Molecular and Cellular Networks in The Suprachiasmatic Nuclei
Abstract
:1. Introduction
2. General Principles of Cell-Autonomous Circadian Timekeeping
3. Cell-Autonomous Circadian Timekeeping in SCN Neurons
4. The Origin of Circadian Precision and Robustness within The SCN
5. Complexity of Circadian Entrainment in The Integrated SCN
6. Topological and Functional Complexity of The SCN
7. Exploring The SCN Cell Network Organization
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SCN | Suprachiasmatic nuclei |
| TTFL | Transcription-translation feedback loops |
| CCG | Clock-controlled gene |
| VIP | Vasoactive intestinal polypeptide |
| AVP | Arginine Vasopressin |
| GABA | Gamma-aminobutyric acid |
| SPZ | Subparaventricular zone |
| GH | Growth hormone |
| SMS | Somatostatin |
| GHRH | GH-releasing hormone |
References
- Edgar, R.S.; Green, E.W.; Zhao, Y.; van Ooijen, G.; Olmedo, M.; Qin, X.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Woelfle, M.A.; Ouyang, Y.; Phanvijhitsiri, K.; Johnson, C.H. The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr. Biol. 2004, 14, 1481–1486. [Google Scholar] [CrossRef]
- Yerushalmi, S.; Green, R.M. Evidence for the adaptive significance of circadian rhythms. Ecol. Lett. 2009, 12, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Cordina-Duverger, E.; Menegaux, F.; Popa, A.; Rabstein, S.; Harth, V.; Pesch, B.; Bruning, T.; Fritschi, L.; Glass, D.C.; Heyworth, J.S.; et al. Night shift work and breast cancer: A pooled analysis of population-based case-control studies with complete work history. Eur. J. Epidemiol. 2018, 33, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Logan, R.W.; McClung, C.A. Rhythms of life: Circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 2019, 20, 49–65. [Google Scholar] [PubMed]
- Thosar, S.S.; Butler, M.P.; Shea, S.A. Role of the circadian system in cardiovascular disease. J. Clin. Investig. 2018, 128, 2157–2167. [Google Scholar] [CrossRef]
- Watanabe, K.; Sakuraya, A.; Kawakami, N.; Imamura, K.; Ando, E.; Asai, Y.; Eguchi, H.; Kobayashi, Y.; Nishida, N.; Arima, H.; et al. Work-related psychosocial factors and metabolic syndrome onset among workers: A systematic review and meta-analysis. Obes. Rev. 2018, 19, 1557–1568. [Google Scholar] [CrossRef]
- Wefers, J.; van Moorsel, D.; Hansen, J.; Connell, N.J.; Havekes, B.; Hoeks, J.; van Marken Lichtenbelt, W.D.; Duez, H.; Phielix, E.; Kalsbeek, A.; et al. Circadian misalignment induces fatty acid metabolism gene profiles and compromises insulin sensitivity in human skeletal muscle. Proc. Natl. Acad. Sci. USA 2018, 115, 7789–7794. [Google Scholar] [CrossRef]
- Hastings, M.H. Physiology. A looser clock to cure jet lag. Science 2013, 342, 52–53. [Google Scholar] [CrossRef]
- Moore, R.Y.; Eichler, V.B. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972, 42, 201–206. [Google Scholar] [CrossRef]
- Stephan, F.K.; Zucker, I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. USA 1972, 69, 1583–1586. [Google Scholar] [CrossRef]
- Herzog, E.D.; Aton, S.J.; Numano, R.; Sakaki, Y.; Tei, H. Temporal precision in the mammalian circadian system: A reliable clock from less reliable neurons. J. Biol. Rhythm. 2004, 19, 35–46. [Google Scholar] [CrossRef]
- Honma, S.; Nakamura, W.; Shirakawa, T.; Honma, K. Diversity in the circadian periods of single neurons of the rat suprachiasmatic nucleus depends on nuclear structure and intrinsic period. Neurosci. Lett. 2004, 358, 173–176. [Google Scholar] [CrossRef]
- Liu, A.C.; Welsh, D.K.; Ko, C.H.; Tran, H.G.; Zhang, E.E.; Priest, A.A.; Buhr, E.D.; Singer, O.; Meeker, K.; Verma, I.M.; et al. Intercellular coupling confers robustness against mutations in the scn circadian clock network. Cell 2007, 129, 605–616. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Isejima, H.; Matsuo, T.; Okura, R.; Yagita, K.; Kobayashi, M.; Okamura, H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 2003, 302, 1408–1412. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef]
- Young, M.W. Time travels: A 40-year journey from drosophila’s clock mutants to human circadian disorders (nobel lecture). Angew. Chem. Int. Ed. Engl. 2018, 57, 11532–11539. [Google Scholar] [CrossRef]
- O’Neill, J.S.; Reddy, A.B. Circadian clocks in human red blood cells. Nature 2011, 469, 498–503. [Google Scholar] [CrossRef]
- Reddy, A.B.; Rey, G. Metabolic and nontranscriptional circadian clocks: Eukaryotes. Annu. Rev. Biochem. 2014, 83, 165–189. [Google Scholar] [CrossRef]
- Wong, D.C.; O’Neill, J.S. Non-transcriptional processes in circadian rhythm generation. Curr. Opin. Physiol. 2018, 5, 117–132. [Google Scholar] [CrossRef]
- Nagoshi, E.; Saini, C.; Bauer, C.; Laroche, T.; Naef, F.; Schibler, U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 2004, 119, 693–705. [Google Scholar] [CrossRef]
- Yagita, K.; Tamanini, F.; van Der Horst, G.T.; Okamura, H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science 2001, 292, 278–281. [Google Scholar] [CrossRef]
- Rey, G.; Cesbron, F.; Rougemont, J.; Reinke, H.; Brunner, M.; Naef, F. Genome-wide and phase-specific DNA-binding rhythms of bmal1 control circadian output functions in mouse liver. PLoS Biol. 2011, 9, e1000595. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Torres, M.; Becquet, D.; Blanchard, M.P.; Guillen, S.; Boyer, B.; Moreno, M.; Franc, J.L.; Francois-Bellan, A.M. Circadian rna expression elicited by 3’-utr iralu-paraspeckle associated elements. Elife 2016, 5. [Google Scholar] [CrossRef]
- Beytebiere, J.R.; Trott, A.J.; Greenwell, B.J.; Osborne, C.A.; Vitet, H.; Spence, J.; Yoo, S.H.; Chen, Z.; Takahashi, J.S.; Ghaffari, N.; et al. Tissue-specific bmal1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev. 2019, 33, 294–309. [Google Scholar] [CrossRef]
- Storch, K.F.; Lipan, O.; Leykin, I.; Viswanathan, N.; Davis, F.C.; Wong, W.H.; Weitz, C.J. Extensive and divergent circadian gene expression in liver and heart. Nature 2002, 417, 78–83. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 359. [Google Scholar] [CrossRef]
- Travnickova-Bendova, Z.; Cermakian, N.; Reppert, S.M.; Sassone-Corsi, P. Bimodal regulation of mperiod promoters by creb-dependent signaling and clock/bmal1 activity. Proc. Natl. Acad. Sci. USA 2002, 99, 7728–7733. [Google Scholar] [CrossRef]
- Yagita, K.; Okamura, H. Forskolin induces circadian gene expression of rper1, rper2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 2000, 465, 79–82. [Google Scholar] [CrossRef]
- Cheon, S.; Park, N.; Cho, S.; Kim, K. Glucocorticoid-mediated period2 induction delays the phase of circadian rhythm. Nucleic Acids Res. 2013, 41, 6161–6174. [Google Scholar] [CrossRef]
- Le Minh, N.; Damiola, F.; Tronche, F.; Schutz, G.; Schibler, U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001, 20, 7128–7136. [Google Scholar] [CrossRef]
- So, A.Y.; Bernal, T.U.; Pillsbury, M.L.; Yamamoto, K.R.; Feldman, B.J. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl. Acad. Sci. USA 2009, 106, 17582–17587. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nakahata, Y.; Tanaka, M.; Yoshida, M.; Soma, H.; Shinohara, K.; Yasuda, A.; Mamine, T.; Takumi, T. Acute physical stress elevates mouse period1 mrna expression in mouse peripheral tissues via a glucocorticoid-responsive element. J. Biol. Chem. 2005, 280, 42036–42043. [Google Scholar] [CrossRef]
- Kornmann, B.; Schaad, O.; Bujard, H.; Takahashi, J.S.; Schibler, U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007, 5, e34. [Google Scholar] [CrossRef]
- Saini, C.; Morf, J.; Stratmann, M.; Gos, P.; Schibler, U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012, 26, 567–580. [Google Scholar] [CrossRef]
- Tamaru, T.; Hattori, M.; Honda, K.; Benjamin, I.; Ozawa, T.; Takamatsu, K. Synchronization of circadian per2 rhythms and hsf1-bmal1: Clock interaction in mouse fibroblasts after short-term heat shock pulse. PLoS ONE 2011, 6, e24521. [Google Scholar] [CrossRef]
- Morf, J.; Rey, G.; Schneider, K.; Stratmann, M.; Fujita, J.; Naef, F.; Schibler, U. Cold-inducible rna-binding protein modulates circadian gene expression posttranscriptionally. Science 2012, 338, 379–383. [Google Scholar] [CrossRef]
- Panda, S.; Antoch, M.P.; Miller, B.H.; Su, A.I.; Schook, A.B.; Straume, M.; Schultz, P.G.; Kay, S.A.; Takahashi, J.S.; Hogenesch, J.B. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002, 109, 307–320. [Google Scholar] [CrossRef]
- Jin, X.; Shearman, L.P.; Weaver, D.R.; Zylka, M.J.; de Vries, G.J.; Reppert, S.M. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 1999, 96, 57–68. [Google Scholar] [CrossRef]
- Hurst, W.J.; Mitchell, J.W.; Gillette, M.U. Synchronization and phase-resetting by glutamate of an immortalized scn cell line. Biochem. Biophys. Res. Commun. 2002, 298, 133–143. [Google Scholar] [CrossRef]
- Cheng, M.Y.; Bullock, C.M.; Li, C.; Lee, A.G.; Bermak, J.C.; Belluzzi, J.; Weaver, D.R.; Leslie, F.M.; Zhou, Q.Y. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 2002, 417, 405–410. [Google Scholar] [CrossRef]
- Deery, M.J.; Maywood, E.S.; Chesham, J.E.; Sladek, M.; Karp, N.A.; Green, E.W.; Charles, P.D.; Reddy, A.B.; Kyriacou, C.P.; Lilley, K.S.; et al. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr. Biol. 2009, 19, 2031–2036. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.H.; Yamazaki, S.; Lowrey, P.L.; Shimomura, K.; Ko, C.H.; Buhr, E.D.; Siepka, S.M.; Hong, H.K.; Oh, W.J.; Yoo, O.J.; et al. Period2::Luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 5339–5346. [Google Scholar] [CrossRef] [PubMed]
- Welsh, D.K.; Logothetis, D.E.; Meister, M.; Reppert, S.M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 1995, 14, 697–706. [Google Scholar] [CrossRef]
- Albus, H.; Bonnefont, X.; Chaves, I.; Yasui, A.; Doczy, J.; van der Horst, G.T.; Meijer, J.H. Cryptochrome-deficient mice lack circadian electrical activity in the suprachiasmatic nuclei. Curr. Biol. 2002, 12, 1130–1133. [Google Scholar] [CrossRef]
- Colwell, C.S. Linking neural activity and molecular oscillations in the scn. Nat. Rev. Neurosci. 2011, 12, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Belle, M.D.; Diekman, C.O.; Forger, D.B.; Piggins, H.D. Daily electrical silencing in the mammalian circadian clock. Science 2009, 326, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Belle, M.D.C.; Diekman, C.O. Neuronal oscillations on an ultra-slow timescale: Daily rhythms in electrical activity and gene expression in the mammalian master circadian clockwork. Eur. J. Neurosci. 2018, 48, 2696–2717. [Google Scholar]
- Kudo, T.; Block, G.D.; Colwell, C.S. The circadian clock gene period1 connects the molecular clock to neural activity in the suprachiasmatic nucleus. Asn Neuro 2015, 7. [Google Scholar] [CrossRef]
- Liu, C.; Weaver, D.R.; Strogatz, S.H.; Reppert, S.M. Cellular construction of a circadian clock: Period determination in the suprachiasmatic nuclei. Cell 1997, 91, 855–860. [Google Scholar] [CrossRef]
- Nakamura, W.; Honma, S.; Shirakawa, T.; Honma, K. Clock mutation lengthens the circadian period without damping rhythms in individual scn neurons. Nat. Neurosci. 2002, 5, 399–400. [Google Scholar] [CrossRef]
- Flourakis, M.; Kula-Eversole, E.; Hutchison, A.L.; Han, T.H.; Aranda, K.; Moose, D.L.; White, K.P.; Dinner, A.R.; Lear, B.C.; Ren, D.; et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell 2015, 162, 836–848. [Google Scholar] [CrossRef]
- Webb, A.B.; Angelo, N.; Huettner, J.E.; Herzog, E.D. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc. Natl. Acad. Sci. USA 2009, 106, 16493–16498. [Google Scholar] [CrossRef]
- Michel, S.; Meijer, J.H. From clock to functional pacemaker. Eur. J. Neurosci. 2019. [Google Scholar] [CrossRef]
- Hastings, M.H.; Maywood, E.S.; Brancaccio, M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 2018, 19, 453–469. [Google Scholar] [CrossRef]
- Ko, C.H.; Yamada, Y.R.; Welsh, D.K.; Buhr, E.D.; Liu, A.C.; Zhang, E.E.; Ralph, M.R.; Kay, S.A.; Forger, D.B.; Takahashi, J.S. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010, 8, e1000513. [Google Scholar] [CrossRef]
- Colwell, C.S.; Michel, S.; Itri, J.; Rodriguez, W.; Tam, J.; Lelievre, V.; Hu, Z.; Liu, X.; Waschek, J.A. Disrupted circadian rhythms in vip- and phi-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 285, R939–R949. [Google Scholar] [CrossRef]
- Harmar, A.J.; Marston, H.M.; Shen, S.; Spratt, C.; West, K.M.; Sheward, W.J.; Morrison, C.F.; Dorin, J.R.; Piggins, H.D.; Reubi, J.C.; et al. The vpac(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 2002, 109, 497–508. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Suzuki, T.; Mizoro, Y.; Kori, H.; Okada, K.; Chen, Y.; Fustin, J.M.; Yamazaki, F.; Mizuguchi, N.; Zhang, J.; et al. Mice genetically deficient in vasopressin v1a and v1b receptors are resistant to jet lag. Science 2013, 342, 85–90. [Google Scholar] [CrossRef]
- Brown, S.A.; Zumbrunn, G.; Fleury-Olela, F.; Preitner, N.; Schibler, U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr. Biol. 2002, 12, 1574–1583. [Google Scholar] [CrossRef]
- Abraham, U.; Granada, A.E.; Westermark, P.O.; Heine, M.; Kramer, A.; Herzel, H. Coupling governs entrainment range of circadian clocks. Mol. Syst. Biol. 2010, 6, 438. [Google Scholar] [CrossRef]
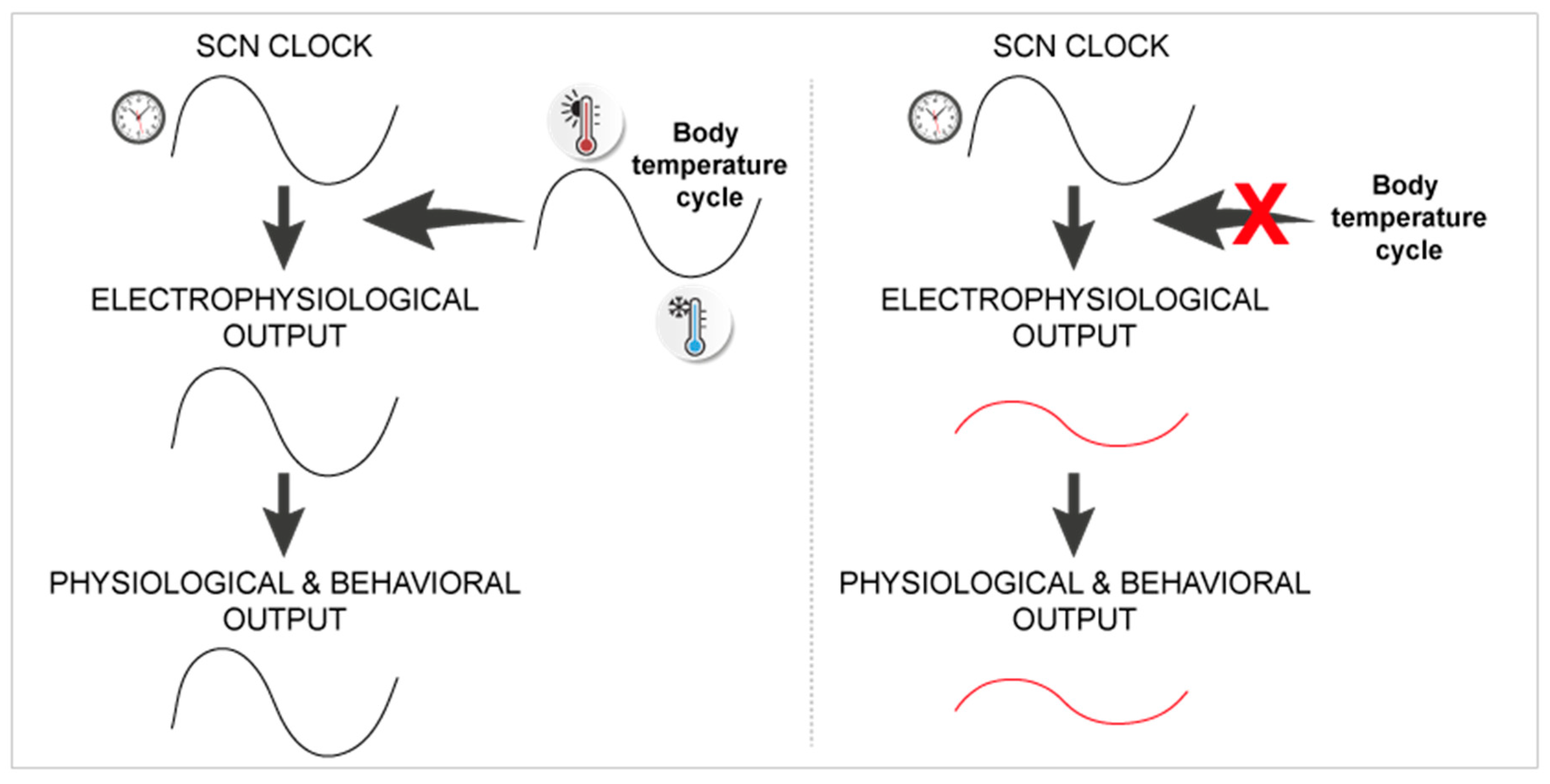
- Buhr, E.D.; Yoo, S.H.; Takahashi, J.S. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 2010, 330, 379–385. [Google Scholar] [CrossRef]
- Herzog, E.D.; Huckfeldt, R.M. Circadian entrainment to temperature, but not light, in the isolated suprachiasmatic nucleus. J. Neurophysiol. 2003, 90, 763–770. [Google Scholar] [CrossRef]
- Ruby, N.F.; Burns, D.E.; Heller, H.C. Circadian rhythms in the suprachiasmatic nucleus are temperature-compensated and phase-shifted by heat pulses in vitro. J. Neurosci. 1999, 19, 8630–8636. [Google Scholar] [CrossRef]
- Ruby, N.F. Rethinking temperature sensitivity of the suprachiasmatic nucleus. J. Biol. Rhythm. 2011, 26, 368–370; author reply 371–363. [Google Scholar] [CrossRef]
- Abitbol, K.; Debiesse, S.; Molino, F.; Mesirca, P.; Bidaud, I.; Minami, Y.; Mangoni, M.E.; Yagita, K.; Mollard, P.; Bonnefont, X. Clock-dependent and system-driven oscillators interact in the suprachiasmatic nuclei to pace mammalian circadian rhythms. PLoS ONE 2017, 12, e0187001. [Google Scholar] [CrossRef]
- Ingenwerth, M.; Noichl, E.; Stahr, A.; Korf, H.W.; Reinke, H.; von Gall, C. Heat shock factor 1 deficiency affects systemic body temperature regulation. Neuroendocrinology 2016, 103, 605–615. [Google Scholar] [CrossRef]
- Reinke, H.; Saini, C.; Fleury-Olela, F.; Dibner, C.; Benjamin, I.J.; Schibler, U. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008, 22, 331–345. [Google Scholar] [CrossRef]
- Hung, H.C.; Kay, S.A.; Weber, F. Hsp90, a capacitor of behavioral variation. J. Biol. Rhythm. 2009, 24, 183–192. [Google Scholar] [CrossRef]
- Xu, F.; Kula-Eversole, E.; Iwanaszko, M.; Hutchison, A.L.; Dinner, A.; Allada, R. Circadian clocks function in concert with heat shock organizing protein to modulate mutant huntingtin aggregation and toxicity. Cell Rep. 2019, 27, 59–70 e54. [Google Scholar] [CrossRef]
- Goh, G.H.; Mark, P.J.; Maloney, S.K. Altered energy intake and the amplitude of the body temperature rhythm are associated with changes in phase, but not amplitude, of clock gene expression in the rat suprachiasmatic nucleus in vivo. Chronobiol. Int. 2016, 1–13. [Google Scholar] [CrossRef]
- Nakamura, T.J.; Nakamura, W.; Yamazaki, S.; Kudo, T.; Cutler, T.; Colwell, C.S.; Block, G.D. Age-related decline in circadian output. J. Neurosci. 2011, 31, 10201–10205. [Google Scholar] [CrossRef]
- McArthur, A.J.; Gillette, M.U.; Prosser, R.A. Melatonin directly resets the rat suprachiasmatic circadian clock in vitro. Brain Res. 1991, 565, 158–161. [Google Scholar] [CrossRef]
- Poirel, V.J.; Boggio, V.; Dardente, H.; Pevet, P.; Masson-Pevet, M.; Gauer, F. Contrary to other non-photic cues, acute melatonin injection does not induce immediate changes of clock gene mrna expression in the rat suprachiasmatic nuclei. Neuroscience 2003, 120, 745–755. [Google Scholar] [CrossRef]
- Agez, L.; Laurent, V.; Pevet, P.; Masson-Pevet, M.; Gauer, F. Melatonin affects nuclear orphan receptors mrna in the rat suprachiasmatic nuclei. Neuroscience 2007, 144, 522–530. [Google Scholar] [CrossRef]
- Best, J.D.; Maywood, E.S.; Smith, K.L.; Hastings, M.H. Rapid resetting of the mammalian circadian clock. J. Neurosci. 1999, 19, 828–835. [Google Scholar] [CrossRef]
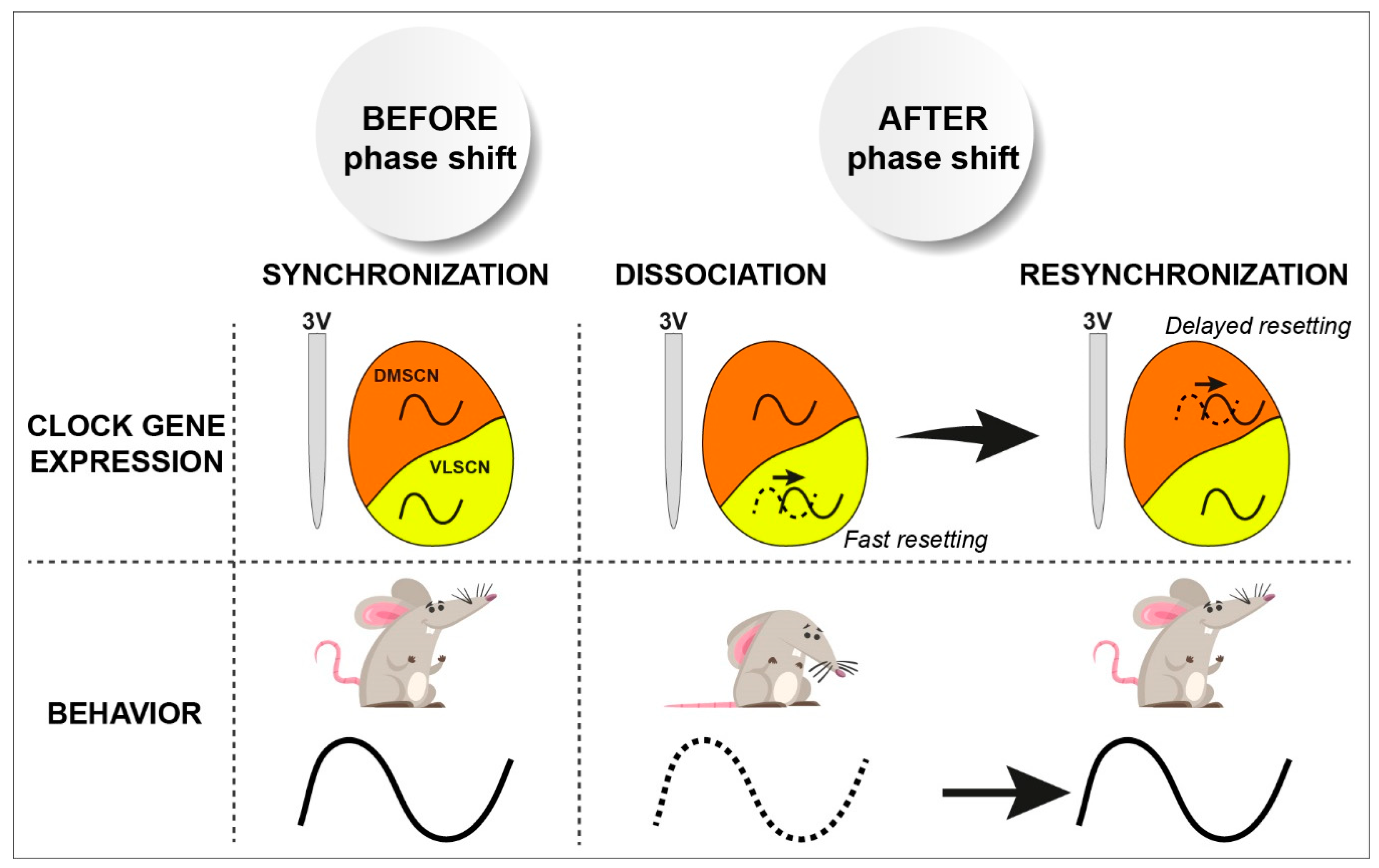
- Vansteensel, M.J.; Yamazaki, S.; Albus, H.; Deboer, T.; Block, G.D.; Meijer, J.H. Dissociation between circadian per1 and neuronal and behavioral rhythms following a shifted environmental cycle. Curr. Biol. 2003, 13, 1538–1542. [Google Scholar] [CrossRef]
- Yamazaki, S.; Numano, R.; Abe, M.; Hida, A.; Takahashi, R.; Ueda, M.; Block, G.D.; Sakaki, Y.; Menaker, M.; Tei, H. Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000, 288, 682–685. [Google Scholar] [CrossRef]
- Takamure, M.; Murakami, N.; Takahashi, K.; Kuroda, H.; Etoh, T. Rapid reentrainment of the circadian clock itself, but not the measurable activity rhythms to a new light-dark cycle in the rat. Physiol. Behav. 1991, 50, 443–449. [Google Scholar] [CrossRef]
- Davidson, A.J.; Castanon-Cervantes, O.; Leise, T.L.; Molyneux, P.C.; Harrington, M.E. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur. J. Neurosci. 2009, 29, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Nagano, M.; Adachi, A.; Nakahama, K.; Nakamura, T.; Tamada, M.; Meyer-Bernstein, E.; Sehgal, A.; Shigeyoshi, Y. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J. Neurosci. 2003, 23, 6141–6151. [Google Scholar] [CrossRef]
- Nakamura, W.; Yamazaki, S.; Takasu, N.N.; Mishima, K.; Block, G.D. Differential response of period 1 expression within the suprachiasmatic nucleus. J. Neurosci. 2005, 25, 5481–5487. [Google Scholar] [CrossRef]
- Albus, H.; Vansteensel, M.J.; Michel, S.; Block, G.D.; Meijer, J.H. A gabaergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr. Biol. 2005, 15, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Abrahamson, E.E.; Moore, R.Y. Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001, 916, 172–191. [Google Scholar] [CrossRef]
- Leak, R.K.; Moore, R.Y. Topographic organization of suprachiasmatic nucleus projection neurons. J. Comp. Neurol. 2001, 433, 312–334. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, K.; Honma, S.; Katsuno, Y.; Abe, H.; Honma, K. Circadian rhythms in the release of vasoactive intestinal polypeptide and arginine-vasopressin in organotypic slice culture of rat suprachiasmatic nucleus. Neurosci. Lett. 1994, 170, 183–186. [Google Scholar] [CrossRef]
- Moffitt, J.R.; Bambah-Mukku, D.; Eichhorn, S.W.; Vaughn, E.; Shekhar, K.; Perez, J.D.; Rubinstein, N.D.; Hao, J.; Regev, A.; Dulac, C.; et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 2018, 362. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Zhu, H.; O’Sullivan, S.; Ogunnaike, B.A.; Weaver, D.R.; Schwaber, J.S.; Vadigepalli, R. Single-cell transcriptional analysis reveals novel neuronal phenotypes and interaction networks involved in the central circadian clock. Front. Neurosci. 2016, 10, 481. [Google Scholar] [CrossRef]
- Romanov, R.A.; Zeisel, A.; Bakker, J.; Girach, F.; Hellysaz, A.; Tomer, R.; Alpar, A.; Mulder, J.; Clotman, F.; Keimpema, E.; et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 2017, 20, 176–188. [Google Scholar] [CrossRef]
- Lee, I.T.; Chang, A.S.; Manandhar, M.; Shan, Y.; Fan, J.; Izumo, M.; Ikeda, Y.; Motoike, T.; Dixon, S.; Seinfeld, J.E.; et al. Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 2015, 85, 1086–1102. [Google Scholar] [CrossRef]
- Mieda, M.; Ono, D.; Hasegawa, E.; Okamoto, H.; Honma, K.; Honma, S.; Sakurai, T. Cellular clocks in avp neurons of the scn are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 2015, 85, 1103–1116. [Google Scholar] [CrossRef]
- Low-Zeddies, S.S.; Takahashi, J.S. Chimera analysis of the clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell 2001, 105, 25–42. [Google Scholar] [CrossRef]
- Smyllie, N.J.; Chesham, J.E.; Hamnett, R.; Maywood, E.S.; Hastings, M.H. Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. USA 2016, 113, 3657–3662. [Google Scholar] [CrossRef]
- Prolo, L.M.; Takahashi, J.S.; Herzog, E.D. Circadian rhythm generation and entrainment in astrocytes. J. Neurosci. 2005, 25, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Girardet, C.; Lebrun, B.; Cabirol-Pol, M.J.; Tardivel, C.; Francois-Bellan, A.M.; Becquet, D.; Bosler, O. Brain-derived neurotrophic factor/trkb signaling regulates daily astroglial plasticity in the suprachiasmatic nucleus: Electron-microscopic evidence in mouse. Glia 2013, 61, 1172–1177. [Google Scholar] [CrossRef]
- Barca-Mayo, O.; Pons-Espinal, M.; Follert, P.; Armirotti, A.; Berdondini, L.; De Pietri Tonelli, D. Astrocyte deletion of bmal1 alters daily locomotor activity and cognitive functions via gaba signalling. Nat. Commun. 2017, 8, 14336. [Google Scholar] [CrossRef] [PubMed]
- Tso, C.F.; Simon, T.; Greenlaw, A.C.; Puri, T.; Mieda, M.; Herzog, E.D. Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr. Biol. 2017, 27, 1055–1061. [Google Scholar] [CrossRef]
- Brancaccio, M.; Patton, A.P.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron 2017, 93, 1420–1435 e1425. [Google Scholar] [CrossRef]
- Brancaccio, M.; Edwards, M.D.; Patton, A.P.; Smyllie, N.J.; Chesham, J.E.; Maywood, E.S.; Hastings, M.H. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 2019, 363, 187–192. [Google Scholar] [CrossRef]
- Aschoff, J. Circadian rhythms in man. Science 1965, 148, 1427–1432. [Google Scholar] [CrossRef]
- Cambras, T.; Weller, J.R.; Angles-Pujoras, M.; Lee, M.L.; Christopher, A.; Diez-Noguera, A.; Krueger, J.M.; de la Iglesia, H.O. Circadian desynchronization of core body temperature and sleep stages in the rat. Proc. Natl. Acad. Sci. USA 2007, 104, 7634–7639. [Google Scholar] [CrossRef]
- Abrahamson, E.E.; Moore, R.Y. Lesions of suprachiasmatic nucleus efferents selectively affect rest-activity rhythm. Mol. Cell. Endocrinol. 2006, 252, 46–56. [Google Scholar] [CrossRef]
- Vujovic, N.; Gooley, J.J.; Jhou, T.C.; Saper, C.B. Projections from the subparaventricular zone define four channels of output from the circadian timing system. J. Comp. Neurol. 2015, 523, 2714–2737. [Google Scholar] [CrossRef]
- Bur, I.M.; Cohen-Solal, A.M.; Carmignac, D.; Abecassis, P.Y.; Chauvet, N.; Martin, A.O.; van der Horst, G.T.; Robinson, I.C.; Maurel, P.; Mollard, P.; Bonnefont, X. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J. Biol. Chem. 2009, 284, 9066–9073. [Google Scholar] [CrossRef]
- Davies, J.S.; Carter, D.A.; Wells, T. Photic stimulation inhibits growth hormone secretion in rats: A hypothalamic mechanism for transient entrainment. Endocrinology 2004, 145, 2950–2958. [Google Scholar] [CrossRef]
- Willoughby, J.O.; Martin, J.B. The suprachiasmatic nucleus synchronizes growth hormone secretory rhythms with the light-dark cycle. Brain Res. 1978, 151, 413–417. [Google Scholar] [CrossRef]
- Yulyaningsih, E.; Loh, K.; Lin, S.; Lau, J.; Zhang, L.; Shi, Y.; Berning, B.A.; Enriquez, R.; Driessler, F.; Macia, L.; et al. Pancreatic polypeptide controls energy homeostasis via npy6r signaling in the suprachiasmatic nucleus in mice. Cell Metab. 2014, 19, 58–72. [Google Scholar] [CrossRef]
- Glad, C.A.; Kitchen, E.E.; Russ, G.C.; Harris, S.M.; Davies, J.S.; Gevers, E.F.; Gabrielsson, B.G.; Wells, T. Reverse feeding suppresses the activity of the gh axis in rats and induces a preobesogenic state. Endocrinology 2011, 152, 869–882. [Google Scholar] [CrossRef]
- Freeman, G.M., Jr.; Krock, R.M.; Aton, S.J.; Thaben, P.; Herzog, E.D. Gaba networks destabilize genetic oscillations in the circadian pacemaker. Neuron 2013, 78, 799–806. [Google Scholar] [CrossRef]
- Liu, C.; Reppert, S.M. Gaba synchronizes clock cells within the suprachiasmatic circadian clock. Neuron 2000, 25, 123–128. [Google Scholar] [CrossRef]
- Maywood, E.S.; Chesham, J.E.; O’Brien, J.A.; Hastings, M.H. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. USA 2011, 108, 14306–14311. [Google Scholar] [CrossRef]
- Brancaccio, M.; Maywood, E.S.; Chesham, J.E.; Loudon, A.S.; Hastings, M.H. A gq-ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 2013, 78, 714–728. [Google Scholar] [CrossRef]
- Enoki, R.; Kuroda, S.; Ono, D.; Hasan, M.T.; Ueda, T.; Honma, S.; Honma, K. Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. USA 2012, 109, 21498–21503. [Google Scholar] [CrossRef]
- Enoki, R.; Oda, Y.; Mieda, M.; Ono, D.; Honma, S.; Honma, K.I. Synchronous circadian voltage rhythms with asynchronous calcium rhythms in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. USA 2017, 114, E2476–E2485. [Google Scholar] [CrossRef]
- Hong, J.H.; Jeong, B.; Min, C.H.; Lee, K.J. Circadian waves of cytosolic calcium concentration and long-range network connections in rat suprachiasmatic nucleus. Eur. J. Neurosci. 2012, 35, 1417–1425. [Google Scholar] [CrossRef]
- Kruse, F.; Junker, J.P.; van Oudenaarden, A.; Bakkers, J. Tomo-seq: A method to obtain genome-wide expression data with spatial resolution. Methods Cell Biol. 2016, 135, 299–307. [Google Scholar]
- Abel, J.H.; Meeker, K.; Granados-Fuentes, D.; St John, P.C.; Wang, T.J.; Bales, B.B.; Doyle, F.J., 3rd; Herzog, E.D.; Petzold, L.R. Functional network inference of the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. USA 2016, 113, 4512–4517. [Google Scholar] [CrossRef]
- Taylor, S.R.; Wang, T.J.; Granados-Fuentes, D.; Herzog, E.D. Resynchronization dynamics reveal that the ventral entrains the dorsal suprachiasmatic nucleus. J. Biol. Rhythm. 2017, 32, 35–47. [Google Scholar] [CrossRef]
- Hafner, M.; Koeppl, H.; Gonze, D. Effect of network architecture on synchronization and entrainment properties of the circadian oscillations in the suprachiasmatic nucleus. PLoS Comput. Biol. 2012, 8, e1002419. [Google Scholar] [CrossRef] [PubMed]
- Inouye, S.T.; Kawamura, H. Persistence of circadian rhythmicity in a mammalian hypothalamic "island" containing the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. USA 1979, 76, 5962–5966. [Google Scholar] [CrossRef]
- Meijer, J.H.; Schaap, J.; Watanabe, K.; Albus, H. Multiunit activity recordings in the suprachiasmatic nuclei: In vivo versus in vitro models. Brain Res. 1997, 753, 322–327. [Google Scholar] [CrossRef]
- Tsuji, T.; Tsuji, C.; Ludwig, M.; Leng, G. The rat suprachiasmatic nucleus: The master clock ticks at 30 hz. J. Physiol. 2016, 594, 3629–3650. [Google Scholar] [CrossRef]
- Ono, D.; Honma, S.; Nakajima, Y.; Kuroda, S.; Enoki, R.; Honma, K.I. Dissociation of per1 and bmal1 circadian rhythms in the suprachiasmatic nucleus in parallel with behavioral outputs. Proc. Natl. Acad. Sci. USA 2017, 114, E3699–E3708. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Kobayashi, M.; Mitsui, S.; Ishida, Y.; van der Horst, G.T.; Suzuki, M.; Shibata, S.; Okamura, H. View of a mouse clock gene ticking. Nature 2001, 409, 684. [Google Scholar] [CrossRef]
- Mei, L.; Fan, Y.; Lv, X.; Welsh, D.K.; Zhan, C.; Zhang, E.E. Long-term in vivo recording of circadian rhythms in brains of freely moving mice. Proc. Natl. Acad. Sci. USA 2018, 115, 4276–4281. [Google Scholar] [CrossRef]
- Jones, J.R.; Simon, T.; Lones, L.; Herzog, E.D. Scn vip neurons are essential for normal light-mediated resetting of the circadian system. J. Neurosci. 2018, 38, 7986–7995. [Google Scholar] [CrossRef]
- Colwell, C.S. Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur. J. Neurosci. 2000, 12, 571–576. [Google Scholar] [CrossRef]
- Ghosh, K.K.; Burns, L.D.; Cocker, E.D.; Nimmerjahn, A.; Ziv, Y.; Gamal, A.E.; Schnitzer, M.J. Miniaturized integration of a fluorescence microscope. Nat. Methods 2011, 8, 871–878. [Google Scholar] [CrossRef]
- Ziv, Y.; Burns, L.D.; Cocker, E.D.; Hamel, E.O.; Ghosh, K.K.; Kitch, L.J.; El Gamal, A.; Schnitzer, M.J. Long-term dynamics of ca1 hippocampal place codes. Nat. Neurosci. 2013, 16, 264–266. [Google Scholar] [CrossRef]
- Chen, K.S.; Xu, M.; Zhang, Z.; Chang, W.C.; Gaj, T.; Schaffer, D.V.; Dan, Y. A hypothalamic switch for rem and non-rem sleep. Neuron 2018, 97, 1168–1176. [Google Scholar] [CrossRef]
- Jennings, J.H.; Ung, R.L.; Resendez, S.L.; Stamatakis, A.M.; Taylor, J.G.; Huang, J.; Veleta, K.; Kantak, P.A.; Aita, M.; Shilling-Scrivo, K.; et al. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell 2015, 160, 516–527. [Google Scholar] [CrossRef]
- Remedios, R.; Kennedy, A.; Zelikowsky, M.; Grewe, B.F.; Schnitzer, M.J.; Anderson, D.J. Social behaviour shapes hypothalamic neural ensemble representations of conspecific sex. Nature 2017, 550, 388–392. [Google Scholar] [CrossRef]
- Kohl, J.; Babayan, B.M.; Rubinstein, N.D.; Autry, A.E.; Marin-Rodriguez, B.; Kapoor, V.; Miyamishi, K.; Zweifel, L.S.; Luo, L.; Uchida, N.; et al. Functional circuit architecture underlying parental behaviour. Nature 2018, 556, 326–331. [Google Scholar] [CrossRef]
- Guo, F.; Chen, X.; Rosbash, M. Temporal calcium profiling of specific circadian neurons in freely moving flies. Proc. Natl. Acad. Sci. USA 2017, 114, E8780–E8787. [Google Scholar] [CrossRef]
- Li, M.T.; Cao, L.H.; Xiao, N.; Tang, M.; Deng, B.; Yang, T.; Yoshii, T.; Luo, D.G. Hub-organized parallel circuits of central circadian pacemaker neurons for visual photoentrainment in drosophila. Nat. Commun. 2018, 9, 4247. [Google Scholar] [CrossRef]
- Liang, X.; Holy, T.E.; Taghert, P.H. Synchronous drosophila circadian pacemakers display nonsynchronous ca(2)(+) rhythms in vivo. Science 2016, 351, 976–981. [Google Scholar] [CrossRef]
- Liang, X.; Holy, T.E.; Taghert, P.H. A series of suppressive signals within the drosophila circadian neural circuit generates sequential daily outputs. Neuron 2017, 94, 1173–1189. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Cheikh Hussein, L.; Mollard, P.; Bonnefont, X. Molecular and Cellular Networks in The Suprachiasmatic Nuclei. Int. J. Mol. Sci. 2019, 20, 2052. https://doi.org/10.3390/ijms20082052
El Cheikh Hussein L, Mollard P, Bonnefont X. Molecular and Cellular Networks in The Suprachiasmatic Nuclei. International Journal of Molecular Sciences. 2019; 20(8):2052. https://doi.org/10.3390/ijms20082052
Chicago/Turabian StyleEl Cheikh Hussein, Lama, Patrice Mollard, and Xavier Bonnefont. 2019. "Molecular and Cellular Networks in The Suprachiasmatic Nuclei" International Journal of Molecular Sciences 20, no. 8: 2052. https://doi.org/10.3390/ijms20082052
APA StyleEl Cheikh Hussein, L., Mollard, P., & Bonnefont, X. (2019). Molecular and Cellular Networks in The Suprachiasmatic Nuclei. International Journal of Molecular Sciences, 20(8), 2052. https://doi.org/10.3390/ijms20082052