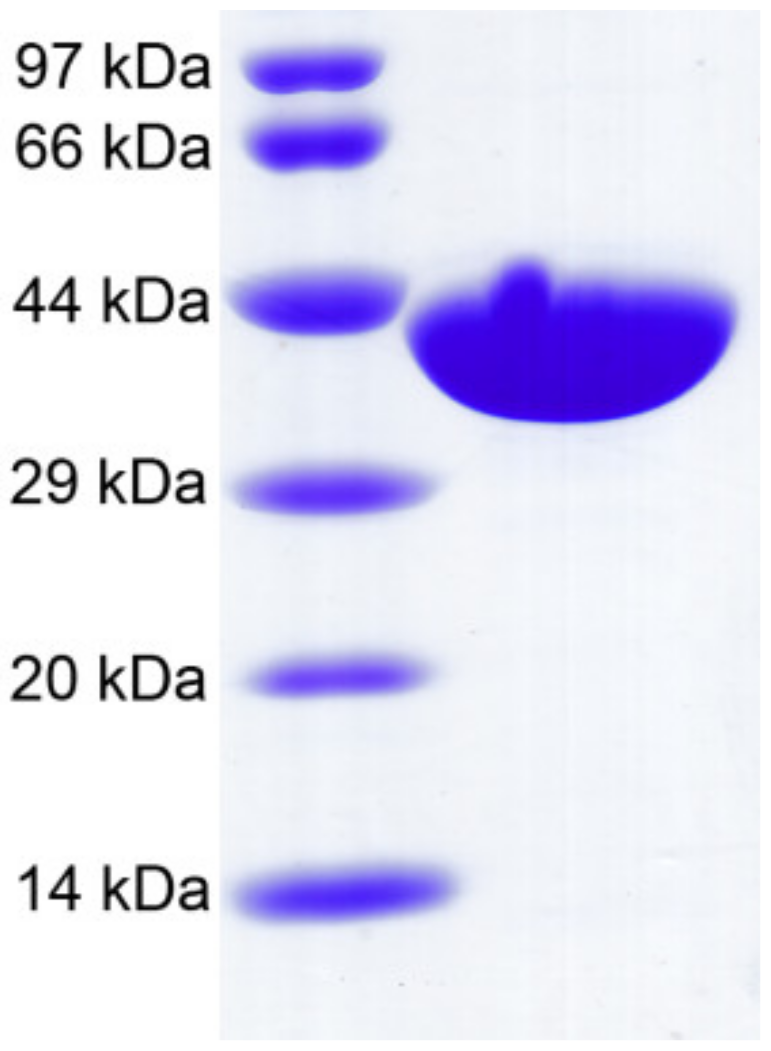
2.3. Recombinant CaAga1 Production, Purification, and Agarase Activity
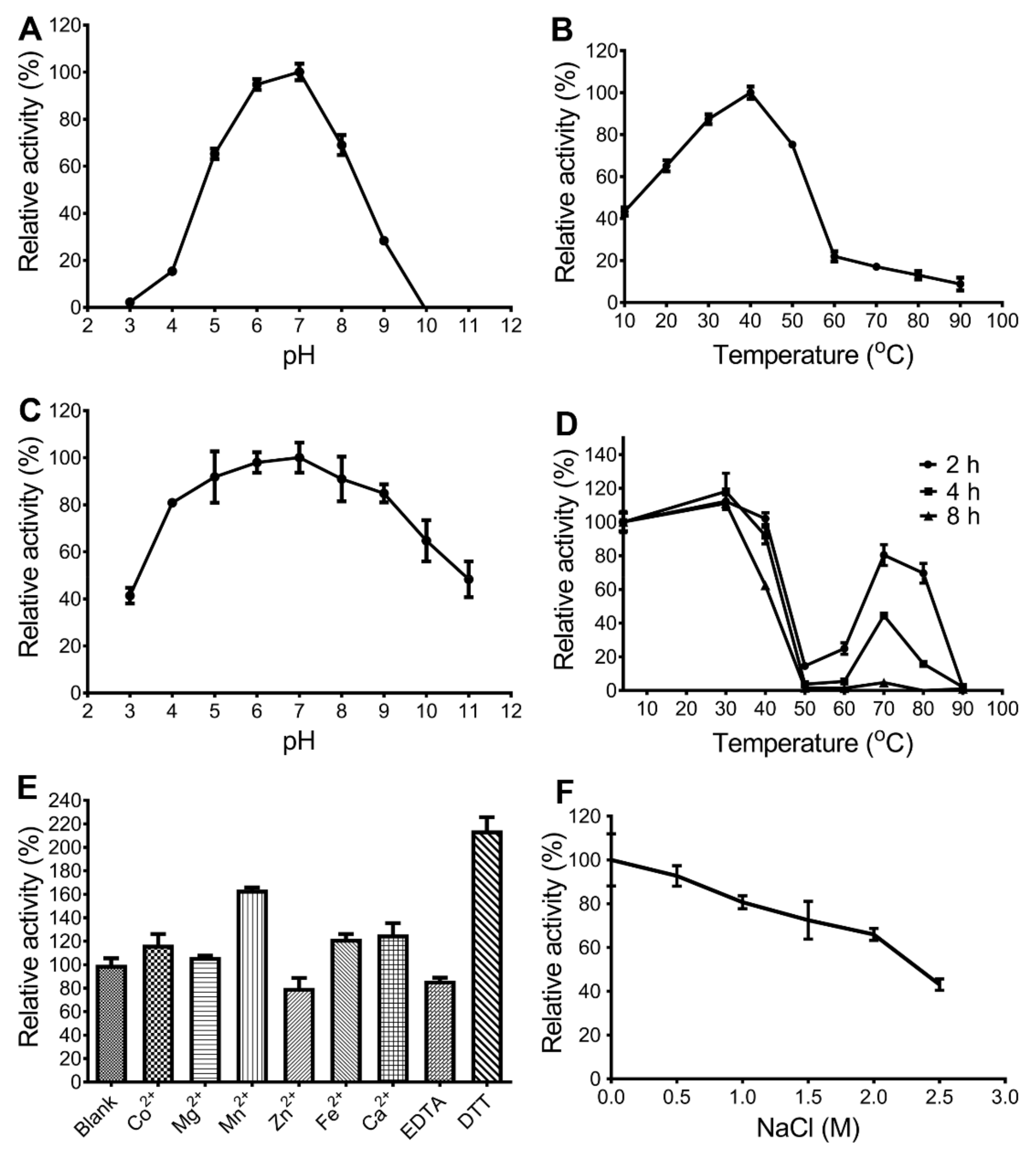
2.4 Biochemical Characterization of CaAga1
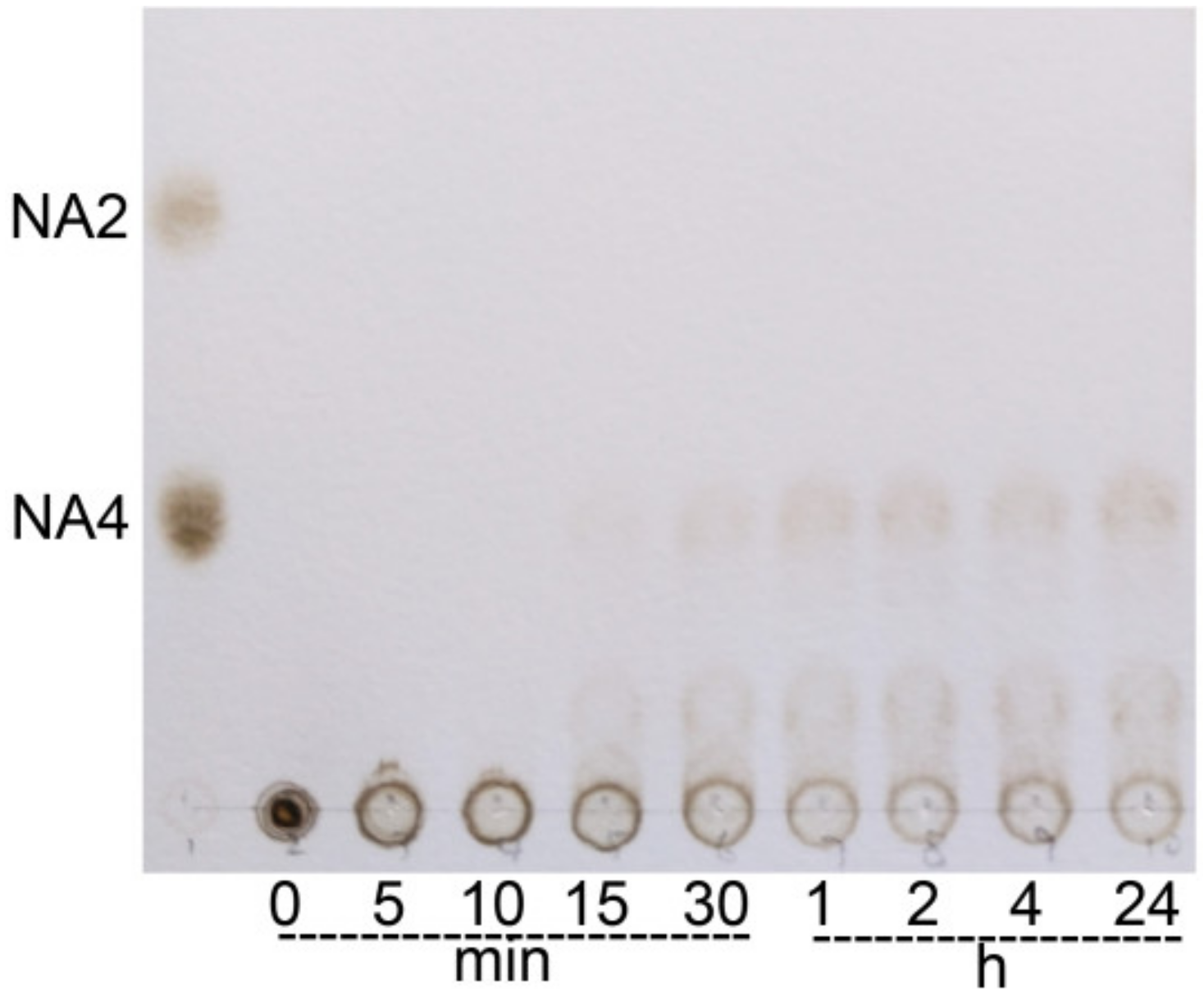
2.5 Hydrolytic Products of CaAga1
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Amino Acid Sequence Analysis
4.3. Cloning and Expression of Agarases
4.4. Preparation of Crude Agar from Red Algae
4.5. Standard Agarase Assay
4.6. Biochemical Characterization
4.7 Analysis of Hydrolysis Products
Author Contributions
Funding
Conflicts of Interest
References
- Renn, D. Biotechnology and the red seaweed polysaccharide industry: Status, needs and prospects. Trends Biotechnol. 1997, 15, 9–14. [Google Scholar] [CrossRef]
- Fu, X.T.; Kim, S.M. Agarase: Review of major sources, categories, purification method, enzyme characteristics and applications. Mar. Drugs 2010, 26, 200–218. [Google Scholar] [CrossRef] [PubMed]
- Jahromi, S.T.; Barzkar, N. Future direction in marine bacterial agarases for industrial applications. Appl. Microbiol. Biotechnol. 2018, 102, 6847–6863. [Google Scholar] [CrossRef] [PubMed]
- Yun, E.J.; Yu, S.; Kim, K.H. Current knowledge on agarolytic enzymes and the industrial potential of agar-derived sugars. Appl. Microbiol. Biotechnol. 2017, 101, 5581–5589. [Google Scholar] [CrossRef]
- Correc, G.; Hehemann, J.H.; Czjzek, M.; Helbert, W. Structural analysis of the degradation products of porphyran digested by Zobellia galactanivorans β-porphyranase A. Carbohydr. Polym. 2011, 83, 277–283. [Google Scholar] [CrossRef]
- Yanagisawa, M.; Kawai, S.; Murata, K. Strategies for the production of high concentrations of bioethanol from seaweeds: Production of high concentrations of bioethanol from seaweeds. Bioengineered 2013, 4, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.R.; Zhang, M.; Zhong, M.; Hu, Z. Synergistic enzymatic saccharification and fermentation of agar for biohydrogen production. Bioresour. Technol. 2017, 241, 369–373. [Google Scholar] [CrossRef]
- Chi, W.J.; Chang, Y.K.; Hong, S.K. Agar degradation by microorganisms and agar-degrading enzymes. Appl. Microbiol. Biotechnol. 2012, 94, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Gong, Q.; Wang, Y.; Ma, Y.; Li, J.; Yu, W. Prebiotic effects of neoagaro-oligosaccharides prepared by enzymatic hydrolysis of agarose. Anaerobe 2006, 12, 260–266. [Google Scholar] [CrossRef]
- Jang, M.K.; Lee, D.G.; Kim, N.Y.; Yu, K.H.; Jang, H.J.; Lee, S.W.; Jang, H.J.; Lee, Y.J.; Lee, S.H. Purification and characterization of neoagarotetraose from hydrolyzed agar. J. Microbiol. Biotechnol. 2009, 19, 1197–1200. [Google Scholar] [PubMed]
- Kang, O.L.; Ghani, M.; Hassan, O.; Rahmati, S.; Ramli, N. Novel agarooligosaccharide production through enzymatic hydrolysis: Physicochemical properties and antioxidant activities. Food Hydrocolloid 2014, 42, 304–308. [Google Scholar] [CrossRef]
- Enoki, T.; Okuda, S.; Kudo, Y.; Takashima, F.; Sagawa, H.; Kato, I. Oligosaccharides from agar inhibit pro-inflammatory mediator release by inducing heme oxygenase 1. Biosci. Biotechnol. Biochem. 2010, 74, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, M.; Rownd, T.H. A rapid method for extracting DNA from agarose gels. Plasmid 1978, 1, 557–562. [Google Scholar] [CrossRef]
- Araki, T.; Lu, Z.; Morishita, T. Optimization of parameters for isolation of protoplasts from Gracilaria verrucosa (Rhodophyta). J. Mar. Biotechnol. 1998, 6, 193–197. [Google Scholar]
- Kim, J.H.; Yun, E.J.; Seo, N.; Yu, S.; Kim, D.H.; Cho, K.M.; An, H.J.; Kim, J.H.; Choi, I.G.; Kim, K.H. Enzymatic liquefaction of agarose above the sol–gel transition temperature using a thermostable endo-type β-agarase, Aga16B. Appl. Microbiol. Biotechnol. 2017, 101, 1111–1120. [Google Scholar] [CrossRef]
- Abt, B.; Lu, M.; Misra, M.; Han, C.; Nolan, M.; Lucas, S.; Hammon, N.; Deshpande, S.; Cheng, J.F.; Tapia, R.; et al. Complete genome sequence of Cellulophaga algicola type strain (IC166T). Stand Genomic Sci. 2011, 2, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Ramos, K.R.M.; Valdehuesa, K.N.G.; Nisola, G.M.; Lee, W.K.; Chung, W.J. Identification and characterization of a thermostable endolytic β-agarase Aga2 from a newly isolated marine agarolytic bacteria Cellulophaga omnivescoria W5C. N. Biotechnol. 2018, 40, 261–267. [Google Scholar] [CrossRef]
- Takagi, E.; Hatada, Y.; Akita, M.; Ohta, Y.; Yokoi, G.; Miyazaki, T.; Nishikawa, A.; Tonozuka, T. Crystal structure of the catalytic domain of a GH16 β-agarase from a deep-sea bacterium, Microbulbifer thermotolerans JAMB-A94. Biosci. Biotechnol. Biochem. 2015, 79, 625–632. [Google Scholar] [CrossRef]
- Lee, Y.R.; Jung, S.; Chi, W.J.; Bae, C.H.; Jeong, B.C.; Hong, S.K.; Lee, C.R. Biochemical characterization of a novel GH86 β-agarase producing neoagarohexaose from Gayadomonas joobiniege G7. J. Microbiol. Biotechnol. 2018, 28, 284–292. [Google Scholar] [CrossRef]
- Jung, S.; Lee, C.R.; Chi, W.J.; Bae, C.H.; Hong, S.K. Biochemical characterization of a novel cold-adapted GH39 β-agarase, AgaJ9, from an agar-degrading marine bacterium Gayadomonas joobiniege G7. Appl. Microbiol. Biotechnol. 2017, 101, 1965–1974. [Google Scholar] [CrossRef]
- Li, G.; Sun, M.; Wu, J.; Ye, M.; Ge, X.; Wei, W.; Li, H.; Hu, F. Identification and biochemical characterization of a novel endotype β-agarase AgaW from Cohnella sp. strain LGH. Appl. Microbiol. Biotechnol. 2015, 99, 10019–10029. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.T.; Pan, C.H.; Lin, H.; Kim, S.M. Gene cloning, expression, and characterization of a beta-agarase, agaB34, from Agarivorans albus YKW-34. J. Microbiol. Biotechnol. 2009, 19, 257–264. [Google Scholar]
- Dong, Q.; Ruan, L.W.; Shi, H.A. β-agarase with high pH stability from Flammeovirga sp. SJP92. Carbohydr. Res. 2016, 432, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Hou, Y.P.; Jin, M.; Zeng, R.Y.; Lin, H.T. Expression and characterization of a novel thermostable and pH-stable β-agarase from Deep-Sea bacterium Flammeovirga Sp. OC4. J. Agric. Food Chem. 2016, 64, 7251–7258. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Zhang, L.; Miao, S.; Zhang, Y.; Zeng, S.; Zheng, B. Preliminary characterization of a novel β-agarase from Thalassospira profundimonas. Springerplus 2016, 5, 1086. [Google Scholar] [CrossRef]
- Hou, Y.; Chen, X.; Chan, Z.; Zeng, R. Expression and characterization of a thermostable and pH-stable β-agarase encoded by a new gene from Flammeovirga pacifica WPAGA1. Process Biochem. 2015, 50, 1068–1075. [Google Scholar] [CrossRef]
- Oh, C.; Nikapitiya, C.; Lee, Y.; Whang, I.; Kang, D.H.; Heo, S.J.; Choi, Y.U.; Lee, J. Molecular cloning, characterization and enzymatic properties of a novel β-agarase from a marine isolate Psudoalteromonas sp. AG52BRAZ. J. Microbiol. 2010, 41, 876–889. [Google Scholar]
- Ohta, Y.; Hatada, Y.; Nogi, Y.; Miyazaki, M.; Li, Z.; Akita, M.; Hidaka, Y.; Goda, S.; Ito, S.; Horikoshi, K. Enzymatic properties and nucleotide and amino acid sequences of a thermostable β-agarase from a novel species of deep-sea Microbulbifer. Appl. Biochem. Biotechnol. 2004, 64, 505–514. [Google Scholar] [CrossRef]
- Di, W.; Qu, W.; Zeng, R. Cloning, expression, and characterization of thermal-stable and pH-stable agarase from mangrove sediments. J. Basic. Microbiol. 2018, 58, 302–309. [Google Scholar] [CrossRef]
- Long, M.; Yu, Z.; Xu, X. A novel β-agarase with high pH stability from marine Agarivorans sp. LQ48. Mar. Biotechnol. 2010, 12, 62–69. [Google Scholar] [CrossRef]
- Song, T.; Cao, Y.; Xu, H.; Zhang, W.; Fei, B.; Qiao, D.; Cao, Y. Purification and characterization of a novel β-agarase of Paenibacillus sp. SSG-1 isolated from soil. J. Biosci. Bioeng. 2014, 118, 125–129. [Google Scholar] [CrossRef]
- Li, R.K.; Chen, Z.; Ying, X.J.; Ng, T.B.; Ye, X.Y. A novel GH16 beta-agarase isolated from a marine bacterium, Microbulbifer sp. BN3 and its characterization and high-level expression in Pichia pastoris. Int. J. Biol. Macromol. 2018, 119, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- An, K.; Shi, X.; Cui, F.; Cheng, J.; Liu, N.; Zhao, X.; Zhang, X.H. Characterization and overexpression of a glycosyl hydrolase family 16 beta-agarase YM01-1 from marine bacterium Catenovulum agarivorans YM01T. Protein Expr. Purif. 2018, 143, 1–8. [Google Scholar] [CrossRef]
- Park, D.Y.; Chi, W.J.; Park, J.S.; Chang, Y.K.; Hong, S.K. Cloning, Expression, and Biochemical Characterization of a GH16 β-Agarase AgaH71 from Pseudoalteromonas hodoensis H7. Appl. Biochem. Biotechnol. 2014, 175, 733–747. [Google Scholar] [CrossRef]
- Chi, WJ.; Park, D.Y.; Seo, B.Y.; Chang, Y.K.; Lee, S.Y.; Hong, S.K. Cloning, expression, and biochemical characterization of a novel GH16 β-agarase AgaG1 from Alteromonas sp. GNUM-1. Appl. Microbiol. Biotechnol. 2014, 98, 4545–4555. [Google Scholar] [CrossRef]
- Allouch, J.; Jam, M.; Helbert, W.; Barbeyron, T.; Kloareg, B.; Henrissat, B.; Czjzek, M. The three-dimensional structures of two β-agarases. J. Biol. Chem. 2003, 278, 47171–47180. [Google Scholar] [CrossRef]
- Jam, M.; Flament, D.; Allouch, J.; Potin, P.; Thion, L.; Kloareg, B.; Czjzek, M.; Helbert, W.; Michel, G.; Barbeyron, T. The endo-beta-agarases AgaA and AgaB from the marine bacterium Zobellia galactanivorans: Two paralogue enzymes with different molecular organizations and catalytic behaviours. Biochem. J. 2005, 385, 703–713. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 18. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Han, Z.; Shang-Guan, F.; Yang, J. Characterization of a novel cold-active xylanase from Luteimonas species. World J. Microbiol. Biotechnol. 2018, 34, 123. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Prot. Exp. Pur. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Biochem. 1959, 31, 426–428. [Google Scholar]
| Agarase | Glycoside Hydrolase Family | Optimal Temperature | Optimal pH | Thermal Stability (Retaining Activity) | pH Stability (Retaining Activity) | Product | Kinetic Parameter | Reference and Accession Number |
|---|---|---|---|---|---|---|---|---|
| CaAga1 (Cellulophaga algicola DSM 14237) | 16 | 40 | 7 | 90%, 40 °C, 4 h; 80%, 70 °C, 2 h | 80%, pH 4–9, 20 °C, 2 h | NA4, NA6 | Km: 1.19 mg/mL, Vmax: 36.21 U/mg | This study WP_013551224.1 |
| N3-1 (Microbulbifer sp. BN3) | 16 | 50 | 6 | 40%, 30–50 °C, 30 min | 80%, pH 4–9, room temperature, 1 h | NA2, NA4 | [32] AXY54922.1 | |
| AgaM1 (environmental DNA ofmangrove sediments) | 16 | 50 | 7 | 80%, 40–50 °C, 3 h | 80%, pH 5–10, 50 °C, 3 h | NA4, NA6 | Km: 1.82 mg/mL, Vmax: 357.14 U/mg | [29] AYA70425.1 |
| YM01-1 (Catenovulum agarivorans YM01) | 16 | 50 | 7 | <10%, 70 °C, 1 h | 80%, pH 6–9, 4 °C, 12 h | NA2 | Km: 8.69 mg/mL, Vmax: 4350 U/mg | [33] ATI96843.1 |
| Aga2 (Cellulophaga omnivescoria W5C) | 16 | 45 | 8 | NA4, NA6 | Km: 2.59 mg/mL, Vmax: 275.48 U/mg | [17] ATI14840.1 | ||
| Aga16B (Saccharophagus degradans 2-40T) | 16 | 40–60 | 7.5 | 100%, 50 °C, 2 h | NA4, NA6 | [15] Q21LJ2 | ||
| AgaB (Flammeovirga sp. SJP92) | 16 | 45 | 8 | 60%, 55 °C, 30 min | 50%, pH 6–9, room temperature, 1 h | NA4, NA6 | Km: 3.99 mg/mL, Vmax: 700 U/mg | [23] ANN44251.1 |
| Aga4436 (Flammeovirga sp. OC4) | 16 | 50–55 | 6 | 80%, 40 °C, 144 h | 70%, pH 3–10, 50 °C, 1 h | NA4, NA6 | [24] AJW82062.1. | |
| Aga21 (Pseudoalteromonas sp. NJ21) | 16 | 30 | 8 | 20%, 40 °C, 1 h | NA2 | [21] | ||
| AgaH71 (Pseudoalteromonas hodoensis) | 16 | 45 | 6 | 90%, 45 °C, 1 h | NA2, NA4, NA6 | Km: 28.33 mg/mL, Vmax: 88.25 U/mg | [34] AIF29515.1 | |
| AgaG1 (Alteromonas sp. GNUM-1) | 16 | 40 | 7 | 70%, 45 °C, 30 min | NA2 | Km: 3.74 mg/mL, Vmax: 23.8 U/mg | [35] AGW43026.1 | |
| AgaA (Agarivorans sp. LQ48) | 16 | 40 | 7 | 95%, 40 °C, 1 h | 95%, pH 3–11, 20 °C, 1 h | NA4, NA6 | Km: 43.9 mg/mL, Vmax: 909.1 U/mg | [30] ACM50513.1 |
| AgaP (Pseudoalteromonas sp. AG4) | 16 | 55 | 5.5 | 75%, 55 °C, 1 h | NA4 | [27] ADD60418.1 | ||
| AgaB34 (Agarivorans albus YKW-34) | 16 | 30 | 7 | 80%, 50 °C, 1 h | 70%, pH 5–9, 40 °C, 1 h | NA4 | Km: 0.24 mg/mL, Vmax: 50 U/mg | [22] ABW77762.1 |
| ZgAgaA (Zobellia galactanivorans) | 16 | 6 | NA2 (minor), NA4, NA6 | Km: 2 mg/mL, kcat: 150 s−1 | [37] AAF21820.1 | |||
| ZgAgaB (Zobellia galactanivorans) | 16 | 7 | NA2, NA4, | Km: 1 mg/mL, kcat: 100 s-1 | [37] AAF21821.1 | |||
| AgaA7 (deep-sea Microbulbifer) | 16 | 50 | 7 | 50%, 50 °C, 502 min | 50%, pH 3.5–9.5, 40 °C, 30 min | NA4 | [28] BAC99022.1 | |
| AgaJ9 (Gayadomonas joobiniege G7) | 39 | 25 | 5 | <20%, 40 °C, 30 min | NA2, NA4, | Km: 1.43 mg/mL, Vmax: 10.7 U/mg | [20] WP_017446561.1 | |
| AgaJ5 (Gayadomonas joobiniege G7) | 86 | 30 | 4.5 | NA6 | Km: 8.9 mg/mL, Vmax: 188.6 U/mg | [19] WP_017446675.1 | ||
| AgaP4383 (Flammeovirga pacifica WPAGA1) | 86 | 50 | 9 | 100%, 50 °C, 10 h | 90%, pH 5–10, 30 °C, 24 h | NA4, NA6 | Km: 8.53 mg/mL, Vmax: 1.2 U mg | [26] AIA22721.1 |
| Agarase-fst (Thalassospira profundimaris fst-13007) | 45 | 8 | 70%, 50 °C, 1 h | 80%, pH 5–8, 45 °C, 1 h | NA2, NA4, NA6 | [25] | ||
| SSG-1a (Paenibacillus sp. SSG-1) | 50 | 6 | 95%, 40 °C, 1 h | NA8 | [31] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Z.; Zhang, Y.; Yang, J. Biochemical Characterization of a New β-Agarase from Cellulophaga algicola. Int. J. Mol. Sci. 2019, 20, 2143. https://doi.org/10.3390/ijms20092143
Han Z, Zhang Y, Yang J. Biochemical Characterization of a New β-Agarase from Cellulophaga algicola. International Journal of Molecular Sciences. 2019; 20(9):2143. https://doi.org/10.3390/ijms20092143
Chicago/Turabian StyleHan, Zhenggang, Yuxi Zhang, and Jiangke Yang. 2019. "Biochemical Characterization of a New β-Agarase from Cellulophaga algicola" International Journal of Molecular Sciences 20, no. 9: 2143. https://doi.org/10.3390/ijms20092143
APA StyleHan, Z., Zhang, Y., & Yang, J. (2019). Biochemical Characterization of a New β-Agarase from Cellulophaga algicola. International Journal of Molecular Sciences, 20(9), 2143. https://doi.org/10.3390/ijms20092143





