Marek’s Disease Virus Regulates the Ubiquitylome of Chicken CD4+ T Cells to Promote Tumorigenesis
Abstract
1. Introduction
2. Results and Discussion
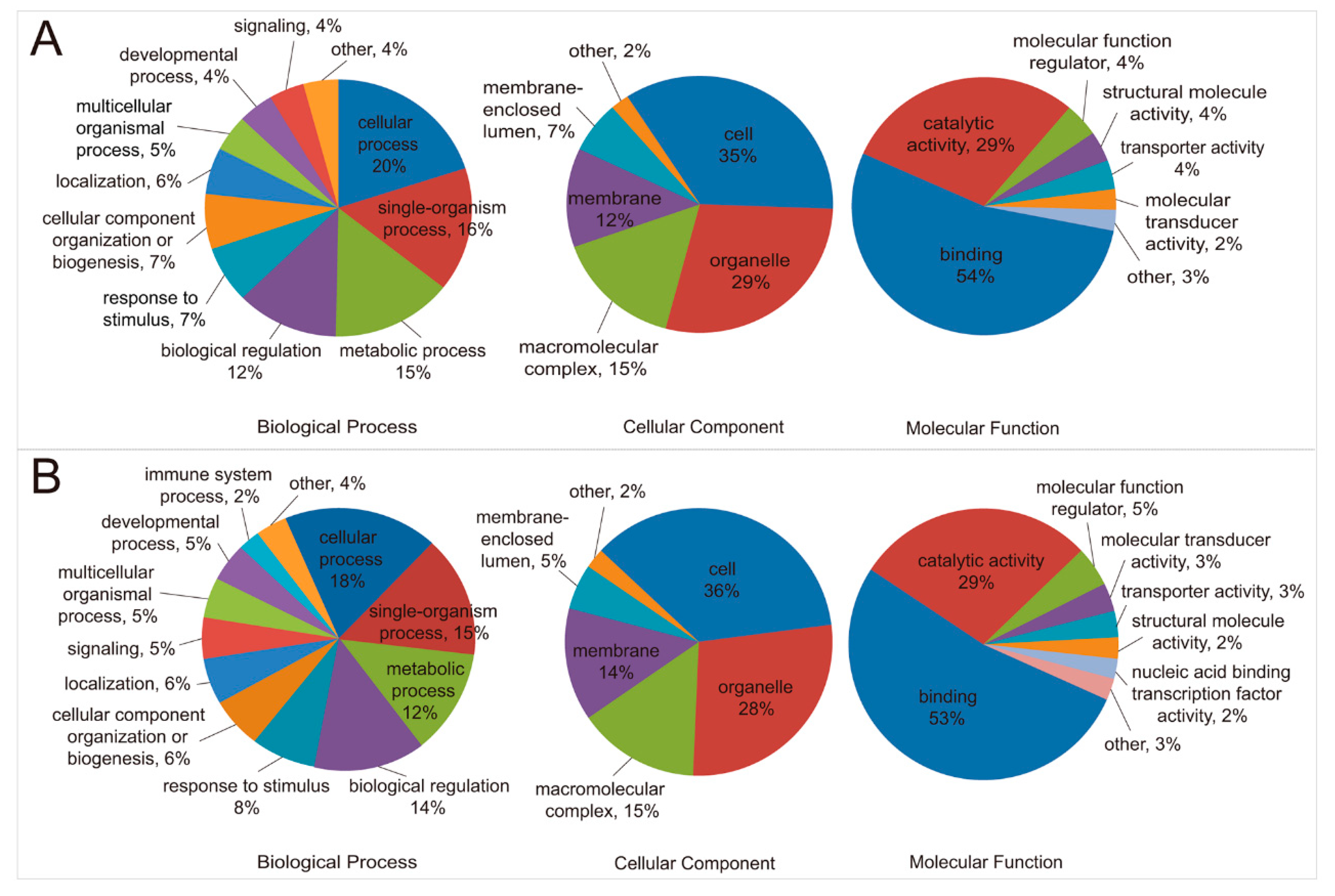
2.1. Quantification of Ubiquitinated Peptides and Annotation of Proteins
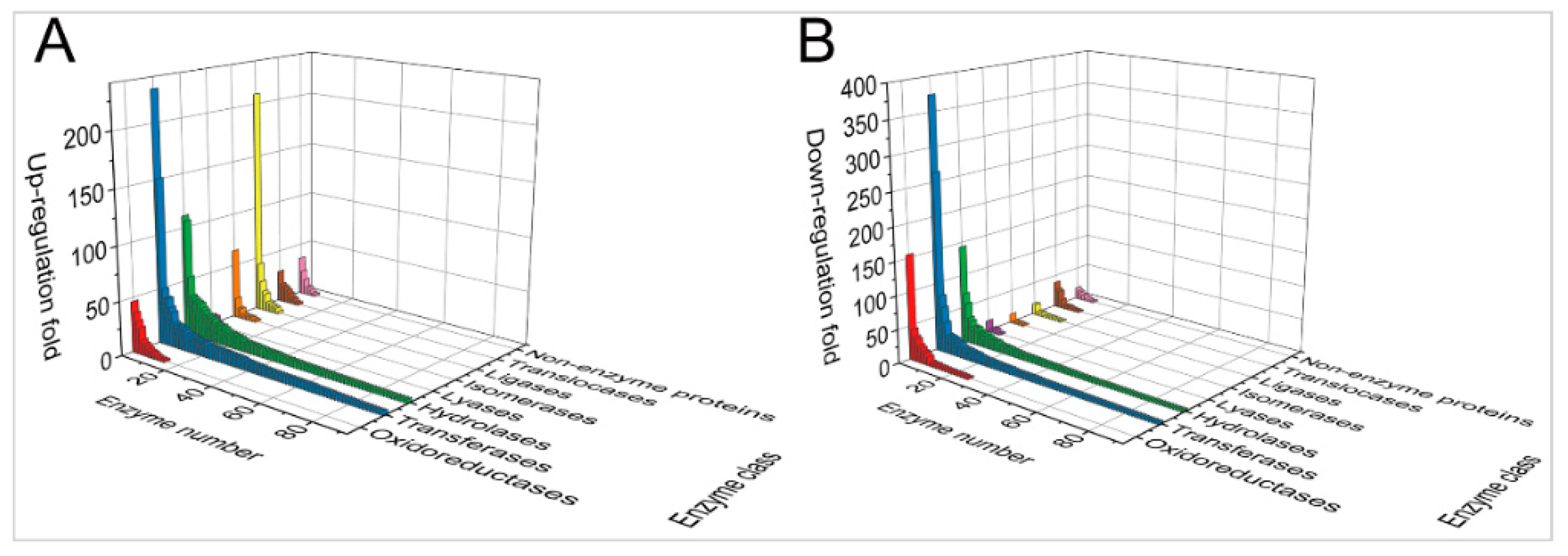
2.2. MDV Up- and Downregulated Ubiquitination of Two Different Sets of Proteins with Similar Ratios
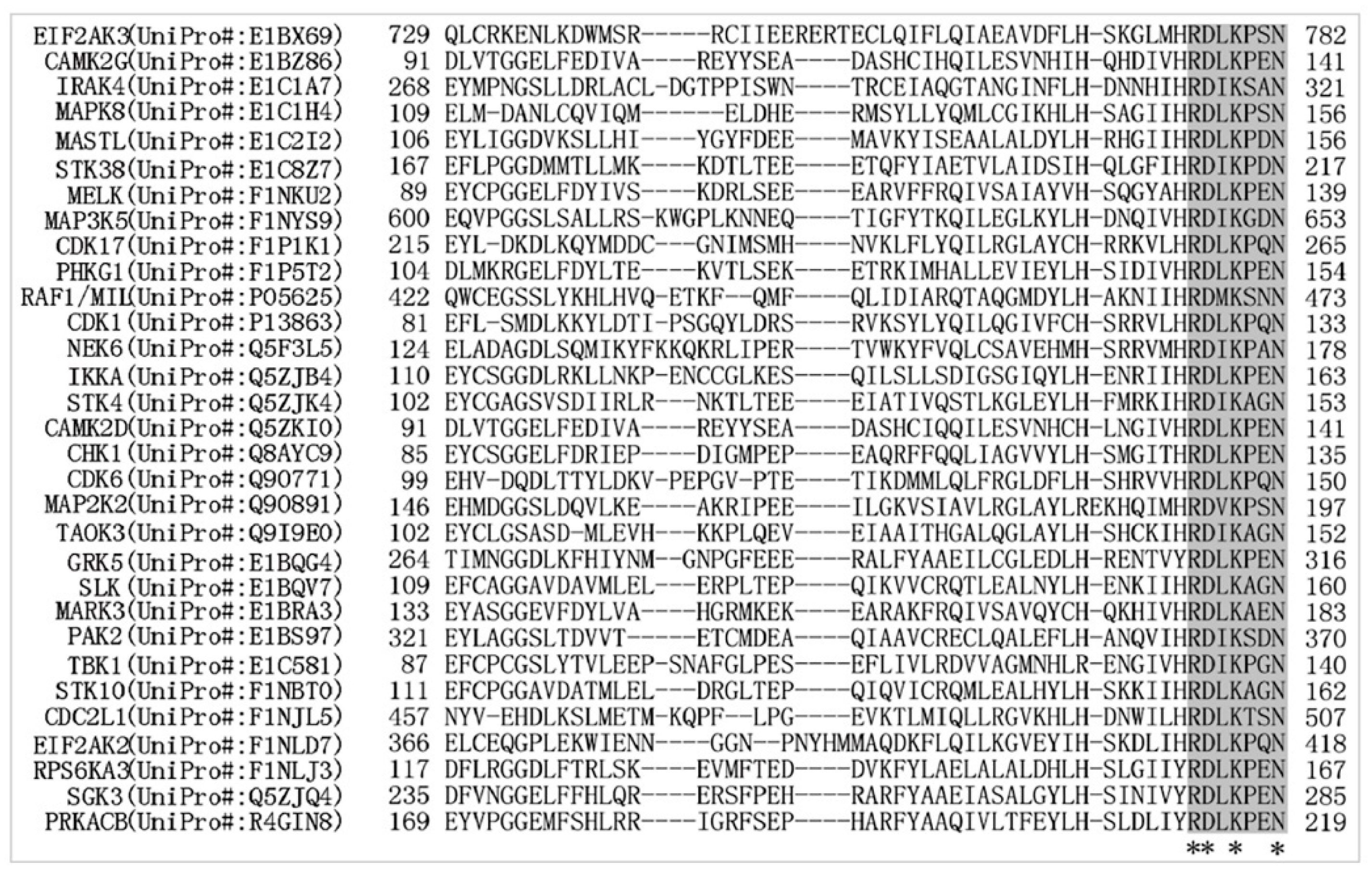
2.3. “RD.K..N” Motif Is a Major Regulatory Target of MDV
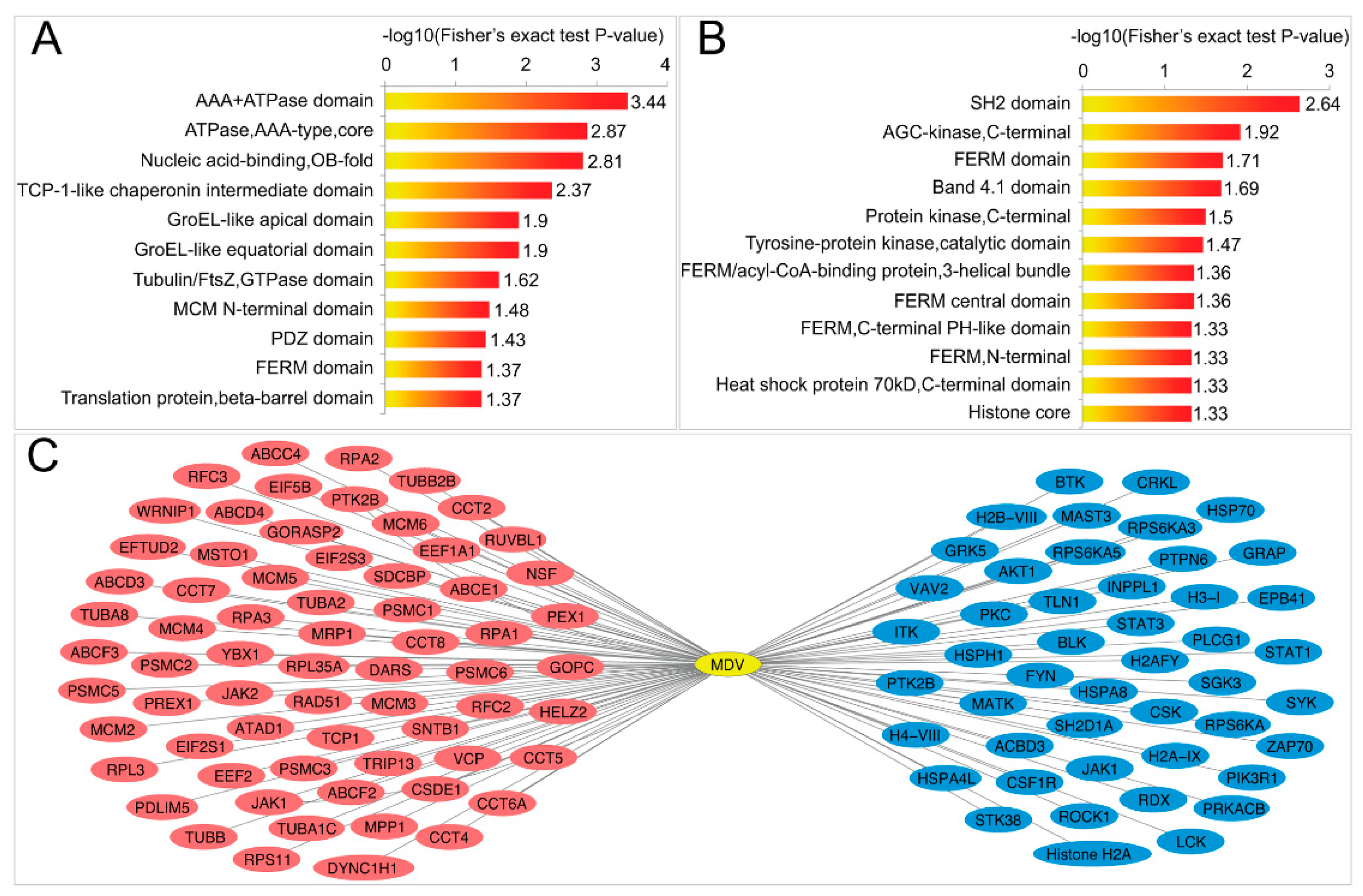
2.4. MDV Up- and Downregulates Two Different Sets of Domains
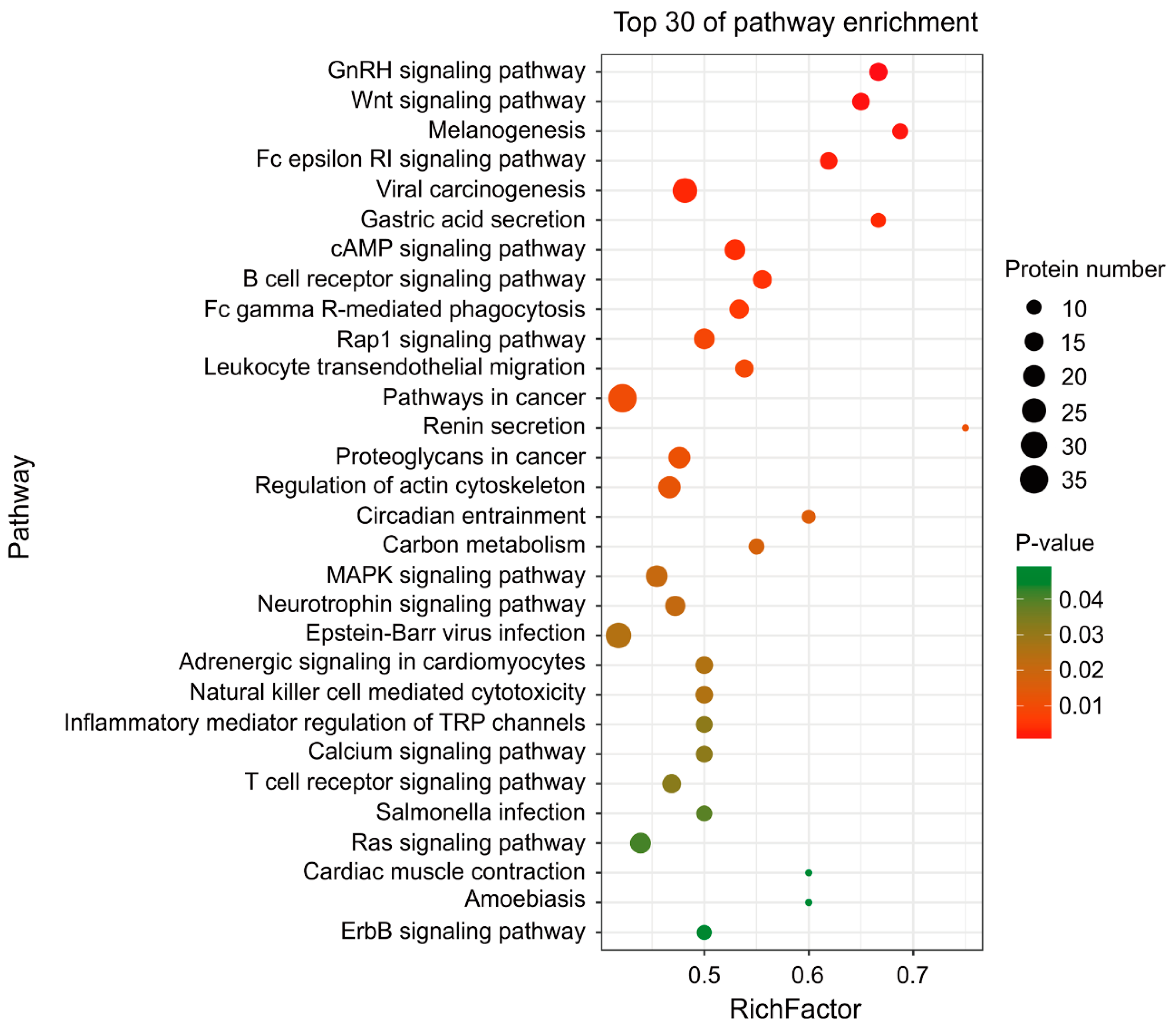
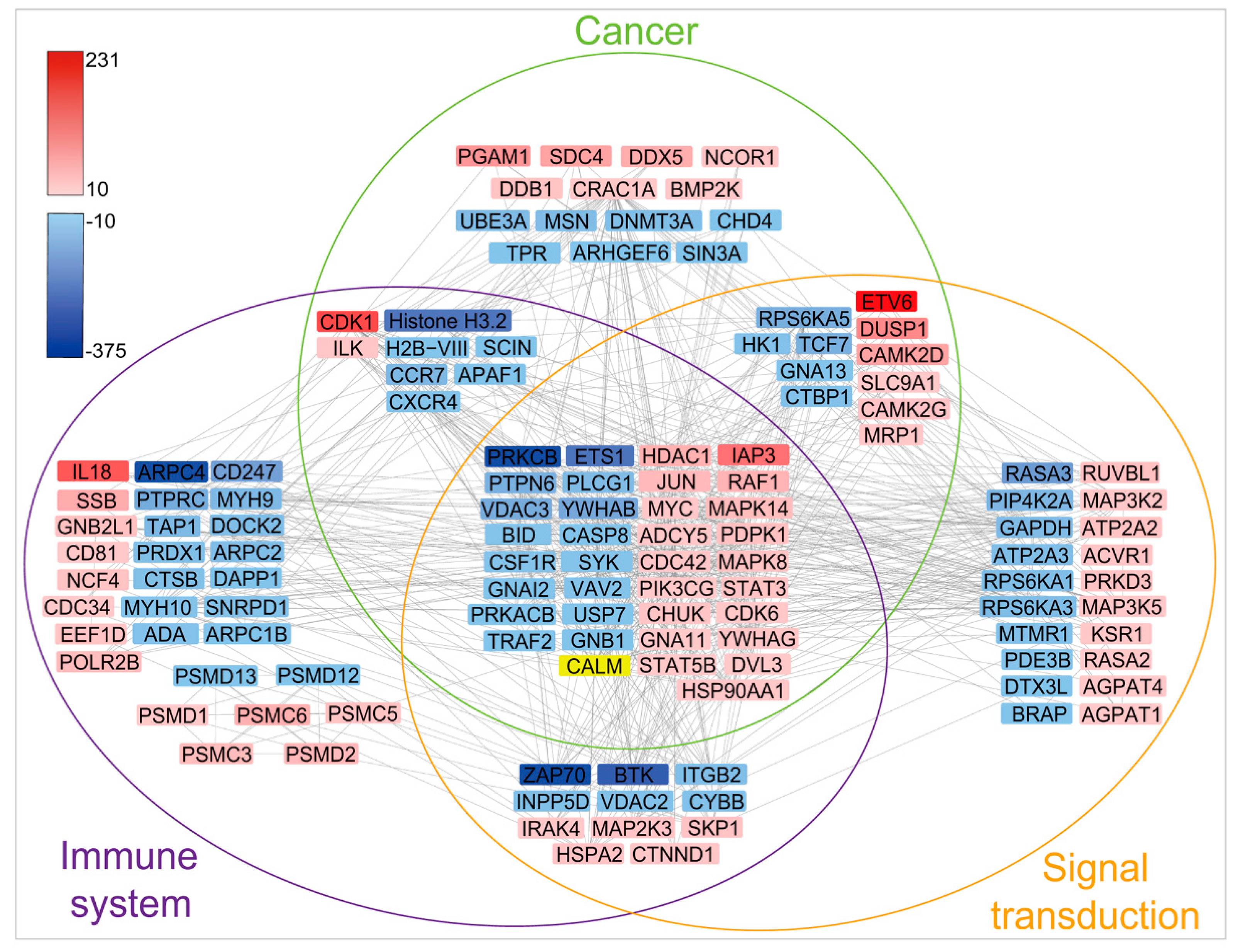
2.5. MDV Mainly Alters Ubiquitination of Proteins Involved in Immune and Cancer Pathways
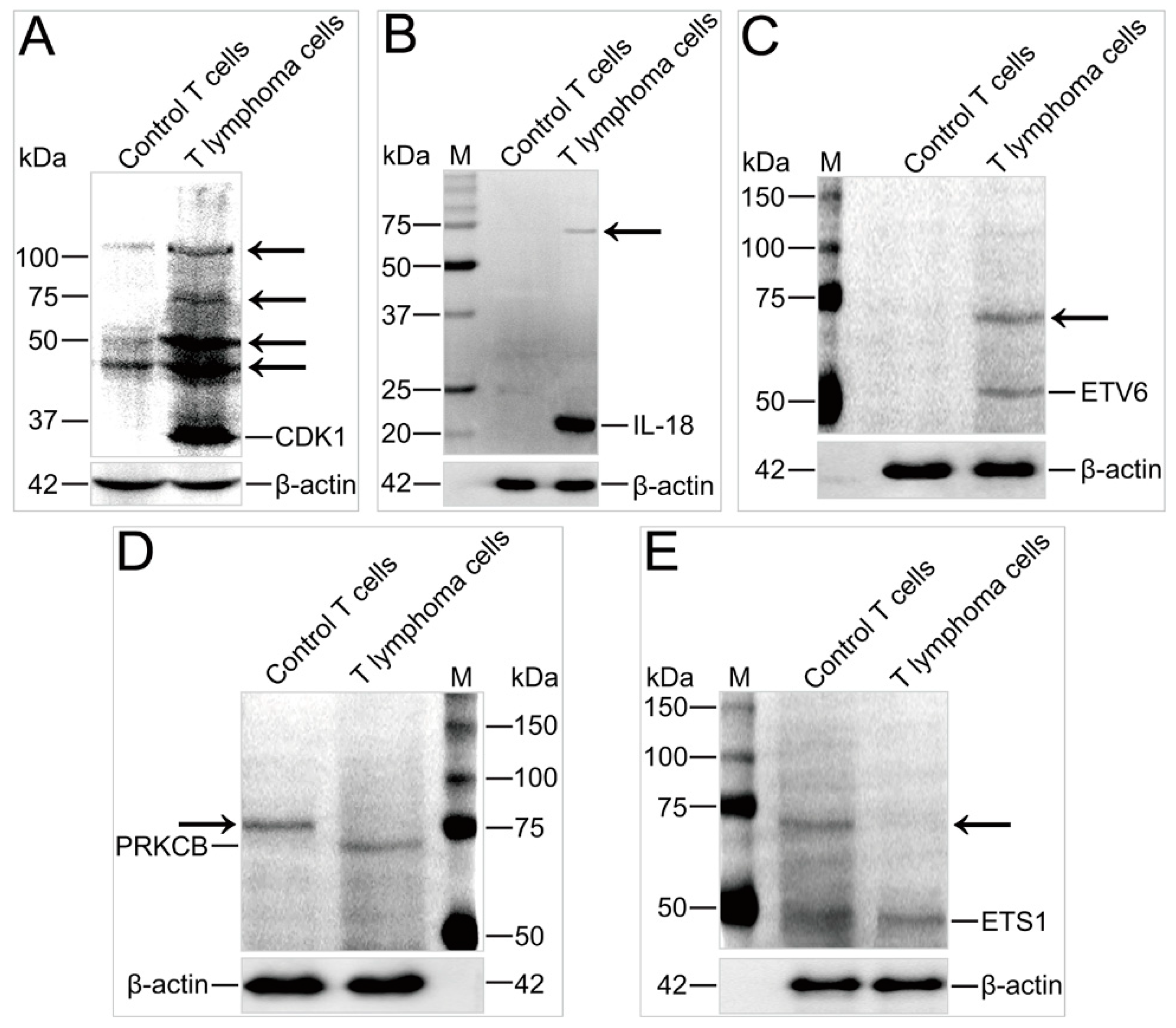
2.6. Ubiquitination of Key Proteins Was Significantly Altered in T Lymphoma Cells
3. Materials and Methods
3.1. Isolation and Characterization of T Lymphocytes
3.2. Protein Extraction and Immunoaffinity Enrichment of Ubiquitinated Peptides
3.3. LC-MS/MS Analysis and Database Search
3.4. Annotation and Classification of Ubiquitinated Proteins
3.5. Profile Analysis of Ubiquitinated Motifs
3.6. Pathway and Social Network Analysis of Ubiquitinated Proteins
3.7. Verification of Unique Proteins with Immunoblotting
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MD | Marek’s disease |
| MDV | Marek’s disease virus |
| Ub | Ubiquitin |
| DUB | Deubiquitinase |
| SPF | Specific pathogen free |
| FITC | Fluorescein isothiocyanate |
| MS/MS | Tandem mass spectrometry |
| FA | Formic acid |
References
- Boodhoo, N.; Gurung, A.; Sharif, S.; Behboudi, S. Marek’s disease in chickens: A review with focus on immunology. Vet. Res. 2016, 47, 119. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, I.M. Marek’s disease vaccines: A solution for today but a worry for tomorrow? Vaccine 2008, 26, C31–C41. [Google Scholar] [CrossRef] [PubMed]
- Churchill, A.E.; Payne, L.N.; Chubb, R.C. Immunization against Marek’s disease using a live attenuated virus. Nature 1969, 221, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Padhi, A.; Parcells, M.S. Positive Selection Drives Rapid Evolution of the meq Oncogene of Marek’s Disease Virus. PLoS ONE 2016, 11, e0162180. [Google Scholar] [CrossRef] [PubMed]
- Chbab, N.; Egerer, A.; Veiga, I.; Jarosinski, K.W.; Osterrieder, N. Viral control of vTR expression is critical for efficient formation and dissemination of lymphoma induced by Marek’s disease virus (MDV). Vet. Res. 2010, 41, 56. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.T.; Selvaraj, R.K.; Kamil, J.P.; Osterrieder, N.; Kaufer, B.B. Marek’s disease viral interleukin-8 promotes lymphoma formation through targeted recruitment of B cells and CD4+ CD25+ T cells. J. Virol. 2012, 86, 8536–8545. [Google Scholar] [CrossRef]
- Veiga, I.B.; Jarosinski, K.W.; Kaufer, B.B.; Osterrieder, N. Marek’s disease virus (MDV) ubiquitin-specific protease (USP) performs critical functions beyond its enzymatic activity during virus replication. Virology 2013, 437, 110–117. [Google Scholar] [CrossRef]
- McPherson, M.C.; Delany, M.E. Virus and host genomic, molecular, and cellular interactions during Marek’s disease pathogenesis and oncogenesis. Poult. Sci. 2016, 95, 412–429. [Google Scholar] [CrossRef]
- Wozniakowski, G.; Samorek-Salamonowicz, A.E. Molecular evolution of Marek’s disease virus (MDV) field strains in a 40-year time period. Avian Dis. 2014, 58, 550–557. [Google Scholar] [CrossRef]
- Calnek, B.W. Pathogenesis of Marek’s Disease Virus Infection. In Marek’s Disease; Hirai, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 25–55. [Google Scholar]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Porras-Yakushi, T.R.; Hess, S. Recent advances in defining the ubiquitylome. Expert Rev. Proteom. 2014, 11, 477–490. [Google Scholar] [CrossRef]
- Lian, L.; Qu, L.; Chen, Y.; Lamont, S.J.; Yang, N. A systematic analysis of miRNA transcriptome in Marek’s disease virus-induced lymphoma reveals novel and differentially expressed miRNAs. PLoS ONE 2012, 7, e51003. [Google Scholar] [CrossRef]
- Haq, K.; Brisbin, J.T.; Thanthrige-Don, N.; Heidari, M.; Sharif, S. Transcriptome and proteome profiling of host responses to Marek’s disease virus in chickens. Vet. Immunol. Immunopathol. 2010, 138, 292–302. [Google Scholar] [CrossRef]
- Dong, K.; Chang, S.; Xie, Q.; Black-Pyrkosz, A.; Zhang, H. Comparative transcriptomics of genetically divergent lines of chickens in response to Marek’s disease virus challenge at cytolytic phase. PLoS ONE 2017, 12, e0178923. [Google Scholar] [CrossRef]
- Ebner, P.; Versteeg, G.A.; Ikeda, F. Ubiquitin enzymes in the regulation of immune responses. Crit. Rev. Biochem. Mol. 2017, 52, 425–460. [Google Scholar] [CrossRef]
- Gallo, L.H.; Ko, J.; Donoghue, D.J. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle 2017, 16, 634–648. [Google Scholar] [CrossRef]
- Cao, J.; Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2012, 2, 26. [Google Scholar] [CrossRef]
- Sadeghi, L.; Siggens, L.; Svensson, J.P.; Ekwall, K. Centromeric histone H2B monoubiquitination promotes noncoding transcription and chromatin integrity. Nat. Struct. Mol. Biol. 2014, 21, 236. [Google Scholar] [CrossRef]
- Calistri, A.; Munegato, D.; Carli, I.; Parolin, C.; Palù, G. The ubiquitin-conjugating system: Multiple roles in viral replication and infection. Cells 2014, 3, 386–417. [Google Scholar] [CrossRef]
- Schlieker, C.; Korbel, G.A.; Kattenhorn, L.M.; Ploegh, H.L. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 2005, 79, 15582–15585. [Google Scholar] [CrossRef]
- Mengyun, W.; Hongda, Z.; Meitian, W.; Yan, L.; Shanli, W.; Yongxing, A. Expression of UL36 protein of Marek’s disease virus and its activity of deubiquitinase. Anim. Husb. Vet. Med. 2017, 49, 77–82. (In Chinese) [Google Scholar]
- Navarro, M.N.; Cantrell, D.A. Serine-threonine kinases in TCR signaling. Nat. Immunol. 2014, 15, 808. [Google Scholar] [CrossRef]
- Forsburg, S.L. Eukaryotic MCM proteins: Beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004, 68, 109–131. [Google Scholar] [CrossRef]
- Bauler, T.J.; Hendriks, W.J.A.J.; King, P.D. The FERM and PDZ domain-containing protein tyrosine phosphatases, PTPN4 and PTPN3, are both dispensable for T cell receptor signal transduction. PLoS ONE 2008, 3, e4014. [Google Scholar] [CrossRef]
- Chaudhary, A.; Kamischke, C.; Leite, M.; Altura, M.A.; Kinman, L.; Kulasekara, H.; Blanc, M.-P.; Wang, G.; Terhorst, C.; Miller, S.I. β-Barrel outer membrane proteins suppress mTORC2 activation and induce autophagic responses. Sci. Signal. 2018, 11, eaat7493. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Müller, S.; Knapp, S. SH2 domains: Modulators of nonreceptor tyrosine kinase activity. Curr. Opin. Struc. Biol. 2009, 19, 643–649. [Google Scholar] [CrossRef]
- Leroux, A.E.; Schulze, J.O.; Biondi, R.M. AGC kinases, mechanisms of regulation and innovative drug development. Semin. Cancer Biol. 2018, 48, 1–17. [Google Scholar] [CrossRef]
- Duronio, R.J.; Xiong, Y. Signaling pathways that control cell proliferation. CSH Perspect. Biol. 2013, 5, a008904. [Google Scholar] [CrossRef]
- Zhang, H.-X.; Yang, P.-L.; Li, E.-M.; Xu, L.-Y. STAT3beta, a distinct isoform from STAT3. Int. J. Biochem. Cell B 2019, 110, 130–139. [Google Scholar] [CrossRef]
- Wada, T.; Penninger, J.M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 2004, 23, 2838. [Google Scholar] [CrossRef]
- Yasuda, K.; Nakanishi, K.; Tsutsui, H. Interleukin-18 in Health and Disease. Int. J. Mol. Sci. 2019, 20, 649. [Google Scholar] [CrossRef]
- Whiteside, T.L. Down-regulation of ζ-chain expression in T cells: A biomarker of prognosis in cancer? Cancer Immunol. Immunother. 2004, 53, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Takeuchi, T. CD247 variants and single-nucleotide polymorphisms observed in systemic lupus erythematosus patients. Rheumatology 2013, 52, 1551–1555. [Google Scholar]
- Wang, H.; Kadlecek, T.A.; Au-Yeung, B.B.; Goodfellow, H.E.S.; Hsu, L.-Y.; Freedman, T.S.; Weiss, A. ZAP-70: An essential kinase in T-cell signaling. CSH Perspect. Biol. 2010, 2, a002279. [Google Scholar] [CrossRef]
- Ponader, S.; Burger, J.A. Bruton’s tyrosine kinase: From X-linked agammaglobulinemia toward targeted therapy for B-cell malignancies. J. Clin. Oncol. 2014, 32, 1830–1839. [Google Scholar] [CrossRef]
- Hsueh, R.C.; Scheuermann, R.H. Tyrosine kinase activation in the decision between growth, differentiation, and death responses initiated from the B cell antigen receptor. Adv. Immunol. 2000, 75, 283–316. [Google Scholar]
- Schile, A.J.; Garcia-Fernandez, M.; Steller, H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Gene. Dev. 2008, 22, 2256–2266. [Google Scholar] [CrossRef] [PubMed]
- Deveraux, Q.L.; Takahashi, R.; Salvesen, G.S.; Reed, J.C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 1997, 388, 300–304. [Google Scholar] [CrossRef]
- Sun, S.-C. Non-canonical NF-κB signaling pathway. Cell Res. 2011, 21, 71–85. [Google Scholar] [CrossRef]
- Thuille, N.; Gruber, T.; Böck, G.; Leitges, M.; Baier, G. Protein kinase C beta is dispensable for TCR-signaling. Mol. Immunol. 2004, 41, 385–390. [Google Scholar] [CrossRef]
- Fujimitsu, K.; Grimaldi, M.; Yamano, H. Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science 2016, 352, 1121–1124. [Google Scholar] [CrossRef] [PubMed]
- Topka, S.; Vijai, J.; Walsh, M.F.; Jacobs, L.; Maria, A.; Villano, D.; Gaddam, P.; Wu, G.; McGee, R.B.; Quinn, E.; et al. Germline ETV6 mutations confer susceptibility to acute lymphoblastic leukemia and thrombocytopenia. PLoS Genet. 2015, 11, e1005262. [Google Scholar] [CrossRef]
- Sizemore, G.M.; Pitarresi, J.R.; Balakrishnan, S.; Ostrowski, M.C. The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer 2017, 17, 337. [Google Scholar] [CrossRef] [PubMed]
- Göbel, T.W.; Schneider, K.; Schaerer, B.; Mejri, I.; Puehler, F.; Weigend, S.; Staeheli, P.; Kaspers, B. IL-18 stimulates the proliferation and IFN-γ release of CD4+ T Cells in the chicken: Conservation of a Th1-like system in a nonmammalian species. J. Immunol. 2003, 171, 1809–1815. [Google Scholar] [CrossRef]
- Schneider, K.; Puehler, F.; Baeuerle, D.; Elvers, S.; Staeheli, P.; Kaspers, B.; Weining, K.C. cDNA cloning of biologically active chicken interleukin-18. J. Interferon Cytokine Res. 2000, 20, 879–883. [Google Scholar] [CrossRef]
- Liu, B.; Novick, D.; Kim, S.-H.; Rubinstein, M. Production of a biologically active human interleukin 18 requires its prior synthesis as PRO-IL-18. Cytokine 2000, 12, 1519–1525. [Google Scholar] [CrossRef]
- Dittmer, J. The biology of the Ets1 proto-oncogene. Mol. Cancer 2003, 2, 29. [Google Scholar] [CrossRef]
- Garrett-Sinha, L.A. Review of Ets1 structure, function, and roles in immunity. Cell. Mol. Life Sci. 2013, 70, 3375–3390. [Google Scholar] [CrossRef]
- Lumpkin, R.J.; Gu, H.; Zhu, Y.; Leonard, M.; Ahmad, A.S.; Clauser, K.R.; Meyer, J.G.; Bennett, E.J.; Komives, E.A. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat. Commun. 2017, 8, 1171. [Google Scholar] [CrossRef]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef]
- Brown, J.C.; Newcomb, W.W. Herpesvirus capsid assembly: Insights from structural analysis. Curr. Opin. Virol. 2011, 1, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Rock, D.L.; Kutish, G.F. The genome of a very virulent Marek’s disease virus. J. Virol. 2000, 74, 7980–7988. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Gao, H.; Cui, X.; Zhao, Y.; Shi, X.; Li, Q.; Yan, S.; Gao, M.; Wang, M.; Liu, C.; et al. Avirulent Marek’s disease virus type 1 strain 814 vectored vaccine expressing avian influenza (AI) virus H5 haemagglutinin induced better protection than turkey herpesvirus vectored AI vaccine. PLoS ONE 2013, 8, e53340. [Google Scholar] [CrossRef]
- Hathcock, K.S. T cell enrichment by nonadherence to nylon. Curr. Protoc. Immunol. 2001, 30, 3.2.1–3.2.4. [Google Scholar]
- Chen, Y.; Huang, A.; Ao, W.; Wang, Z.; Yuan, J.; Song, Q.; Wei, D.; Ye, H. Proteomic analysis of serum proteins from HIV/AIDS patients with Talaromyces marneffei infection by TMT labeling-based quantitative proteomics. Clin. Proteom. 2018, 15, 40. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liu, J.; Wei, Q.; Wang, R.; Yang, W.; Ma, Y.; Chen, G.; Yu, Y. Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation in Petunias. Plant Physiol. 2017, 173, 668–687. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.L.; Feng, M.G.; Ying, S.H. Qualitative ubiquitome unveils the potential significances of protein lysine ubiquitination in hyphal growth of Aspergillus nidulans. Curr. Genet. 2016, 62, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Udeshi, N.D.; Mertins, P.; Svinkina, T.; Carr, S.A. Large-scale identification of ubiquitination sites by mass spectrometry. Nat. Protoc. 2013, 8, 1950–1960. [Google Scholar] [CrossRef] [PubMed]
- Pundir, S.; Martin, M.J.; O’Donovan, C.; UniProt, C. UniProt Tools. Curr. Protoc. Bioinform. 2016, 53, 1.29.1–1.29.15. [Google Scholar]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bairoch, A. The ENZYME database in 2000. Nucleic Acids Res. 2000, 28, 304–305. [Google Scholar] [CrossRef]
- Tipton, K.F.; Boyce, S.; McDonald, A.G. ExplorEnz: The primary source of the IUBMB enzyme list. Nucleic Acids Res. 2008, 37, D593–D597. [Google Scholar]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztanyi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef]
- Chou, M.F.; Schwartz, D. Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinform. 2002, 35, 13.15.1–13.15.24. [Google Scholar]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhen, H.; Wang, M.; Zhao, C.; Wu, S.; Yu, P.; Lü, Y.; Wang, T.; Ai, Y. Characterization of the deubiquitination activity and substrate specificity of the chicken ubiquitin-specific protease 1/USP associated factor 1 complex. PLoS ONE 2017, 12, e0186535. [Google Scholar]
- Zhen, H.; Deng, C.; Wang, M.; Jin, W.; Wu, L.; Lü, Y.; Yu, Z.; Ai, Y. Expression and polyclonal antibody preparation of UL36USP protein encoded by MDV. Chin. J. Vet. Sci. 2014, 34, 712–716. (In Chinese) [Google Scholar]

| Motif | Motif Score 2 | Foreground | Background | Fold Increase 7 | ||
|---|---|---|---|---|---|---|
| Matches 3 | Size 4 | Matches 5 | Size 6 | |||
| ..........K.L........ 1 | 16 | 1255 | 9647 | 51,190 | 494,441 | 1.26 |
| ..........KL......... | 15.95 | 964 | 8392 | 39,148 | 443,251 | 1.3 |
| ........F.K.......... | 13.75 | 399 | 7428 | 14,597 | 404,103 | 1.49 |
| .......RD.K..N....... | 45.1 | 50 | 7029 | 234 | 389,506 | 11.84 |
| ........I.K.......... | 13.06 | 492 | 6979 | 19,499 | 389,272 | 1.41 |
| ........L.K.......... | 11.96 | 891 | 6487 | 40,364 | 369,773 | 1.26 |
| ........A.K.......... | 12.82 | 595 | 5596 | 25,931 | 329,409 | 1.35 |
| ........D.K.......... | 13.66 | 456 | 5001 | 19,269 | 303,478 | 1.44 |
| ........V.K.......... | 12.78 | 526 | 4545 | 23,908 | 284,209 | 1.38 |
| ..........K...L...... | 11.6 | 496 | 4019 | 23,564 | 260,301 | 1.36 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Wu, S.; Zhou, H.; Wang, M.; Wang, M.; Lü, Y.; Cheng, Z.; Xu, J.; Ai, Y. Marek’s Disease Virus Regulates the Ubiquitylome of Chicken CD4+ T Cells to Promote Tumorigenesis. Int. J. Mol. Sci. 2019, 20, 2089. https://doi.org/10.3390/ijms20092089
Zhou X, Wu S, Zhou H, Wang M, Wang M, Lü Y, Cheng Z, Xu J, Ai Y. Marek’s Disease Virus Regulates the Ubiquitylome of Chicken CD4+ T Cells to Promote Tumorigenesis. International Journal of Molecular Sciences. 2019; 20(9):2089. https://doi.org/10.3390/ijms20092089
Chicago/Turabian StyleZhou, Xiaolu, Shanli Wu, Hongda Zhou, Mengyun Wang, Menghan Wang, Yan Lü, Zhongyi Cheng, Jiacui Xu, and Yongxing Ai. 2019. "Marek’s Disease Virus Regulates the Ubiquitylome of Chicken CD4+ T Cells to Promote Tumorigenesis" International Journal of Molecular Sciences 20, no. 9: 2089. https://doi.org/10.3390/ijms20092089
APA StyleZhou, X., Wu, S., Zhou, H., Wang, M., Wang, M., Lü, Y., Cheng, Z., Xu, J., & Ai, Y. (2019). Marek’s Disease Virus Regulates the Ubiquitylome of Chicken CD4+ T Cells to Promote Tumorigenesis. International Journal of Molecular Sciences, 20(9), 2089. https://doi.org/10.3390/ijms20092089






