Abstract
The colonic epithelium is exposed to a mixture of compounds through diet, among which some are procarcinogens, whereas others have a protective effect. Therefore, the net impact of these compounds on human health depends on the overall balance between all factors involved. Strong scientific evidence has demonstrated the relationship between nitrosamines (NA), heterocyclic amines (HCAs), and polycyclic aromatic hydrocarbons (PAHs), which are the major genotoxins derived from cooking and food processing, and cancer. The mechanisms of the relationship between dietary toxic xenobiotics and cancer risk are not yet well understood, but it has been suggested that differences in dietary habits affect the colonic environment by increasing or decreasing the exposure to mutagens directly and indirectly through changes in the composition and activity of the gut microbiota. Several changes in the proportions of specific microbial groups have been proposed as risk factors for the development of neoplastic lesions and the enrichment of enterotoxigenic microbial strains in stool. In addition, changes in the gut microbiota composition and activity promoted by diet may modify the faecal genotoxicity/cytotoxicity, which can be associated with a higher or lower risk of developing cancer. Therefore, the interaction between dietary components and intestinal bacteria may be a modifiable factor for the development of colorectal cancer in humans and deserves more attention in the near future.
1. Introduction
Both diet and intestinal microbiota are considered to be two major factors that influence colonic health and the incidence of intestinal disorders such as colorectal cancer (CRC) [1]. It is generally accepted that diet acts as an essential factor for health maintenance. However, after several decades of research, the specific dietary compounds implicated in this protective effect have not yet been determined, which presents the opportunity to define an optimal diet.
This review discusses the impact of diet on CRC, the generation of xenobiotics, the interactions between these xenobiotics and gut microbiota, and identifying the factors that contribute to a balance of the factors necessary for a healthy gut. We then discuss the perspectives that can be used to guide our understanding of the contributions of diet and microbiota in protection against CRC.
1.1. Impact of Diet on Colorectal Cancer
Since the beginning of agriculture and animal husbandry approximately 10,000 years ago, humans have been exposed to profound changes both in diet and lifestyle that generally have taken place too rapidly to allow the fixation of genetic adaptations in the population [2]. This has prompted many authors to suggest that the current increase in the prevalence of the so-called “diseases of civilization” may be the result of discordance between the human Palaeolithic gastrointestinal system and modern diets.
Considering scientific evidence regarding the link between diet and health for the most generalized patterns worldwide, Western diets (WDs) are characterized by a high consumption of fatty and sugary foods, salt, sauces, meat and meat products, and processed foods [3]. Strong scientific evidence has indicated a protective role of the Mediterranean diet (MD) against the development of some high-prevalence and non-communicable pathologies in developed countries such as CRC [4,5], while adherence to a Westernized dietary pattern has been recognized as a potential risk factor.
A meta-analysis of thirteen prospective cohort studies concluded that a high-fat diet did not increase the risk of CRC [6], and no reduction in the risk of this disease was found with a low-fat diet after an eight-year follow-up in a randomized clinical trial [7]. Some authors have also evaluated the effect of fruit and vegetable consumption on colon cancer [8]. This risk of CRC increased when the consumption of these two food groups was below 300 g/day [8]. To the contrary, the results from the European Prospective Investigation into Cancer and Nutrition study (EPIC), one of the largest cohort studies in the world, did not support a significant inverse association between the consumption of fruits and vegetables and the occurrence of CRC, suggesting that there was little benefit of increasing the consumption of fruits and vegetables in comparison to the protection associated with an overall balanced diet [9]. The most recent revision of the Continuous Update Project from the World Cancer Research Fund International demonstrated that consumption of 90 g/day of whole grain is associated with a decrease in the risk of colon cancer, mainly attributable to the fibre content of whole grains [8]. Among the different components included within the concept of the WD, meat and meat products have accumulated the strongest scientific evidence in the context of CRC [10,11,12] and have been classified recently by the International Agency for Research on Cancer (IARC) as likely carcinogenic and carcinogenic foods, respectively [13]. A recent systematic review and meta-analysis of cohort studies have associated red meat with a significant increase in the risk of CRC (relative risk (RR) for 100 g/day of increase: 1.22; confidence interval (CI) 95%: 1.06–1.39). Nevertheless, this association was not observed for rectal cancer (RR: 1.13; CI 95%: 0.94–1.34). In addition, whereas the intake of processed meat has been shown to significantly increase the risk of colon cancer (RR: 1.23; CI 95%: 1.11–1.35), this association was marginally significant for rectal cancer (RR: 1.08; CI 95%: 1.00–1.18) [13,14]. It has been estimated that the risk of CRC increases 17% for every 100 g of red meat consumed per day [15], a risk that would be augmented in the case of chemically treated red meats [15,16]. This information is of high importance considering that in most developed countries, as is the case in Spain, the consumption of processed meat has increased in recent years, reaching 8 kg/capita/year in 2017 [17], while the intake of fruits and vegetables during the same period of time has gradually decreased [17]. The different mechanisms by which dietary patterns may be related to health are diverse in nature. In the literature, the net effect of the WD on CRC has been mostly related to the overall balance between low contents of antioxidants, fibre and polyunsaturated fatty acids and a high proportion of foods with a low-density high glycaemic index and rich in animal fats. In addition, in recent decades, major emphasis has been placed on the link between the ingestion of cooked and processed foods and the risk of colon cancer [18].
1.2. Intestinal Microbiota and Human Health
The gastrointestinal tract is inhabited by a dense microbial community known as the ‘microbiota’ that is composed of viruses and members of the three domains of life: bacteria, archaea and eukarya [19]. Metagenomic studies estimate that the total bacteria in our body exceeds approximately 10 times the number of nucleated eukaryotic cells [20], harbouring a genetic potential 100-fold larger than that of the whole human genome [21]. The intestinal bacterial population is mainly composed of members belonging to just two phyla, Bacteroidetes and Firmicutes, both constituting approximately 80–90% of the microorganisms in this habitat. Other subdominant microorganisms, in decreasing order of abundance (less than 10% of total intestinal bacteria), are members of the phyla Actinobacteria, Proteobacteria and Verrucomicrobia, respectively [22]. The intestinal microbiota carry out crucial functions that are beneficial to the host [23] and that could be mainly grouped as metabolic-degradation of non-digestible carbon sources and production of different metabolites such as vitamins and short chain fatty acids (SCFAs), protective-inhibition of pathogen adhesion to intestinal surfaces and trophic-maintenance of the intestinal epithelium integrity and functionality [24].
Intestinal microbial communities vary greatly among individuals, and it is difficult to define a ‘healthy microbiota’. It has been proposed that distinct types of gut microbial communities (‘enterotypes’), driven by diet and defined by their bacterial composition, are mainly characterized by relatively higher levels of a single bacterial genus: Prevotella, Ruminococcus or Bacteroides [22]. However, the vast interindividual variability of the microbiota demonstrated by large-scale metagenomic studies indicates that these differences are distributed in the population as continuous gradients of dominant taxa rather than discrete defined clusters [25,26,27]. Considering the concept of a healthy microbiota, despite this interindividual taxonomic variability, the functions of the gut microbiota remain relatively stable among individuals as there is a functional redundancy among the diverse members of this microbial community. Thus, a “core microbiome” constituted by specific microbial gene family combinations, metabolic modules, and regulatory pathways collectively promoting a stable host-associated ecology, could be ideally defined [28]. Gender, age, body mass, ethnicity, geographic location, and immune status [28] are intrinsic factors that influence the concept of a “healthy microbiota”. However, it must be noted that the influence of the microbiota on other factors is usually not considered, such as the intestinal transit time or previous drug consumption [28]. An imbalance in the composition and functionality of the microbiota occurring in several diseases is known as dysbiosis. The link between disease and microbiota has been repeatedly replicated in experiments with faecal transplantation in mice and the reproduction of the initial altered phenotype [29,30]. However, it is still challenging to determine whether changes in the microbiota are the cause or consequence of the disease [31]. One of the most common events occurring in dysbiosis states is a decrease in intestinal bacterial richness, frequently accompanied by variations in the relative abundance of some microbial genes and functions that differ among pathologies, as has been described in obesity [32], inflammatory bowel disease [33], autism [34], and CRC [35], among others. Specifically, the dysbiosis associated with CRC is generally characterized by an increase in the prevalence of pathogenic or pathogen-associated microorganisms from genera Fusobacterium, Porphyromonas, Peptostreptococcus, Parvimonas and Enterobacter and by a depletion of gram-positive fibre-fermenting Clostridia [35,36]. Furthermore, the consumption of chemically and thermally processed foods and the adherence to a WD have been shown to drive specific changes in gut microbiota composition and activity [37,38] towards the production of metabolites with potential carcinogenic effects [39]. Thus, a comprehensive understanding of how these compounds derived from food processing can interact with the microbiota and microbiome is necessary to determine their true impact on overall gastrointestinal health [40].
2. Food Processing and Xenobiotics
Although the exact mechanism by which meat is related to cancer is unknown, several authors have postulated that the thermal formation of different carcinogens during cooking, such as heterocyclic amines (HCAs) and polycyclic aromatic hydrocarbons (PAHs), the addition of N-nitroso compounds (NOCs) to cured meats, the endogenous NOC formation from haem iron and the generation of lipid and protein oxidation products, are included within the possible mechanisms underlying this association (Figure 1) [41].
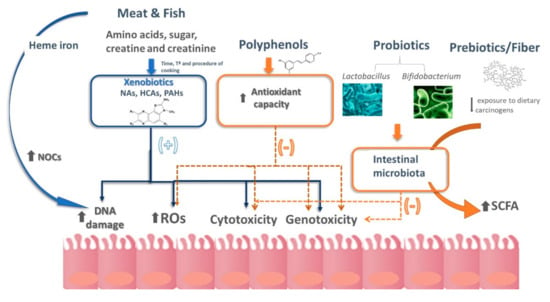
Figure 1.
Schematic representation of the different hypotheses currently available that contribute to explaining the relationship between diet and colorectal cancer. HCAs, heterocyclic amines; NAs, nitrosamines; NOCs, N-nitroso compounds; PAHs, polycyclic aromatic hydrocarbons; ROS, reactive oxygen species; SCFA, short chain fatty acids. More details are explained in the text.
In this regard, haem iron, in addition to being related to the production of NOCs at the intestinal level, has been associated with the generation of aldehydes with cytotoxic and genotoxic properties [42]. Additionally, meat processing involves the addition of nitrites, salt, smoke and the application of different grades of temperature depending on the cooking method [11], all of which are related to an increased risk of colon cancer. From xenobiotics, HCAs have accumulated the strongest scientific evidence as cancer risk factors in epidemiological and interventional studies (Table 1) and were classified by the IARC as potential carcinogens. In recent years, more than twenty-five HCAs have been identified in regular food products [43], formed from creatinine, creatine, hexoses, amino acids and some dipeptides, which are present mainly in the muscle of meats and fish [44,45].

Table 1.
Observational studies in recent years associating heterocyclic aromatic amines and polycyclic aromatic hydrocarbons with colorectal cancer.
HCAs can be classified into two large groups according to their molecular structures and metabolic pathways: aminocarbolines (ACs), or pyrolytic amines, and aminoimidazoazarenes (AIAs), or thermal amines. ACs are formed by the pyrolysis of proteins at temperatures above 300 °C, while AIAs are generated by applying temperatures from 100 to 300 °C to dietary sources of sugars, amino acids and creatinine [49]. As the mutagenic activity of HCAs increases with temperature [50] and with the browning degree of cooked food, cooking methods such as frying, grilling or roasting lead to the formation of higher amounts of HCAs than boiling, steaming or braising [51]. In this regard, data from the EPIC revealed the existence of a large variation in the intake of these foodstuffs and in the cooking methods among European countries. The Netherlands has been found to be the population with the highest intake of red meat prepared at high temperature (mean intake of 39.4 g/day and 59.7 g/day for women and men, respectively [52].
PAHs are formed in a large variety of foods, including oils, grains and vegetables, after applying a heat treatment for cooking (frying, baking, grilling, etc.) or processing. Among the different types of PAHs classified by the IARC, benzo(a)pyrene (BaP) has been classified as carcinogenic to humans [53]. Nevertheless, given the ubiquity of PAHs in food and their presence as contaminants, it is very difficult to assess to what extent the amount ingested from food may contribute to cancer development. PAHs [54] can be formed by pyrolysis of organic matter at high temperatures, by direct contact of lipid droplets with a heat source, by the smoke produced during cooking, or by the incomplete combustion of coal or wood in barbecues or grills [55,56,57]. The maximum levels of PAHs have been found in smoked foods and grilled meats [57].
As with many other dietary components, the impact of xenobiotics related to food processing on health depends on the dose of intake and the frequency of exposure to the toxic agent/s. In this regard, some authors have highlighted that chronic exposure to contaminants may progressively induce a low-grade inflammatory status in the host, partly mediated by the aryl hydrocarbon receptor, a cytosolic transcription factor activated by different hydrophobic chemicals [58] and present in different mammalian cells. With considerable variation among countries, the amount of HCAs consumed mainly depends on the cooking method, temperature, the meat or fish itself and the nutritional composition of the foodstuffs [59,60]. However, these factors are very difficult to assess accurately through dietary questionnaires for several reasons. First, cooking methods are highly variable over time. Second, there are no standardized tools currently available, such as photographs of scales, to quantify the degree of browning in foods, so there is high variability between studies. Third, the interaction between the different components of the diet is too difficult to determine long-term. In addition, after the intake of red meat, other carcinogenic compounds associated with many processed meats, such as NOCs, can be formed endogenously [59] by the intestinal microbiota and can be activated to act as carcinogens/mutagens [61].
3. Effect of Food Processing-Borne Xenobiotics on the Gut Microbiota
The human colon is exposed to multiple compounds of dietary origin, as well as those resulting from digestion, intestinal microbial metabolism, and host excretory processes. Intestinal microbiota is known to produce faecal metabolites, with genotoxic and mutagenic potential, some of which have been compiled on Table 2.

Table 2.
Cytotoxic/genotoxic mechanisms of endogenous molecules and compounds generated by intestinal bacteria that could be involved in CRC. Direct mechanisms refer to those that promote genotoxic and/or cytotoxic action directly. Indirect mechanisms are those that cause damage at different levels, from which a cytotoxic and/or genotoxic action is derived.
Cytotoxicity is the capability of certain substances to cause cell injury, with deleterious effects on metabolism, structure and/or viability of cells. Genotoxicity is the capability to induce damage to cellular genetic material, altering the DNA sequence or modifying its structure; more specifically, mutagenicity refers to the capacity of some genotoxic agents to produce alterations (mutations) in the DNA sequence. Some potential faecal mutagens can be produced by the intestinal microbiota, including microbial genotoxins [62]. Other compounds are formed endogenously from dietary constituents, such as nitrates, dietary amines and cholesterol, or are synthesized from precursors originating from human metabolism, such as NOCs, fecapentaenes, long chain fatty acids, and secondary bile acids generated by the metabolism of intestinal bacteria [63]. Table 2 summarizes the cytotoxic and genotoxic mechanisms of endogenous molecules and compounds generated by the intestinal bacteria that could be involved in CRC. A group of toxic substances are from exogenous origins and include mycotoxins, plant glycosides, some food additives and notably, two groups of xenobiotics discussed in this review that are formed by pyrolysis during food cooking and processing: HCA and PAH.
Studies of faecal genotoxicity and mutagenicity related to dietary habits have revealed different levels of toxicity associated with different dietary patterns [85], as well as an association between high faecal genotoxicity and an augmented risk of CRC [63]. Although some authors have suggested the possibility of using faecal genotoxicity as a preventive and early marker for the risk of CRC [86,87], more studies are needed to support this proposal. The relationship between cytotoxicity and intestinal disease is currently less clear. Moreover, it is necessary to elucidate the causal role of the different genotoxic and cytotoxic compounds on CRC and the influence of dietary patterns, intestinal microbiota, host physiology and lifestyle on the resulting toxicity in the intestinal environment. In this sense, some studies indicate that the presence of co-mutagenic, inhibitory or potentiating factors could modify the toxicity of genotoxic and cytotoxic compounds and hence the resulting intestinal toxicity [87,88,89].
3.1. Impact of Xenobiotics on Gut Microbiota
The human gut microbiota interacts with food xenobiotics in dual ways: xenobiotics influence the microbiota, and in turn, the microbiota can also metabolize and transform xenobiotics, altering their toxicity. Few studies are currently available that evaluate the impact of food xenobiotics on the gut microbiota and the consequences on the host immune system and metabolism. Ribière et al. (2016) found that oral exposure to benzo(a)pyrene led to moderate inflammation in ileal and colonic mucosa [90] and induced changes in the gut microbiota composition, without affecting the alpha-diversity index, in a murine model. Among dominant intestinal taxa, bacterial families such as Bacteroidaceae, Porphyromonadaceae and Paraprevotellaceae showed a significant increase in their relative abundance, whereas Lactobacillaceae and Verrucomicrobiaceae (only represented by Akkermansia muciniphila) decreased; in contrast, among the less abundant microorganisms, the Actinobacteria class (mainly represented by the genus Bifidobacterium) and some members of the Coriobacteriaceae, Rikenellaceae, and Desulfovibrionaceae families displayed increased abundance after mice were exposed to benzo(a)pyrene. Interestingly, Defois et al., using in vitro faecal models, demonstrated important changes in the metabolome and transcriptome of the human gut microbiota upon exposure to a variety of food contaminants without affecting the structural composition of the microbial community thus highlighting important changes in microbial metabolic activity [91]. Changes in the volatolome affected sulphur, phenolic and ester compounds. The transcriptome revealed an increase in lipid metabolism processes, cell wall/plasma membrane/periplasmic space and DNA repair and replication systems, whereas the transcription of genes related to glycolysis/gluconeogenesis and bacterial chemotaxis towards simple carbohydrates as well as ribosome, translation and nucleic acid binding was downregulated [91,92].
3.2. Impact of the Gut Microbiota on the Toxicity of Xenobiotics
The intestinal microbiota has the capacity to modify the toxicity of food xenobiotics by direct microbial interference with these compounds and/or by modulating host-microbial interactions. First, some lactic acid bacteria (LAB) and other microorganisms present in the human gut can directly bind or metabolize diet-derived HCAs or other xenobiotics [93,94,95,96], contributing either to the sequestration and excretion of these compounds in faeces or to their transformation into less toxic compounds, which potentially helps to prevent DNA damage and generation and progression of pre-neoplastic lesions [97]. The gut microbiota can also metabolize xenobiotics transforming them into chemically derived molecules with enhanced mutagenic activity [98]. Thus, additional studies are necessary to elucidate the range of bacteria capable of carrying out each of these transformations of different xenobiotics originating during food processing, the metabolic effects of such biotransformation on the microbiota and host, and the relevance of these transformations to intestinal disease and carcinogenesis.
The modification of the toxicity of xenobiotics can also occur via host-microbiota interactions. The most known of these interactions is the exacerbation of the toxicity of xenobiotics through enterohepatic cycling. Xenobiotics are often conjugated to glucuronic acid in the liver (one of the pathways of phase II detoxification in the human body), stored in the gallbladder, and released into the intestine with bile during digestion. When the conjugated xenobiotic enters the intestine, the microbial β-glucuronidases can cleave the deactivated glucuronidated molecule and release the unconjugated xenobiotic, turning it back again into a toxic molecule [99]. The de-glucuronidation of xenobiotics by microbial β-glucuronidases, such as those found in many enterobacteria and in some microorganisms from the Clostridium and Bacteroides groups [98], is a phenomenon already demonstrated to occur with some HCAs [97,100], but it remains unknown whether it could be a general detoxification mechanism that also affects other HCAs and PAHs. Another possible way of increasing the toxicity of xenobiotics in the gut is the alteration of host gene expression by the microbiota. Cytochrome P450 comprises different hepatic enzymes that participate in phase I of detoxification. One of these enzymes, CYP1A1 (aryl-4 monooxygenase), has been linked to the intestinal detoxification of benzo(a)pyrene depending on TLR2, a host cell membrane receptor triggered by bacterial lipoproteins and other cell wall components; TLR2-deficient mice had reduced ability to clear benzo(a)pyrene and developed colon polyps after dietary supplementation with this compound [101]. The results of the study of Do and colleagues suggest the interesting possibility that the gut microbiota could modulate the host xenobiotic metabolism through TLR2 signalling [97].
Conventional tests available for the study of genotoxicity, mutagenicity and cytotoxicity of faecal waters are mostly based on those developed to routinely characterize potential hazards of chemicals, as indicated in the Organization for Economic Cooperation and Development (OECD) Health Effects Test Guidelines [102,103]. Some tests are available for determining genotoxicity in vitro, in vivo and/or ex vivo; the comet test is the most commonly used, but others, such as the micronucleus assay (MN), sister chromatid exchange assay (SCE) or the SOS chromotest are also used [104]. The Ames test is by far the most extensively applied to assess in vitro mutagenicity and is based on the capacity to cause reverse mutations in defective genes for essential amino acids (such as histidine or tryptophan) in auxotrophic strains of Salmonella Typhimurium and Escherichia coli.
Intestinal cytotoxicity could be evaluated in adenocarcinoma cell lines, generally Caco-2 and HT-29. Some assays determine the effect of exposure to toxic agents on cell proliferation capacity [101]. Cytotoxicity can also be evaluated by determining mitochondrial function or membrane integrity [105]. Among the most commonly used are the dye exclusion test and the MTT assay [105]. Real-time electronic sensing (xCELLigence system; Roche, Basel, Switzerland) monitors variations in the impedance of cultures of carcinogenic cell lines at the proliferation or confluence states using gold microelectronic sensor arrays [106,107].
In spite of the classical methods available, the interpretation of faecal mutagenicity/genotoxicity related to intestinal disease and CRC risk is hindered by the limitation of analytical power when applied to the study of human samples, by the inaccuracies and errors of the experimental techniques used, and because the in vitro and in vivo tests may not adequately reflect what happens in the complex human intestinal ecosystem. Recent advances in the culture of pluripotent stem cells and primary tissues have made possible the culture, differentiation and self-assembly of many cell types and structures from true organs in three dimensions to form the so-called “intestinal organoids”. These organoids could allow the construction of human or animal-specific models for the study of intestinal toxicity [108]. This cutting-edge scientific research tool still presents some limitations, as with any technology. The most remarkable is non-vascularization due to limitations in nutrient supply in culture media, the limited presence of stromal cells, including those from the immune system, and the lack of reliable means to synchronize the shape, size and viability of organoids in culture [109]. Beyond the limitations of the analytical and predictive power of the techniques available to determine levels of intestinal toxicity and the necessary improvement of their accuracy, the emerging omics technologies (metagenomics, RNA-seq, metabolomics, and culturomics) are complementary and powerful skills that could help to shed light on the specific metabolic cell routes altered by toxic compounds, contributing to the exploration of new biomarkers for genotoxicity assessment related to intestinal disease and cancer risk.
4. Creating a Balance between Xenobiotics and a Healthy Gut
As mentioned previously in this review, some gastrointestinal microbes are able to generate toxic compounds themselves and/or to convert pre-carcinogens into carcinogens, whereas some commensal bacteria, probiotics, prebiotics and dietary compounds can act as inhibitors or attenuators of the genotoxicity of xenobiotics. Therefore, the resulting damage at the intestinal level will depend on the interaction among these three factors: the intake of potentially toxic xenobiotic formed during food cooking and processing, the global subject´s diet and the host intestinal microbiota profile. The optimum diet for the prevention of CRC has not yet been defined, but it is well accepted that diets rich in fat and meat and poor in vegetables and fibre tend to increase faecal genotoxicity and the risk of CRC. Apart from this traditional approach, the existence of an interactive effect between phenolic antioxidants and HCA-induced carcinogenesis was proposed twenty years ago based on the capacity of phenolics to inhibit the metabolic activation of HCAs [18] and influence cell proliferation, DNA repair enzymes and apoptosis [110] in the host, among other mechanisms [18]. Since then, diverse phytochemicals from different vegetables and vegetable drinks, such as flavonoids from tea [111,112], citrus [113], fruits [114,115], or aromatic species [116], have been reported to inhibit the production of HCAs [117].
5. Future Perspectives
It is clear that wider cohort studies are needed to obtain a clear perspective of the long-term impact that the continued intake of xenobiotics formed during food processing has on CRC, considering the tendency of increased consumption of processed foods and the progressive ageing of the population in industrialized countries, which augments the time of exposure. At the same time, some food ingredients may contribute to minimizing the risk associated with this chronic exposure to xenobiotics. The following sections will analyse both perspectives facing the impact of xenobiotics on CRC and highlight the need for more research in the field of xenobiotics derived from food processing.
5.1. Probiotics and Prebiotics to Counteract the Effect of Pro-Carcinogenic Compounds
Probiotics are defined as live microorganisms that, when administered in adequate amounts, confer a health benefit to the host [118]. In the recent definition of prebiotics proposed by Bindels et al. in 2015 [119], these are considered “nondigestible compounds that, through their metabolization by microorganisms in the gut, modulate composition and/or activity of the gut microbiota and thus confer a beneficial physiological effect on the host”. This definition includes not only nondigestible oligosaccharides but also most dietary fibres. Both probiotics and prebiotics are included in functional food products and supplements or are part of natural foods and have been the subject of extensive research in recent years. There is currently interest in improving our understanding of the interaction mechanisms between probiotics and prebiotics with intestinal procarcinogenic compounds from diet.
Several probiotic strains from the Bifidobacterium and Lactobacillus genera have shown beneficial effects in murine models of colon cancer by promoting the inhibition of aberrant crypt foci formation (early predictors of tumour incidence), suppressing tumour growth, inducing cell apoptosis, or ameliorating inflammation [120,121,122]. These events were associated in most cases with a reduction in intestinal genotoxicity and cytotoxicity [89]. Direct physical binding is the most common method for the removal of carcinogenic compounds by LAB and probiotics (mainly from the Lactobacillus and Bifidobacterium genera) [123]; generally, peptidoglycan is the cell molecule linked to this effect, although the exopolysaccharides produced by some microorganisms can also act as binders of mutagens [124]. Probiotic bacteria tend to maintain high viability in vitro in the presence of food xenobiotics and bind to these compounds; this could facilitate the removal of aggregated particles from the human body, although this mechanism has not yet been specifically proven in the human gut [125]. Another way by which LAB and probiotics can reduce the genotoxicity/mutagenicity is the metabolic modification/inactivation of procarcinogenic molecules [126,127,128]. The antioxidant capacity of some LAB and probiotics may reduce the cytotoxic effect and oxidative damage of some procarcinogens [129]. Probiotics can also contribute to decreasing toxicity through the modulation of the intestinal microbiota composition and its metabolic activity by reducing the microbial production of enzymes involved in the re-activation in the gut of dietary carcinogens and other toxic compounds, such as β-glucuronidases [130]. The interaction with the host immune system, stimulation of host enzymes (such as glutathione transferases, reductases and peroxidases linked to phase I of detoxification primarily) related to the inactivation of carcinogens, modulation of the host’s immune response and regulation of apoptosis are also mechanisms by which LAB ingested with foods or present in the intestine can modify the effect of procarcinogens in the human body [125].
Epidemiological evidence links dietary fibre with colonic health [131]. The mechanisms by which dietary fibres and prebiotics exert a protective effect against cancer can be diverse, and most of them are likely mediated by the intestinal microbiota. Among the physical mechanisms, the increase in bulking associated with high fibre consumption potentiates sequestration of carcinogens by fibre, favouring the elimination of toxic compounds and reducing host exposure. Prebiotics may also exert a protective effect through changes in the composition and activity of the intestinal microbiota by enhancing the production of host xenobiotic-deactivating enzymes, and/or reducing the production, by the intestinal bacteria, of procarcinogenic-producing/activating enzymes (i.e., β-glucuronidases). The protective effect could also be exerted by the activation of xenobiotic-modifying enzymes of the host through the intestinal microbiota, in which at least two mechanisms could be potentially involved: (1) the microbial production of SCFAs, mainly butyrate, by colonic fermentation of prebiotics, can activate the hepatic phase II enzymes that detoxify toxic compounds by conjugation, and (2) the release of phenolic compounds from vegetable fibres (including polysaccharides, lignins and oligosaccharides) in the human colon can act as modulators of the production of xenobiotic-deactivating enzymes by the host [132]. In addition, prebiotics can also modify the host immune response against procarcinogens and can also alter the gene expression of intestinal host tissues through microbiota-host relationships. In this way, the modification by prebiotics of the intestinal microbiota can promote the activation of some enzymatic glycosidic activities in colonocytes or can modify local or systemic responses of the host immune system by enhancing the production of proinflamatory/regulatory cytokines and/or immunoglogulins [132,133].
Despite the attractiveness of probiotics and prebiotics as food agents for CRC prevention, the efficacy of probiotics in maintaining colonic health or in preventing colon cancer needs to be well established in controlled clinical trials. More evidence exists for the link between dietary fibre/prebiotics and colonic health, but most of the mechanisms of action remain speculative and need to be experimentally proven.
5.2. Longitudinal Studies on Long Term Impact of Xenobiotics Derived from Food Processing
More longitudinal studies are needed to reach a consensus on the effect of xenobiotics on health since the net impact of these compounds will depend on the amount ingested as well as the frequency and time of exposure. With this aim in mind, it is necessary to prioritize the development of standardized questionnaires for the collection of data on dietary intake suitable for this purpose, with a limited number of items but allowing quantification in foods the degree of cooking, temperature applied, and other possible variables. In fact, the adequate assessment of the intake of xenobiotics formed while cooking food is a very difficult task since it is necessary for the subject surveyed to remember details over time, such as temperature of cooking, method of preparation, and degree of toasting. These parameters are subjected to high daily variation, and the information is very difficult to retrieve from some population groups, as the people (children, elderly, workers, hospitalized patients, etc.) that usually eat outside home. In this sense, taking advantage of the rapid progress and widespread use of new technologies, the development of computer applications, including photos or validated models, that are easy to use by the people interviewed, could greatly facilitate this task in the near future. Although the conversion/extrapolation of the intake of cooked foods to particular toxic compounds can be done through composition tables, such as those developed by the EPIC group [134], the use of biological markers, representative of the actual intake of the subject, could be of great relevance to validate and complete the reported information. The interpretation of the impact of food-associated xenobiotics on health should be made with caution, taking into account the global diet of the subject and other factors involved in CRC development. The setup of the most adequate methodology to analyse the cytotoxicity or genotoxicity of the diet could allow the exploration of the existence of potential synergisms or antagonisms between the different components assessed. This finding would be very useful when establishing nutritional strategies for health promotion or for the development of functional foods, incorporating specific dietary components capable of counteracting the potential carcinogenic effect of xenobiotics associated with food processing.
6. Conclusions
In the last decade, major emphasis has been placed on the link between the ingestion of thermally processed foods and the risk of CRC. Xenobiotics with potential carcinogenic activity can be generated from meat and fish components through some cooking procedures with high temperatures. These compounds include mainly NOCs, PAHs and HCAs. HCAs have accumulated the strongest scientific evidence as a CRC risk factor in epidemiological and interventional studies. The presence of xenobiotics from food processing contributes to an increase in local genotoxicity, which has been related to an increase in CRC risk. The intestinal microbiota can modify the toxicity of dietary xenobiotics by direct microbial interference with these compounds and/or by modulating host-microbiota interactions through diverse mechanisms. However, diet and dietary components could interact either with the intestinal microbiota or with xenobiotics, contributing to altered gut toxicity. Therefore, the interaction between dietary components and intestinal microbiota could be considered a modifiable factor for the development of CRC in humans. Thus, more studies are needed to determine the long-term impact of xenobiotics derived from food processing on CRC. Specific nutrients, probiotics and prebiotics/fibres have the capacity to reduce intestinal toxicity; their mechanisms of action and efficacy should be carefully evaluated for use as food agents for CRC prevention.
Author Contributions
All authors contributed to writing of the original draft, and review.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ACs | Aminocarbolines |
| AIAs | Aminoimidazoazarenes |
| ATP | Adenosine triphosphate |
| B(a)P | Benzo(a) pyrene |
| CI | Confidence interval |
| CRC | Colorectal cancer |
| CYP1A1 | Aryl-4 monooxigenase |
| CYP450 | Cytochrome P450 |
| DiMeIQx | 2-Amino-3,4,8-trimethylimidazo[4,5-f] quinoxaline |
| EPIC | European Prospective Investigation into Cancer |
| HCAs | Heterocyclic amines |
| IARC | The International Agency for Research on Cancer |
| LAB | Lactic acid bacteria |
| MD | Mediterranean Diet |
| MeIQ | 2-Amino-3,4-dimethylimidazo[4,5-f] quinoline |
| MeIQx | 2-Amino-3,8-dimethylimidazo[4,5-f] quinoxaline |
| MN | Micronucleous assay |
| MTT | (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) |
| NAs | Nitrosamines |
| NOCs | N-Nitroso compounds |
| OECD | Organization for Economic Cooperation and Development |
| PAHs | Polycyclic aromatic hydrocarbons |
| PhIP | 2-Amino-1-methyl-6-phenylimidazo[4,5-b] pyridine |
| ROS | Reactive oxygen species |
| RR | Relative risk |
| SCE | Sister Chromatid Exchange assay |
| SCFA | Short chain fatty acids |
| WD | Western Diet |
References
- Viera, C.; Rodrigues, M.; Russo, E.; Amedei, A. Role of diet and gut microbiota on colorectal cancer immunomodulation. World J. Gastroenterol. 2019, 25, 151–162. [Google Scholar] [CrossRef]
- Cunnane, S.C. Origins and evolution of the Western diet: Implications of iodine and seafood intakes for the human brain. Am. J. Clin. Nutr. 2005, 82, 483. [Google Scholar] [CrossRef][Green Version]
- Naja, F.; Hwalla, N.; Itani, L.; Karam, S.; Mehio Sibai, A.; Nasreddine, L. A Western dietary pattern is associated with overweight and obesity in a national sample of Lebanese adolescents (13–19 years): A cross-sectional study. Br. J. Nutr. 2015, 114, 1909–1919. [Google Scholar] [CrossRef]
- Di Daniele, N.; Noce, A.; Vidiri, M.F.; Moriconi, E.; Marrone, G.; Annicchiarico-Petruzzelli, M.; D’Urso, G.; Tesauro, M.; Rovella, V.; De Lorenzo, A. Impact of Mediterranean diet on metabolic syndrome, cancer and longevity. Oncotarget 2017, 8, 8947–8979. [Google Scholar] [CrossRef] [PubMed]
- Farinetti, A.; Zurlo, V.; Manenti, A.; Coppi, F.; Mattioli, A. Mediterranean diet and colorectal cancer: A systematic review. Nutrition 2017, 43–44, 82–88. [Google Scholar] [CrossRef]
- Liu, L.; Zhuang, W.; Wang, R.-Q.; Mukherjee, R.; Xiao, S.-M.; Chen, Z.; Wu, X.-T.; Zhou, Y.; Zhang, H.-Y. Is dietary fat associated with the risk of colorectal cancer? A meta-analysis of 13 prospective cohort studies. Eur. J. Nutr. 2011, 50, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Beresford, S.A.A.; Johnson, K.C.; Ritenbaugh, C.; Lasser, N.L.; Snetselaar, L.G.; Black, H.R.; Anderson, G.L.; Assaf, A.R.; Bassford, T.; Bowen, D.; et al. Low-fat dietary pattern and risk of colorectal cancer. JAMA 2006, 295, 643–654. [Google Scholar] [CrossRef]
- Vieira, A.R.; Abar, L.; Chan, D.S.M.; Vingeliene, S.; Polemiti, E.; Stevens, C.; Greenwood, D.; Norat, T. Foods and beverages and colorectal cancer risk: A systematic review and meta-analysis of cohort studies, an update of the evidence of the WCRF-AICR Continuous Update Project. Ann. Oncol. 2017, 28, 1788–1802. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Ros, R.; Barupal, D.K.; Rothwell, J.A.; Jenab, M.; Fedirko, V.; Romieu, I.; Aleksandrova, K.; Overvad, K.; Kyrø, C.; Tjønneland, A.; et al. Dietary flavonoid intake and colorectal cancer risk in the European prospective investigation into cancer and nutrition (EPIC) cohort. Int. J. Cancer 2017, 140, 1836–1844. [Google Scholar] [CrossRef]
- De Stefani, E.; Boffetta, P.; Deneo-Pellegrini, H.; Ronco, A.L.; Aune, D.; Acosta, G.; Brennan, P.; Mendilaharsu, M.; Ferro, G. Meat intake, meat mutagens and risk of lung cancer in Uruguayan men. Cancer Causes Control. 2009, 20, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R. Meat and cancer. Meat Sci. 2010, 84, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Wie, G.-A.; Cho, Y.-A.; Kang, H.; Ryu, K.-A.; Yoo, M.-K.; Kim, Y.-A.; Jung, K.-W.; Kim, J.; Lee, J.-H.; Joung, H. Red meat consumption is associated with an increased overall cancer risk: A prospective cohort study in Korea. Br. J. Nutr. 2014, 112, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Bouvard, V.; Loomis, D.; Guyton, K.Z.; Grosse, Y.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Mattock, H.; Straif, K.; Stewart, B.W.; et al. Carcinogenicity of consumption of red and processed meat. Lancet Oncol. 2015, 16, 1599–1600. [Google Scholar] [CrossRef]
- Aune, D.; Chan, D.S.M.; Vieira, A.R.; Navarro Rosenblatt, D.A.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Red and processed meat intake and risk of colorectal adenomas: A systematic review and meta-analysis of epidemiological studies. Cancer Causes Control. 2013, 24, 611–627. [Google Scholar] [CrossRef]
- Chiang, V.S.C.; Quek, S.Y. The relationship of red meat with cancer: Effects of thermal processing and related physiological mechanisms. Crit. Rev. Food Sci. Nutr. 2017, 57, 1153–1173. [Google Scholar] [CrossRef] [PubMed]
- Catsburg, C.E.; Gago-Dominguez, M.; Yuan, J.M.; Castelao, J.E.; Cortessis, V.K.; Pike, M.C.; Stern, M.C. Dietary sources of N-nitroso compounds and bladder cancer risk: Findings from the Los Angeles bladder cancer study. Int. J. Cancer 2014, 134, 125–135. [Google Scholar] [CrossRef]
- Ministerio de Agricultura y Pesca Alimentación y Medio Ambiente (MAPAMA). Informe del Consumo de Alimentación en España; MAPAMA: Madrid, Spain, 2016; pp. 1–225.
- Hirose, M.; Takahashi, S.; Ogawa, K.; Futakuchi, M.; Shirai, T. Phenolics: Blocking agents for heterocyclic amine-induced carcinogenesis. Food Chem. Toxicol. 1999, 37, 985–992. [Google Scholar] [CrossRef]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLOS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Malagelada, J.-R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Knights, D.; Ward, T.L.; McKinlay, C.E.; Miller, H.; Gonzalez, A.; McDonald, D.; Knight, R. Rethinking “Enterotypes” . Cell Host Microbe 2014, 16, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G.; et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 50, 61–66. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Integrative HMP (iHMP) Research Network Consortium. The integrative human microbiome project: Dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 2014, 16, 276–289. [Google Scholar] [CrossRef]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 6150–6172. [Google Scholar] [CrossRef]
- De Palma, G.; Lynch, M.D.J.; Lu, J.; Dang, V.T.; Deng, Y.; Jury, J.; Umeh, G.; Miranda, P.M.; Pigrau Pastor, M.; Sidani, S.; et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 2017, 9, eaaf6397. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Qin, J.; Balzola, F.; Bernstein, C.; Ho, G.T.; Lees, C.; Bork, P.; Dusko, E.; Wang, J. A human gut microbial gene catalogue established by metagenomic sequencing: Commentary. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Krajmalnik-Brown, R.; Lozupone, C.; Kang, D.-W.; Adams, J.B. Gut bacteria in children with autism spectrum disorders: Challenges and promise of studying how a complex community influences a complex disease. Microb. Ecol. Heal. Dis. 2015, 26, 26914. [Google Scholar] [CrossRef]
- Ahn, J.; Sinha, R.; Pei, Z.; Dominianni, C.; Wu, J.; Shi, J.; Goedert, J.J.; Hayes, R.B.; Yang, L. Human gut microbiome and risk for colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 1907–1911. [Google Scholar] [CrossRef]
- Duvallet, C.; Gibbons, S.M.; Gurry, T.; Irizarry, R.A.; Alm, E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017, 8, 1784. [Google Scholar] [CrossRef]
- O’Keefe, S.J.D. Diet, microorganisms and their metabolites and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D. Thermal processing of food reduces gut microbiota diversity of the host and triggers adaptation of the microbiota: Evidence from two vertebrates. Microbiome 2018, 6, 99–113. [Google Scholar] [CrossRef]
- Katsidzira, L.; Laubscher, R.; Gangaidzo, I.T.; Swart, R.; Makunike-Mutasa, R.; Manyanga, T.; Thomson, S.; Ramesar, R.; Matenga, J.A.; Rusakaniko, S. Dietary patterns and colorectal cancer risk in Zimbabwe: A population based case-control study. Cancer Epidemiol. 2018, 57, 33–38. [Google Scholar] [CrossRef]
- Shirai, T.; Tamano, S.; Sano, M.; Masui, T.; Hasegawa, R.; Ito, N. Carcinogenicity of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in rats: Dose-response studies. Princess Takamatsu Symp. 1995, 23, 232–239. [Google Scholar]
- Kim, E.; Coelho, D.; Blachier, F. Review of the association between meat consumption and risk of colorectal cancer. Nutr. Res. 2013, 33, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Bastide, N.; Pierre, F.; Corpet, D. Heme iron from meat and risk of colorectal cancer: A meta-analysis and a review of the mechanisms involved. Cancer Prev. Res. 2011, 4, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Viegas, O.; Novo, P.; Pinto, E.; Pinho, O.; Ferreira, I. Effect of charcoal types and grilling conditions on formation of heterocyclic aromatic amines (HAs) and polycyclic aromatic hydrocarbons (PAHs) in grilled muscle foods. Food Chem. Toxicol. 2012, 50, 2128–2134. [Google Scholar] [CrossRef] [PubMed]
- Chiavarini, M.; Bertarelli, G.; Minelli, L.; Fabiani, R. Dietary intake of meat cooking-related mutagens(HCAs) and risk of colorectal adenoma and cancer: A systematic review and meta-analysis. Nutrients 2017, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Martín-Calero, A.; Tejral, G.; Ayala, J.; González, V.; Afonso, A. Suitability of ionic liquids as mobile-phase additives in HPLC with fluorescence and UV detection for the determination of heterocyclic aromatic amines. J. Sep. Sci. 2010, 33, 182–190. [Google Scholar] [CrossRef]
- Etemadi, A.; Abnet, C.C.; Graubard, B.I.; Beane-Freeman, L.; Freedman, N.D.; Liao, L.; Dawsey, S.M.; Sinha, R. Anatomical subsite can modify the association between meat and meat compounds and risk of colorectal adenocarcinoma: Findings from three large US cohorts. Int. J. Cancer 2018, 143, 2261–2270. [Google Scholar] [CrossRef]
- Martínez Góngora, V.; Matthes, K.L.; Castaño, P.R.; Linseisen, J.; Rohrmann, S. Dietary heterocyclic amine intake and colorectal adenoma risk: A systematic review and meta-analysis. Cancer Epidemiol. Biomark. Prev. 2018, 28, 99–109. [Google Scholar] [CrossRef]
- Helmus, D.S.; Thompson, C.L.; Zelenskiy, S.; Tucker, T.C.; Li, L. Red Meat-derived heterocyclic amines increase risk of colon cancer: A population-based case-control study. Nutr. Cancer 2013, 65, 1141–1150. [Google Scholar] [CrossRef]
- Khan, M.; Busquets, R.; Santos, F.; Puignou, L. New method for the analysis of heterocyclic amines in meat extracts using pressurised liquid extraction and liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2008, 1194, 155–160. [Google Scholar] [CrossRef]
- Kondjoyan, A.; Chevolleau, S.; Portanguen, S.; Molina, J.; Ikonic, P.; Clerjon, S.; Debrauwer, L. Realation between crust development and heterocyclic aromatic amine formation when air-roasting a meat cylinder. Food Chem. 2016, 213, 641–646. [Google Scholar] [CrossRef]
- Buła, M.; Przybylski, W.; Jaworska, D.; Kajak-Siemaszko, K. Formation of heterocyclic aromatic amines in relation to pork quality and heat treatment parameters. Food Chem. 2019, 276, 511–516. [Google Scholar] [CrossRef]
- Rohrmann, S.; Nimptsch, K.; Sinha, R.; Willett, W.C.; Giovannucci, E.L.; Platz, E.A.; Wu, K. Intake of Meat mutagens and risk of prostate cancer in a cohort of U.S. health professionals. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1557–1563. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, B.; Chu, C.; Carlin, D.J. Polycyclic aromatic hydrocarbons: From metabolism to lung cancer. Toxicol. Sci. 2015, 145, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Larsson, B.; Sahlberg, P.; Eriksson, A.; Busk, L. Polycyclic Aromatic Hydrocarbons in Grilled Food. J. Agric. Food Chem. 1983, 31, 867–873. [Google Scholar] [CrossRef]
- Alomirah, H.; Al-Zenki, S.; Al-Hooti, S.; Zaghloul, S.; Sawaya, W.; Ahmed, N.; Kannan, K. Concentrations and dietary exposure to polycyclic aromatic hydrocarbons (PAHs) from grilled and smoked foods. Food Control 2011, 22, 2028–2035. [Google Scholar] [CrossRef]
- Flores-Balcázar, C.; Rosales-Pérez, S.; Galván-Salazar, G.; López-Navarro, O. Nutrientes de la dieta y apoptosis como mecanismos reguladores del cáncer. iMedPub J. 2015, 11. [Google Scholar] [CrossRef]
- Pérez-Morales, G.; Morales, P.; Haza, A. Polycyclic aromatic hidrocarbons (PAHS) I: Toxicity, population exposure and involved foods. RCCV 2016, 10, 1–15. [Google Scholar] [CrossRef]
- Tamaki, A.; Hayashi, H.; Nakajima, H.; Takii, T.; Katagiri, D.; Miyazawa, K.; Hirose, K.; Onozaki, K. Polycyclic aromatic hydrocarbon increases mRNA level for interleukin 1 beta in human fibroblast-like synoviocyte line via aryl hydrocarbon receptor. Biol. Pharm. Bull. 2004, 27, 407–410. [Google Scholar] [CrossRef]
- Joshi, A.D.; Kim, A.; Lewinger, J.P.; Ulrich, C.M.; Potter, J.D.; Cotterchio, M.; Le Marchand, L.; Stern, M.C. Meat intake, cooking methods, dietary carcinogens, and colorectal cancer risk: Findings from the Colorectal Cancer Family Registry. Cancer Med. 2015, 4, 936–952. [Google Scholar] [CrossRef] [PubMed]
- Busquets, R.; Bordas, M.; Toribio, F.; Puignou, L.; Galceran, M.T. Occurrence of heterocyclic amines in several home-cooked meat dishes of the Spanish diet. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2004, 802, 79–86. [Google Scholar] [CrossRef]
- Trichopoulou, A.; Martínez-González, M.A.; Tong, T.Y.N.; Forouhi, N.G.; Khandelwal, S.; Prabhakaran, D.; Mozaffarian, D.; de Lorgeril, M. Definitions and potential health benefits of the Mediterranean diet: Views from experts around the world. BMC Med. 2014, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Druzhinin, V.G.; Matskova, L.V.; Fucic, A. Induction and modulation of genotoxicity by the bacteriome in mammals. Mutat. Res. 2018, 776, 70–77. [Google Scholar] [CrossRef] [PubMed]
- De Kok, T. Evaluation of fecal mutagenicity and colorectal cancer risk. Mutat. Res. Mutat. Res. 2000, 463, 53–101. [Google Scholar] [CrossRef]
- Grasso, F.; Frisan, T. Bacterial genotoxins: Merging the DNA damage response into infection biology. Biomolecules 2015, 5, 1762–1782. [Google Scholar] [CrossRef] [PubMed]
- Jinadasa, R.N.; Bloom, S.E.; Weiss, R.S.; Duhamel, G.E. Cytolethal distending toxin: A conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology 2011, 157, 1851–1875. [Google Scholar] [CrossRef] [PubMed]
- Gagnaire, A.; Nadel, B.; Raoult, D.; Neefjes, J.; Gorvel, J.-P. Collateral damage: Insights into bacterial mechanisms that predispose host cells to cancer. Nat. Rev. Microbiol. 2017, 15, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Schauer, D.B.; Fox, J.G. In vivo virulence properties of bacterial cytolethal-distending toxin. Cell. Microbiol. 2008, 10, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Nougayrede, J.-P. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006, 313, 848–851. [Google Scholar] [CrossRef]
- Cougnoux, A.; Dalmasso, G.; Martinez, R.; Buc, E.; Delmas, J.; Gibold, L.; Sauvanet, P.; Darcha, C.; Déchelotte, P.; Bonnet, M.; et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 2014, 63, 1932–1942. [Google Scholar] [CrossRef]
- Buc, E.; Dubois, D.; Sauvanet, P.; Raisch, J.; Delmas, J.; Darfeuille-Michaud, A.; Pezet, D.; Bonnet, R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE 2013, 8, e56964. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.; Hogan, A.M.; Winter, D.C. Microbial and viral pathogens in colorectal cancer. Lancet Oncol. 2011, 12, 504–512. [Google Scholar] [CrossRef]
- Wu, S.; Morin, P.J.; Maouyo, D.; Sears, C.L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 2003, 124, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its fadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, Y. Escherichia coli cytotoxic necrotizing factors and Bordetella dermonecrotic toxin: The dermonecrosis-inducing toxins activating Rho small GTPases. Toxicon 2001, 39, 1619–1627. [Google Scholar] [CrossRef]
- Nougayrède, J.-P.; Taieb, F.; De Rycke, J.; Oswald, E. Cyclomodulins: Bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 2005, 13, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Barrasa, J.I.; Olmo, N.; Lizarbe, M.A.; Turnay, J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol. In Vitro 2013, 27, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, H.; Chu, E.S.H.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 2017, 152, 1419–1433. [Google Scholar] [CrossRef]
- Huycke, M.M.; Abrams, V.; Moore, D.R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 2002, 23, 529–536. [Google Scholar] [CrossRef]
- Owen, R.W. Generation of reactive oxygen species by the faecal matrix. Gut 2000, 46, 225–232. [Google Scholar] [CrossRef]
- Attene-Ramos, M.S. Evidence that hydrogen sulfide is a genotoxic agent. Mol. Cancer Res. 2006, 4, 9–14. [Google Scholar] [CrossRef]
- Andriamihaja, M.; Lan, A.; Beaumont, M.; Audebert, M.; Wong, X.; Yamada, K.; Yin, Y.; Tomé, D.; Carrasco-Pozo, C.; Gotteland, M.; et al. The deleterious metabolic and genotoxic effects of the bacterial metabolite p-cresol on colonic epithelial cells. Free Radic. Biol. Med. 2015, 85, 219–227. [Google Scholar] [CrossRef]
- Blachier, F.; Beaumont, M.; Andriamihaja, M.; Davila, A.-M.; Lan, A.; Grauso, M.; Armand, L.; Benamouzig, R.; Tomé, D. Changes in the luminal environment of the colonic epithelial cells and physiopathological consequences. Am. J. Pathol. 2017, 187, 476–486. [Google Scholar] [CrossRef]
- Szekely, J.; Gates, K.S. Noncovalent DNA binding and the mechanism of oxidative DNA damage by fecapentaene-12. Chem. Res. Toxicol. 2006, 19, 117–121. [Google Scholar] [CrossRef]
- Mouillé, B.; Delpal, S.; Mayeur, C.; Blachier, F. Inhibition of human colon carcinoma cell growth by ammonia: A non-cytotoxic process associated with polyamine synthesis reduction. Biochim. Biophys. Acta Gen. Subj. 2003, 1624, 88–97. [Google Scholar] [CrossRef]
- Federici, E.; Prete, R.; Lazzi, C.; Pellegrini, N.; Moretti, M.; Corsetti, A.; Cenci, G. Bacterial composition, genotoxicity, and cytotoxicity of fecal samples from individuals consuming omnivorous or vegetarian diets. Front. Microbiol. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Daniela, E.; Sara, S.; Marcella, M.; Giovanni, A.; Meynier, A.; Sophie, V.; Cristina, C.M. Fecal water genotoxicity in healthy free-living young Italian people. Food Chem. Toxicol. 2014, 64, 104–109. [Google Scholar] [CrossRef]
- Vanhaecke, L.; Knize, M.G.; Noppe, H.; De Brabander, H.; Verstraete, W.; Van de Wiele, T. Intestinal bacteria metabolize the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine following consumption of a single cooked chicken meal in humans. Food Chem. Toxicol. 2008, 46, 140–148. [Google Scholar] [CrossRef]
- Kim, J.; Park, S.-H.; Do, K.H.; Kim, D.; Moon, Y. Interference with mutagenic aflatoxin B1-induced checkpoints through antagonistic action of ochratoxin A in intestinal cancer cells: A molecular explanation on potential risk of crosstalk between carcinogens. Oncotarget 2016, 7, 39627–39639. [Google Scholar] [CrossRef]
- Prete, R.; Tofalo, R.; Federici, E.; Ciarrocchi, A.; Cenci, G.; Corsetti, A. Food-associated Lactobacillus plantarum and yeasts inhibit the genotoxic effect of 4-nitroquinoline-1-oxide. Front. Microbiol. 2017, 8, 2349. [Google Scholar] [CrossRef]
- Ribière, C.; Peyret, P.; Parisot, N.; Darcha, C.; Déchelotte, P.J.; Barnich, N.; Peyretaillade, E.; Boucher, D. Oral exposure to environmental pollutant benzo[a]pyrene impacts the intestinal epithelium and induces gut microbial shifts in murine model. Sci. Rep. 2016, 6, 31027. [Google Scholar] [CrossRef]
- Defois, C.; Ratel, J.; Denis, S.; Batut, B.; Beugnot, R.; Peyretaillade, E.; Engel, E.; Peyret, P. Environmental pollutant benzo[a]pyrene impacts the volatile metabolome and transcriptome of the human gut microbiota. Front. Microbiol. 2017, 8, 1562. [Google Scholar] [CrossRef]
- Defois, C.; Ratel, J.; Garrait, G.; Denis, S.; Le Goff, O.; Talvas, J.; Mosoni, P.; Engel, E.; Peyret, P. Food chemicals disrupt human gut microbiota activity and impact intestinal homeostasis as revealed by in vitro systems. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Morotomi, M.; Mutai, M. In vitro binding of potent mutagenic pyrolysates to intestinal bacteria. J. Natl. Cancer Inst. 1986, 77, 195–201. [Google Scholar] [PubMed]
- Orrhage, K.; Sillerström, E.; Gustafsson, J.-Å.; Nord, C.E.; Rafter, J. Binding of mutagenic heterocyclic amines by intestinal and lactic acid bacteria. Mutat. Res. Mol. Mech. Mutagen. 1994, 311, 239–248. [Google Scholar] [CrossRef]
- Vanhaecke, L.; Vercruysse, F.; Boon, N.; Verstraete, W.; Cleenwerck, I.; De Wachter, M.; De Vos, P.; van de Wiele, T. Isolation and characterization of human intestinal bacteria capable of transforming the dietary carcinogen 2-Amino-1-Methyl-6-Phenylimidazo[4,5-b]Pyridine. Appl. Environ. Microbiol. 2008, 74, 1469–1477. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.; Empl, M.T.; Schwab, C.; Fekry, M.I.; Engels, C.; Schneider, M.; Lacroix, C.; Steinberg, P.; Sturla, S.J. Gut microbial transformation of the dietary imidazoquinoxaline mutagen MelQx reduces its cytotoxic and mutagenic potency. Toxicol. Sci. 2017, 159, 266–276. [Google Scholar] [CrossRef]
- Carmody, R.N.; Turnbaugh, P.J. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Investig. 2014, 124, 4173–4181. [Google Scholar] [CrossRef] [PubMed]
- Kassie, F. Intestinal microflora plays a crucial role in the genotoxicity of the cooked food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline (IQ). Carcinogenesis 2001, 22, 1721–1725. [Google Scholar] [CrossRef]
- Humblot, C.; Murkovic, M.; Rigottier-Gois, L.; Bensaada, M.; Bouclet, A.; Andrieux, C.; Anba, J.; Rabot, S. Glucuronidase in human intestinal microbiota is necessary for the colonic genotoxicity of the food-borne carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline in rats. Carcinogenesis 2007, 28, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Kaderlik, K.R.; Mulder, G.J.; Turesky, R.J.; Lang, N.P.; Teitel, C.H.; Chiarelli, M.P.; Kadlubar, F.F. Glucuronidation of N -hydroxy heterocyclic amines by human and rat liver microsomes. Carcinogenesis 1994, 15, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Do, K.N.; Fink, L.N.; Jensen, T.E.; Gautier, L.; Parlesak, A. TLR2 Controls intestinal carcinogen detoxication by CYP1A1. PLoS ONE 2012, 7, e32309. [Google Scholar] [CrossRef]
- Organisation for Economic Co-operation and Development. Genetic Toxicology Guidance Document; OECD: Paris, France, 2015; pp. 1–58. [Google Scholar]
- Thybaud, V.; Lorge, E.; Levy, D.D.; van Benthem, J.; Douglas, G.R.; Marchetti, F.; Moore, M.M.; Schoeny, R. Main issues addressed in the 2014-2015 revisions to the OECD Genetic Toxicology Test Guidelines. Environ. Mol. Mutagen. 2017, 58, 284–295. [Google Scholar] [CrossRef]
- Turkez, H.; Arslan, M.E.; Ozdemir, O. Genotoxicity testing: Progress and prospects for the next decade. Expert Opin. Drug Metab. Toxicol. 2017, 13, 1089–1098. [Google Scholar] [CrossRef]
- Iglesias, T.; López de Cerain, A.; Irache, J.; Martín-Arbella, N.; Wilcox, M.; Pearson, J.; Azqueta, A. Evaluation of the cytotoxicity, genotoxicity and mucus permeation capacity of several surface modified poly(anhydride) nanoparticles designed for oral drug delivery. Int. J. Pharm. 2017, 517, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Róka, E.; Ujhelyi, Z.; Deli, M.; Bocsik, A.; Fenyvesi, É.; Szente, L.; Fenyvesi, F.; Vecsernyés, M.; Váradi, J.; Fehér, P.; et al. Evaluation of the cytotoxicity of α-cyclodextrin derivatives on the Caco-2 Cell line and human erythrocytes. Molecules 2015, 20, 20269–20285. [Google Scholar] [CrossRef]
- Valdés, L.; Gueimonde, M.; Ruas-Madiedo, P. Monitoring in real time the cytotoxic effect of Clostridium difficile upon the intestinal epithelial cell line HT29. J. Microbiol. Methods 2015, 119, 66–73. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fair, K.L.; Colquhoun, J.; Hannan, N.R.F. Intestinal organoids for modelling intestinal development and disease. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170217. [Google Scholar] [CrossRef]
- Kaushik, G.; Ponnusamy, M.P.; Batra, S.K. Concise review: Current status of three-dimensional organoids as preclinical models. Stem Cells 2018, 36, 1329–1340. [Google Scholar] [CrossRef]
- Huang, W.-Y.; Cai, Y.-Z.; Zhang, Y. Natural phenolic compounds from medicinal herbs and dietary plants: Potential use for cancer prevention. Nutr. Cancer 2009, 62, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, A.; Anderson, D.; de Pascual-Teresa, S.; Santos-Buelga, C.; Clifford, M.N.; Ioannides, C. Evaluation of the antigenotoxic potential of monomeric and dimeric flavanols, and black tea polyphenols against heterocyclic amine-induced DNA damage in human lymphocytes using the Comet assay. Mutat. Res. 2002, 515, 39–56. [Google Scholar] [CrossRef]
- Quelhas, I.; Petisca, C.; Viegas, O.; Melo, A.; Pinho, O.; Ferreira, I.M.P.L.V.O. Effect of green tea marinades on the formation of heterocyclic aromatic amines and sensory quality of pan-fried beef. Food Chem. 2010, 122, 98–104. [Google Scholar] [CrossRef]
- Bear, W.L.; Teel, R.W. Effects of citrus flavonoids on the mutagenicity of heterocyclic amines and on cytochrome P450 1A2 activity. Anticancer Res. 2000, 20, 3609–3614. [Google Scholar] [PubMed]
- Edenharder, R.; Sager, J.W.; Glatt, H.; Muckel, E.; Platt, K.L. Protection by beverages, fruits, vegetables, herbs, and flavonoids against genotoxicity of 2-acetylaminofluorene and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in metabolically competent V79 cells. Mutat. Res. 2002, 521, 57–72. [Google Scholar] [CrossRef]
- Cheng, K.-W.; Wu, Q.; Zheng, Z.P.; Peng, X.; Simon, J.E.; Chen, F.; Wang, M. Inhibitory effect of fruit extracts on the formation of heterocyclic amines. J. Agric. Food Chem. 2007, 55, 10359–10365. [Google Scholar] [CrossRef] [PubMed]
- Sepahpour, S.; Selamat, J.; Khatib, A.; Manap, M.Y.A.; Abdull Razis, A.F.; Hajeb, P. Inhibitory effect of mixture herbs/spices on formation of heterocyclic amines and mutagenic activity of grilled beef. Food Addit. Contam. Part A 2018, 35, 1911–1927. [Google Scholar] [CrossRef] [PubMed]
- Balogh, Z.; Gray, J.I.; Gomaa, E.A.; Booren, A.M. Formation and inhibition of heterocyclic aromatic amines in fried ground beef patties. Food Chem. Toxicol. 2000, 38, 395–401. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Bindels, L.B.; Delzenne, N.M.; Cani, P.D.; Walter, J. Towards a more comprehensive concept for prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 1998, 19, 281–285. [Google Scholar] [CrossRef]
- Chen, C.-C.; Lin, W.-C.; Kong, M.-S.; Shi, H.N.; Walker, W.A.; Lin, C.-Y.; Huang, C.-T.; Lin, Y.-C.; Jung, S.-M.; Lin, T.-Y. Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra-intestinal tissue. Br. J. Nutr. 2012, 107, 1623–1634. [Google Scholar] [CrossRef]
- Gamallat, Y.; Meyiah, A.; Kuugbee, E.D.; Hago, A.M.; Chiwala, G.; Awadasseid, A.; Bamba, D.; Zhang, X.; Shang, X.; Luo, F.; et al. Lactobacillus rhamnosus induced epithelial cell apoptosis, ameliorates inflammation and prevents colon cancer development in an animal model. Biomed. Pharmacother. 2016, 83, 536–541. [Google Scholar] [CrossRef]
- Lili, Z.; Junyan, W.; Hongfei, Z.; Baoqing, Z.; Bolin, Z. Detoxification of cancerogenic compounds by lactic acid bacteria strains. Crit. Rev. Food Sci. Nutr. 2017, 1–16. [Google Scholar] [CrossRef]
- Tsuda, H.; Hara, K.; Miyamoto, T. Binding of mutagens to exopolysaccharide produced by Lactobacillus plantarum mutant strain 301102S. J. Dairy Sci. 2008, 91, 2960–2966. [Google Scholar] [CrossRef]
- Kumar, M.; Kumar, A.; Nagpal, R.; Mohania, D.; Behare, P.; Verma, V.; Kumar, P.; Poddar, D.; Aggarwal, P.K.; Henry, C.J.K.; et al. Cancer-preventing attributes of probiotics: An update. Int. J. Food Sci. Nutr. 2010, 61, 473–496. [Google Scholar] [CrossRef]
- Nowak, A.; Libudzisz, Z. Ability of probiotic Lactobacillus casei DN 114001 to bind or/and metabolise heterocyclic aromatic amines in vitro. Eur. J. Nutr. 2009, 48, 419–427. [Google Scholar] [CrossRef]
- Nowak, A.; Śliżewska, K.; Otlewska, A. Antigenotoxic activity of lactic acid bacteria, prebiotics, and products of their fermentation against selected mutagens. Regul. Toxicol. Pharmacol. 2015, 73, 938–946. [Google Scholar] [CrossRef]
- Beer, F.; Urbat, F.; Steck, J.; Huch, M.; Bunzel, D.; Bunzel, M.; Kulling, S.E. Metabolism of foodborne heterocyclic aromatic amines by Lactobacillus reuteri DSM 20016. J. Agric. Food Chem. 2017, 65, 6797–6811. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhang, X.; Covasa, M. Emerging roles of lactic acid bacteria in protection against colorectal cancer. World J. Gastroenterol. 2014, 20, 7878–7886. [Google Scholar] [CrossRef]
- Nowak, A.; Śliżewska, K.; Błasiak, J.; Libudzisz, Z. The influence of Lactobacillus casei DN 114 001 on the activity of faecal enzymes and genotoxicity of faecal water in the presence of heterocyclic aromatic amines. Anaerobe 2014, 30, 129–136. [Google Scholar] [CrossRef]
- Gianfredi, V.; Salvatori, T.; Villarini, M.; Moretti, M.; Nucci, D.; Realdon, S. Is dietary fibre truly protective against colon cancer? A systematic review and meta-analysis. Int. J. Food Sci. Nutr. 2018, 69, 904–915. [Google Scholar] [CrossRef]
- Lim, C.C.; Ferguson, L.R.; Tannock, G.W. Dietary fibres as “prebiotics”: Implications for colorectal cancer. Mol. Nutr. Food Res. 2005, 49, 609–619. [Google Scholar] [CrossRef]
- Mahowald, M.A.; Rey, F.E.; Seedorf, H.; Turnbaugh, P.J.; Fulton, R.S.; Wollam, A.; Shah, N.; Wang, C.; Magrini, V.; Wilson, R.K.; et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 2009, 106, 5859–5864. [Google Scholar] [CrossRef]
- Jakszyn, P.; Agudo, A.; Ibáñez, R.; García-Closas, R.; Pera, G.; Amiano, P.; González, C.A. Development of a food database of nitrosamines, heterocyclic amines, and polycyclic aromatic hydrocarbons. J. Nutr. 2004, 134, 2011–2014. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).