Benefits and Drawbacks of Harboring Plasmid pP32BP2, Identified in Arctic Psychrophilic Bacterium Psychrobacter sp. DAB_AL32B
Abstract
1. Introduction
2. Results and Discussion
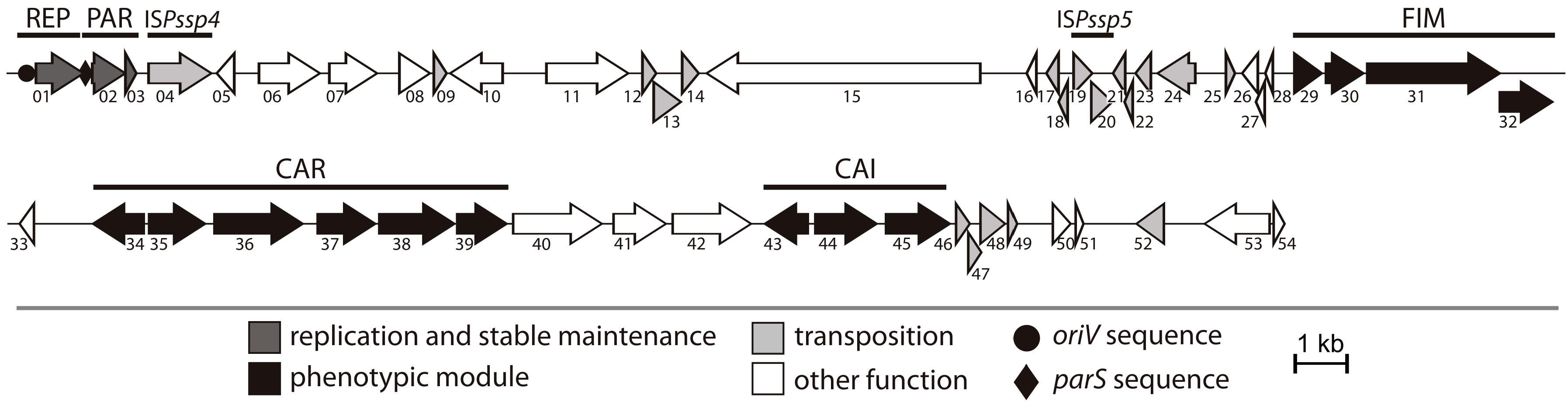
2.1. General Features of Plasmid pP32BP2
2.2. REP and PAR Modules—Plasmid Replication and Active Partitioning
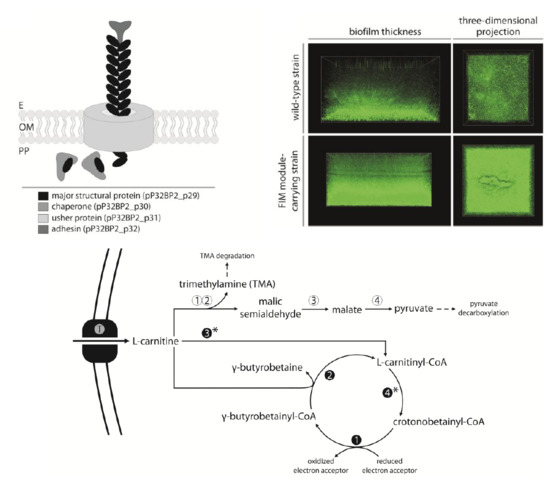
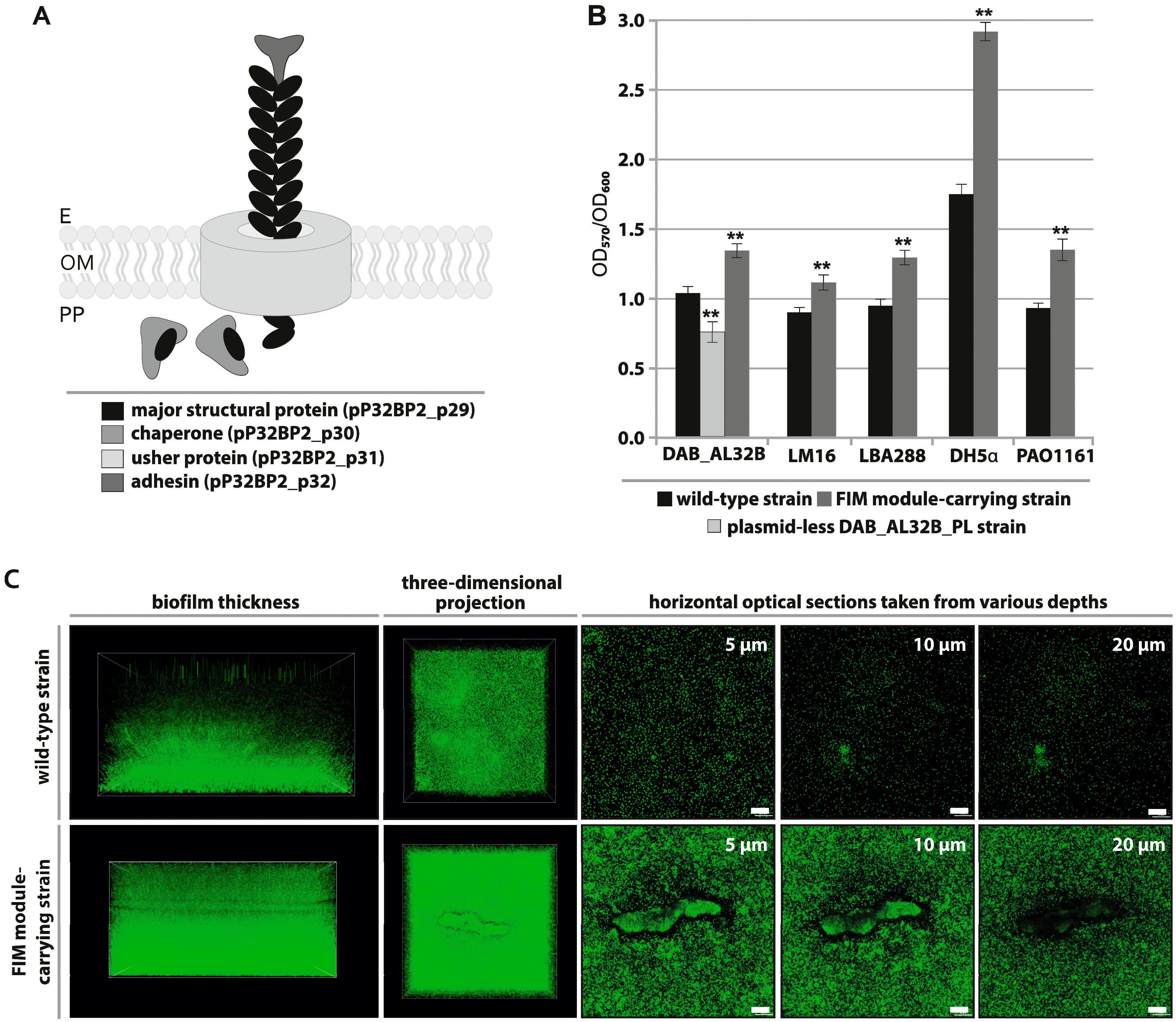
2.3. FIM Module—Cell Adhesion to Solid Surfaces and Biofilm Formation
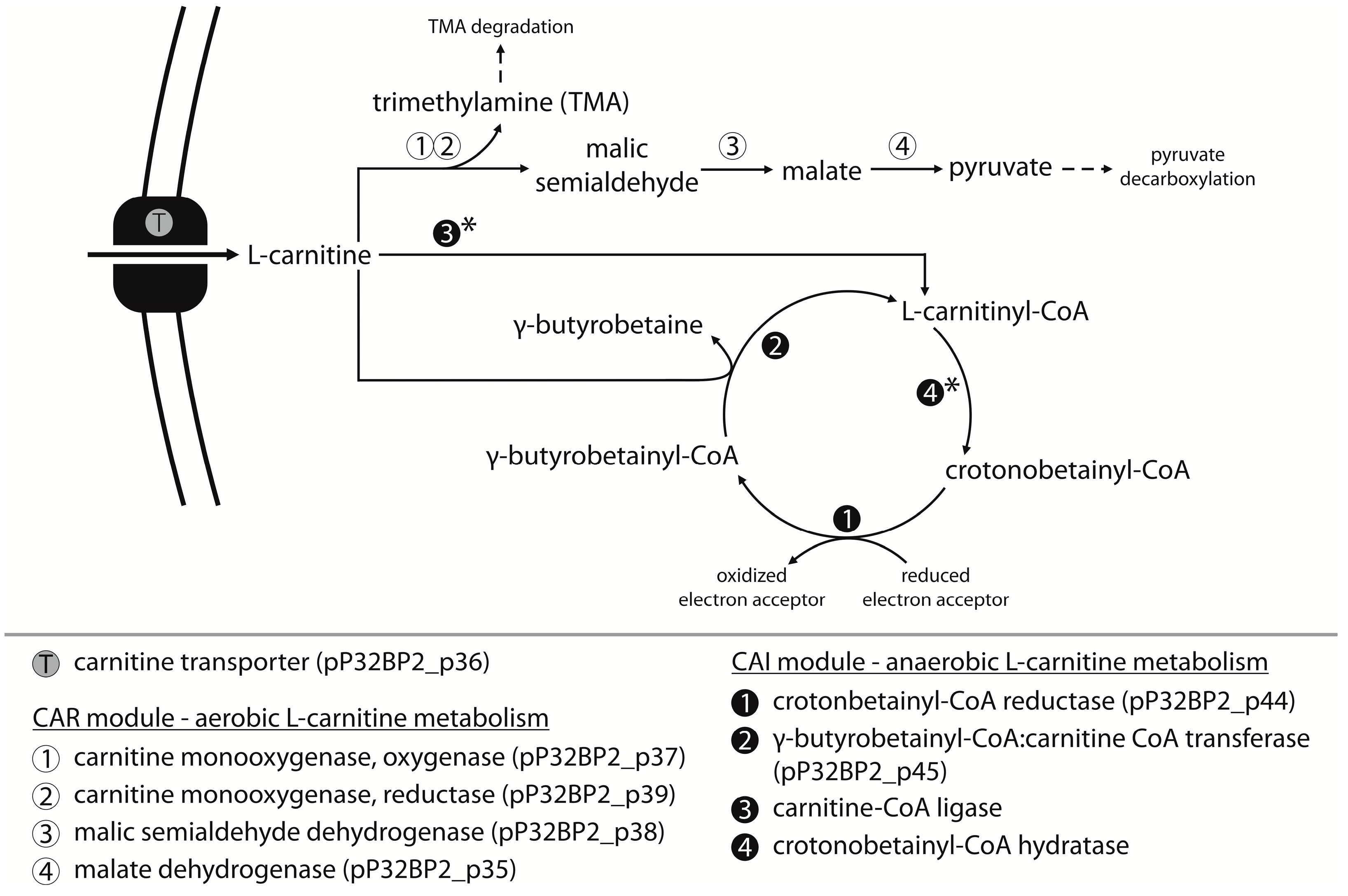
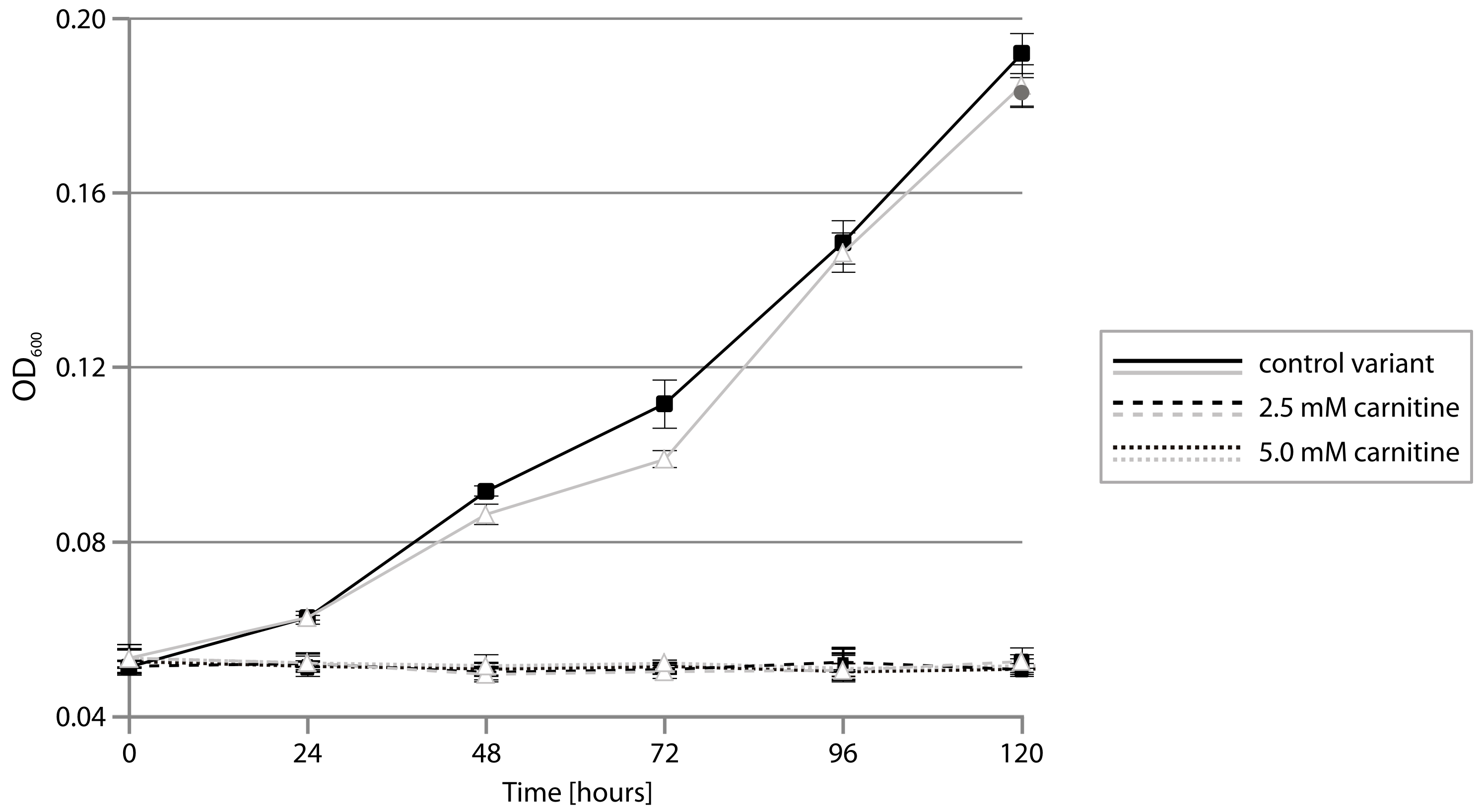
2.4. CAR Module—Aerobic Metabolism of Carnitine
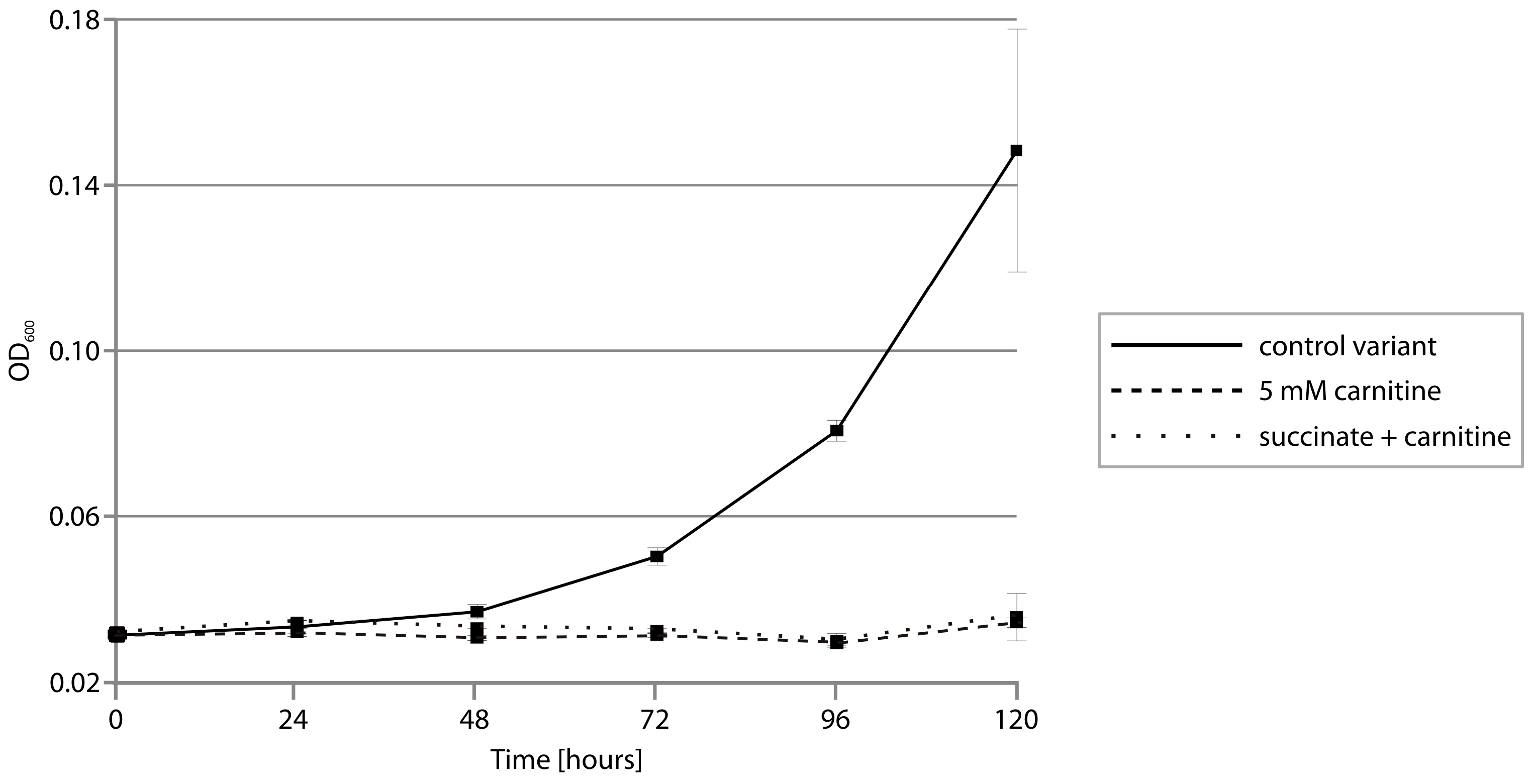
2.5. CAI Module—Anaerobic Metabolism of Carnitine
3. Materials and Methods
3.1. Bacterial Strains, Plasmids and Culture Conditions
3.2. DNA Manipulations and Introduction of Plasmid DNA into Bacterial Cells
3.3. Construction of Plasmid-Less Psychrobacter sp. DAB_AL32B
3.4. DNA Sequencing
3.5. Bioinformatic Analyses
3.6. Testing Bacterial Adherence to Artificial Surfaces
3.7. Live Cell Confocal Microscopy (Biofilm Analysis)
3.8. Testing Psychrobacter spp. Growth on Carnitine as the Sole Carbon Source Under Aerobic Conditions
3.9. Determination of Trimethylamine Concentration
3.10. Testing Anaerobic Growth of Psychrobacter spp.
3.11. Nucleotide Sequence Accession Numbers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bozal, N.; Montes, M.J.; Tudela, E.; Guinea, J. Characterization of several Psychrobacter strains isolated from Antarctic environments and description of Psychrobacter luti sp. nov. and Psychrobacter fozii sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P. The genus Psychrobacter. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 920–930. [Google Scholar]
- Romanenko, L.A.; Schumann, P.; Rohde, M.; Lysenko, A.M.; Mikhailov, V.V.; Stackebrandt, E. Psychrobacter submarinus sp. nov. and Psychrobacter marincola sp. nov., psychrophilic halophiles from marine environments. Int. J. Syst. Evol. Microbiol. 2002, 52, 1291–1297. [Google Scholar] [CrossRef]
- Bowman, J.P.; Cavanagh, J.; Austin, J.J.; Sanderson, K. Novel Psychrobacter species from Antarctic ornithogenic soils. Int. J. Syst. Bacteriol. 1996, 46, 841–848. [Google Scholar] [CrossRef]
- Rodrigues, D.F.; Da, C.J.E.; Ayala-Del-Rio, H.L.; Pellizari, V.H.; Gilichinsky, D.; Sepulveda-Torres, L.; Tiedje, J.M. Biogeography of two cold-adapted genera: Psychrobacter and Exiguobacterium. ISME J. 2009, 3, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Denner, E.B.; Mark, B.; Busse, H.J.; Turkiewicz, M.; Lubitz, W. Psychrobacter proteolyticus sp. nov., a psychrotrophic, halotolerant bacterium isolated from the Antarctic krill Euphausia superba Dana, excreting a cold-adapted metalloprotease. Syst. Appl. Microbiol. 2001, 24, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.-X.; Yu, Y.; Liu, Y.; Li, H.-R. Psychrobacter glaciei sp. nov., isolated from the ice core of an Arctic glacier. Int. J. Syst. Evol. Microbiol. 2016, 66, 1792–1798. [Google Scholar] [CrossRef] [PubMed]
- Dziewit, L.; Cegielski, A.; Romaniuk, K.; Uhrynowski, W.; Szych, A.; Niesiobedzki, P.; Zmuda-Baranowska, M.J.; Zdanowski, M.K.; Bartosik, D. Plasmid diversity in arctic strains of Psychrobacter spp. Extremophiles 2013, 17, 433–444. [Google Scholar] [CrossRef][Green Version]
- Yassin, A.F.; Busse, H.-J. Psychrobacter lutiphocae sp. nov., isolated from the faeces of a seal. Int. J. Syst. Evol. Microbiol. 2009, 59, 2049–2053. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ortiz-Alcantara, J.M.; Segura-Candelas, J.M.; Garces-Ayala, F.; Gonzalez-Duran, E.; Rodriguez-Castillo, A.; Alcantara-Perez, P.; Wong-Arambula, C.; Gonzalez-Villa, M.; Leon-Avila, G.; Garcia-Chequer, A.J.; et al. Fatal Psychrobacter sp. infection in a pediatric patient with meningitis identified by metagenomic next-generation sequencing in cerebrospinal fluid. Arch. Microbiol. 2016, 198, 129–135. [Google Scholar] [CrossRef]
- Bonwitt, J.; Tran, M.; Droz, A.; Gonzalez, A.; Glover, W.A. Psychrobacter sanguinis wound infection associated with marine environment exposure, Washington, USA. Emerg. Infect. Dis. 2018, 24, 1942–1944. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, J.S.N.; Correia, A.; Henriques, I. Molecular analysis of the diversity of genus Psychrobacter present within a temperate estuary. FEMS Microbiol. Ecol. 2013, 84, 451–460. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bjørkevoll, I.; Olsen, R.L.; Skjerdal, O.T. Origin and spoilage potential of the microbiota dominating genus Psychrobacter in sterile rehydrated salt-cured and dried salt-cured cod (Gadus morhua). Int. J. Food Microbiol. 2003, 84, 175–187. [Google Scholar] [CrossRef]
- Ponder, M.A.; Gilmour, S.J.; Bergholz, P.W.; Mindock, C.A.; Hollingsworth, R.; Thomashow, M.F.; Tiedje, J.M. Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol. Ecol. 2005, 53, 103–115. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Casella, P.; Caruso, C.; Mangano, S.; Bruni, V.; De Domenico, M.; Michaud, L. Occurrence and characterization of psychrotolerant hydrocarbon-oxidizing bacteria from surface seawater along the Victoria Land coast (Antarctica). Polar Biol. 2010, 33, 929–943. [Google Scholar] [CrossRef]
- Røberg, S.; Østerhus, J.I.; Landfald, B. Dynamics of bacterial community exposed to hydrocarbons and oleophilic fertilizer in high-Arctic intertidal beach. Polar Biol. 2011, 34, 1455–1465. [Google Scholar] [CrossRef][Green Version]
- Dziewit, L.; Bartosik, D. Plasmids of psychrophilic and psychrotolerant bacteria and their role in adaptation to cold environments. Front. Microbiol. 2014, 5, 596. [Google Scholar] [CrossRef] [PubMed]
- Ciok, A.; Budzik, K.; Zdanowski, M.K.; Gawor, J.; Grzesiak, J.; Decewicz, P.; Gromadka, R.; Bartosik, D.; Dziewit, L. Plasmids of psychrotolerant Polaromonas spp. isolated from arctic and antarctic glaciers–diversity and role in adaptation to polar environments. Front. Microbiol. 2018, 9, 1285. [Google Scholar] [CrossRef]
- Romaniuk, K.; Golec, P.; Dziewit, L. Insight into the diversity and possible role of plasmids in the adaptation of psychrotolerant and metalotolerant Arthrobacter spp. to extreme Antarctic environments. Front. Microbiol. 2018, 9, 3144. [Google Scholar] [CrossRef]
- Petrova, M.; Kurakov, A.; Shcherbatova, N.; Mindlin, S. Genetic structure and biological properties of the first ancient multiresistance plasmid pKLH80 isolated from a permafrost bacterium. Microbiology 2014, 160, 2253–2263. [Google Scholar] [CrossRef]
- Moghadam, M.S.; Albersmeier, A.; Winkler, A.; Cimmino, L.; Rise, K.; Hohmann-Marriott, M.F.; Kalinowski, J.; Ruckert, C.; Wentzel, A.; Lale, R. Isolation and genome sequencing of four Arctic marine Psychrobacter strains exhibiting multicopper oxidase activity. BMC Genomics 2016, 17, 117. [Google Scholar] [CrossRef]
- Ciok, A.; Dziewit, L. Exploring the genome of Arctic Psychrobacter sp. DAB_AL32B and construction of novel Psychrobacter-specific cloning vectors of an increased carrying capacity. Arch. Microbiol. 2019. [Google Scholar] [CrossRef]
- Lasek, R.; Dziewit, L.; Ciok, A.; Decewicz, P.; Romaniuk, K.; Jedrys, Z.; Wibberg, D.; Schlüter, A.; Pühler, A.; Bartosik, D. Genome content, metabolic pathways and biotechnological potential of the psychrophilic Arctic bacterium Psychrobacter sp. DAB_AL43B, a source and a host of novel Psychrobacter-specific vectors. J. Biotechnol. 2017, 263, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Tutino, M.L.; Duilio, A.; Moretti, M.A.; Sannia, G.; Marino, G. A rolling-circle plasmid from Psychrobacter sp. TA144: evidence for a novel rep subfamily. Biochem. Biophys. Res. Commun. 2000, 274, 488–495. [Google Scholar] [CrossRef]
- Lasek, R.; Dziewit, L.; Bartosik, D. Plasmid pP62BP1 isolated from an Arctic Psychrobacter sp. strain carries two highly homologous type II restriction-modification systems and a putative organic sulfate metabolism operon. Extremophiles 2012, 16, 363–376. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dziewit, L.; Czarnecki, J.; Wibberg, D.; Radlinska, M.; Mrozek, P.; Szymczak, M.; Schlüter, A.; Pühler, A.; Bartosik, D. Architecture and functions of a multipartite genome of the methylotrophic bacterium Paracoccus aminophilus JCM 7686, containing primary and secondary chromids. BMC Genomics 2014, 15, 124. [Google Scholar] [CrossRef] [PubMed]
- Czarnecki, J.; Dziewit, L.; Kowalski, L.; Ochnio, M.; Bartosik, D. Maintenance and genetic load of plasmid pKON1 of Paracoccus kondratievae, containing a highly efficient toxin-antitoxin module of the hipAB family. Plasmid 2015, 80, 45–53. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Konieczny, I.; Bury, K.; Wawrzycka, A.; Wegrzyn, K. Iteron Plasmids. Microbiol Spectr 2014, 2. [Google Scholar] [CrossRef]
- Tutino, M.L.; Duilio, A.; Parrilli, R.; Remaut, E.; Sannia, G.; Marino, G. A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperature. Extremophiles 2001, 5, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Nuccio, S.P.; Baumler, A.J. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 2007, 71, 551–575. [Google Scholar] [CrossRef]
- Waksman, G.; Hultgren, S.J. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 2009, 7, 765–774. [Google Scholar] [CrossRef]
- Hinsa-Leasure, S.M.; Koid, C.; Tiedje, J.M.; Schultzhaus, J.N. Biofilm formation by Psychrobacter arcticus and the role of a large adhesin in attachment to surfaces. Appl. Environ. Microbiol. 2013, 79, 3967–3973. [Google Scholar] [CrossRef]
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, C.C.C.R. Biofilms: Microbial Strategies for Surviving UV Exposure. Adv. Exp. Med. Biol. 2017, 996, 233–239. [Google Scholar] [CrossRef]
- Sutherland, I.W. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 2001, 147, 3–9. [Google Scholar] [CrossRef]
- Schroll, C.; Barken, K.B.; Krogfelt, K.A.; Struve, C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.; Garcillan-Barcia, M.P.; Francia, M.V.; Rocha, E.P.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef]
- Fondi, M.; Fani, R. The horizontal flow of the plasmid resistome: clues from inter-generic similarity networks. Environ. Microbiol. 2010, 12, 3228–3242. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Amato, P.; Hennebelle, R.; Magand, O.; Sancelme, M.; Delort, A.-M.; Barbante, C.; Boutron, C.; Ferrari, C. Bacterial characterization of the snow cover at Spitzberg, Svalbard. FEMS Microbiol. Ecol. 2007, 59, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Amores-Arrocha, H.; Malard, L.A.; Els, N.; Sattler, B.; Pearce, D.A. Characterisation of Arctic bacterial communities in the air above Svalbard. Biology (Basel) 2017, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Møller, A.K.; Søborg, D.A.; Abu Al-Soud, W.; Sørensen, S.J.; Kroer, N. Bacterial community structure in High-Arctic snow and freshwater as revealed by pyrosequencing of 16S rRNA genes and cultivation. Polar Res. 2013, 32, 17390. [Google Scholar] [CrossRef]
- Saier, M.H.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Dziewit, L.; Czarnecki, J.; Prochwicz, E.; Wibberg, D.; Schlüter, A.; Pühler, A.; Bartosik, D. Genome-guided insight into the methylotrophy of Paracoccus aminophilus JCM 7686. Front. Microbiol. 2015, 6, 852. [Google Scholar] [CrossRef] [PubMed]
- Murdock, L.; Burke, T.; Coumoundouros, C.; Culham, D.E.; Deutch, C.E.; Ellinger, J.; Kerr, C.H.; Plater, S.M.; To, E.; Wright, G.; et al. Analysis of strains lacking known osmolyte accumulation mechanisms reveals contributions of osmolytes and transporters to protection against abiotic stress. Appl. Environ. Microbiol. 2014, 80, 5366–5378. [Google Scholar] [CrossRef]
- Czarnecki, J.; Dziewit, L.; Puzyna, M.; Prochwicz, E.; Tudek, A.; Wibberg, D.; Schlüter, A.; Pühler, A.; Bartosik, D. Lifestyle-determining extrachromosomal replicon pAMV1 and its contribution to the carbon metabolism of the methylotrophic bacterium Paracoccus aminovorans JCM 7685. Environ. Microbiol. 2017, 19, 4536–4550. [Google Scholar] [CrossRef]
- Zhu, Y.; Jameson, E.; Crosatti, M.; Schafer, H.; Rajakumar, K.; Bugg, T.D.H.; Chen, Y. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. 2014, 111, 4268–4273. [Google Scholar] [CrossRef]
- Bernal, V.; Sevilla, Á.; Cánovas, M.; Iborra, J.L. Production of L-carnitine by secondary metabolism of bacteria. Microb. Cell Fact. 2007, 6, 1–17. [Google Scholar] [CrossRef]
- Jameson, E.; Quareshy, M.; Chen, Y. Methodological considerations for the identification of choline and carnitine-degrading bacteria in the gut. Methods 2018, 149, 42–48. [Google Scholar] [CrossRef]
- Engemann, C.; Elssner, T.; Pfeifer, S.; Krumbholz, C.; Maier, T.; Kleber, H.-P. Identification and functional characterisation of genes and corresponding enzymes involved in carnitine metabolism of Proteus sp. Arch. Microbiol. 2005, 183, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Lee, C.-H.; Yeo, S.-H.; Oh, T.-K. Psychrobacter aquimaris sp. nov. and Psychrobacter namhaensis sp. nov., isolated from sea water of the South Sea in Korea. Int. J. Syst. Evol. Microbiol. 2005, 55, 1007–1013. [Google Scholar] [CrossRef]
- Dziewit, L.; Pyzik, A.; Szuplewska, M.; Matlakowska, R.; Mielnicki, S.; Wibberg, D.; Schlüter, A.; Pühler, A.; Bartosik, D. Diversity and role of plasmids in adaptation of bacteria inhabiting the Lubin copper mine in Poland, an environment rich in heavy metals. Front. Microbiol. 2015, 6, 152. [Google Scholar] [CrossRef]
- Hooykaas, P.J.; den Dulk-Ras, H.; Schilperoort, R.A. Molecular mechanism of Ti plasmid mobilization by R plasmids: isolation of Ti plasmids with transposon-insertions in Agrobacterium tumefaciens. Plasmid 1980, 4, 64–75. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Bartosik, A.A.; Glabski, K.; Jecz, P.; Mikulska, S.; Fogtman, A.; Koblowska, M.; Jagura-Burdzy, G. Transcriptional profiling of ParA and ParB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS ONE 2014, 9, e87276. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, D.; Bialkowska, A.; Baj, J.; Wlodarczyk, M. Construction of mobilizable cloning vectors derived from pBGS18 and their application for analysis of replicator region of a pTAV202 mini-derivative of Paracoccus versutus pTAV1 plasmid. Acta Microbiol. Pol. 1997, 46, 387–392. [Google Scholar]
- Kovach, M.E.; Phillips, R.W.; Elzer, P.H.; Roop, R.M., 2nd; Peterson, K.M. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 1994, 16, 800–802. [Google Scholar]
- Spratt, B.G.; Hedge, P.J.; te Heesen, S.; Edelman, A.; Broome-Smith, J.K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 1986, 41, 337–342. [Google Scholar] [CrossRef]
- Ditta, G.; Stanfield, S.; Corbin, D.; Helinski, D.R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 1980, 77, 7347–7351. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular cloning: A laboratory manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Birnboim, H.C.; Doly, J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979, 7, 1513–1523. [Google Scholar] [CrossRef]
- Bartosik, D.; Szymanik, M.; Wysocka, E. Identification of the partitioning site within the repABC-type replicon of the composite Paracoccus versutus plasmid pTAV1. J. Bacteriol. 2001, 183, 6234–6243. [Google Scholar] [CrossRef]
- Bakermans, C.; Sloup, R.E.; Zarka, D.G.; Tiedje, J.M.; Thomashow, M.F. Development and use of genetic system to identify genes required for efficient low-temperature growth of Psychrobacter arcticus 273-4. Extremophiles 2009, 13, 21–30. [Google Scholar] [CrossRef]
- Kushner, S.R. An improved method for transformation of E. coli with ColE1 derived plasmids. In Genetic engineering; Boyer, H.B., Nicosia, S., Eds.; Elsevier/North-Holland: Amsterdam, The Netherlands, 1978; pp. 17–23. [Google Scholar]
- Irani, V.R.; Rowe, J.J. Enhancement of transformation in Pseudomonas aeruginosa PAO1 by Mg2+ and heat. Biotechniques 1997, 22, 54–56. [Google Scholar] [CrossRef]
- Dziewit, L.; Bartosik, D. Comparative analyses of extrachromosomal bacterial replicons, identification of chromids, and experimental evaluation of their indispensability. Methods Mol. Biol. 2015, 1231, 15–29. [Google Scholar] [CrossRef]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: towards a more sustainable future. Nucleic. Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 2437. [Google Scholar] [CrossRef]
- Kwiatek, A.; Bacal, P.; Wasiluk, A.; Trybunko, A.; Adamczyk-Poplawska, M. The dam replacing gene product enhances Neisseria gonorrhoeae FA1090 viability and biofilm formation. Front. Microbiol. 2014, 5, 712. [Google Scholar] [CrossRef]
- Ikawa, M.; Schaper, T.D.; Dollard, C.A.; Sasner, J.J. Utilization of Folin-Ciocalteu phenol reagent for the detection of certain nitrogen compounds. J. Agric. Food Chem. 2003, 51, 1811–1815. [Google Scholar] [CrossRef] [PubMed]
| Gene(s) | Transposable Element | Family/GROUP | Size (bp) | IR Sequence 1,2,3 | DR Sequence 1 |
|---|---|---|---|---|---|
| pP32BP2_p04 | ISPssp4 | IS256/- | 1302 | IRL: GGGGGTTTCCTAAAAAC TGTGTAACTGC IRR: GAGACCATCCCGAATTC TGTGTAACTGC | CTTAAAAA |
| pP32BP2_p19-20 | ISPssp5 | IS5/IS427 | 837 | IRL: GGGTGTGTCATCAATTA IRR: GGGCGTGTCATCAATTA | TA |
| Plasmid Name | Characteristics 1 | Reference |
|---|---|---|
| pABW1 | Kmr; 4.5 kb; ori pMB1; Mob+; oriT RK2; lacZα; MCS | [58] |
| pABW1-REPPAR | pABW1 carrying REP and PAR modules of pP32BP2 (PCR- amplified with primers L232BREP and R232BREP) | This work |
| pBBR1MCS-2 | Kmr; 5.1 kb; ori pBBR1; Mob+; oriT RK2; lacZα; MCS | [59] |
| pBBR-Ps | pBBR1MCS-2 carrying REP module of pP32BP2 (PCR-amplified with primers L32REP and R32REP) | This work |
| pBBR-Ps-CAR | pBBR-Ps carrying CAR module of pP32BP2 (coordinates 31,262–44,421) inserted between SacI and SalI sites | This work |
| pBBR-FIM | pBBR1MCS-2 carrying FIM module of pP32BP2 (coordinates 24,246–31,265) inserted between BamHI and SalI sites | This work |
| pBBR-Ps-FIM | pBBR-Ps carrying FIM module of pP32BP2 (coordinates 24,246–31,265) inserted between SacI and SalI sites | This work |
| pBGS18 | Kmr; 3.7 kb; ori pMB1; lacZα; MCS | [60] |
| pRK2013 | Kmr; helper plasmid carrying genes for conjugal transfer of RK2 | [61] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciok, A.; Cegielski, A.; Bartosik, D.; Dziewit, L. Benefits and Drawbacks of Harboring Plasmid pP32BP2, Identified in Arctic Psychrophilic Bacterium Psychrobacter sp. DAB_AL32B. Int. J. Mol. Sci. 2019, 20, 2015. https://doi.org/10.3390/ijms20082015
Ciok A, Cegielski A, Bartosik D, Dziewit L. Benefits and Drawbacks of Harboring Plasmid pP32BP2, Identified in Arctic Psychrophilic Bacterium Psychrobacter sp. DAB_AL32B. International Journal of Molecular Sciences. 2019; 20(8):2015. https://doi.org/10.3390/ijms20082015
Chicago/Turabian StyleCiok, Anna, Adrian Cegielski, Dariusz Bartosik, and Lukasz Dziewit. 2019. "Benefits and Drawbacks of Harboring Plasmid pP32BP2, Identified in Arctic Psychrophilic Bacterium Psychrobacter sp. DAB_AL32B" International Journal of Molecular Sciences 20, no. 8: 2015. https://doi.org/10.3390/ijms20082015
APA StyleCiok, A., Cegielski, A., Bartosik, D., & Dziewit, L. (2019). Benefits and Drawbacks of Harboring Plasmid pP32BP2, Identified in Arctic Psychrophilic Bacterium Psychrobacter sp. DAB_AL32B. International Journal of Molecular Sciences, 20(8), 2015. https://doi.org/10.3390/ijms20082015