Optimizing PCR Detection of West Nile Virus from Body Fluid Specimens to Delineate Natural History in an Infected Human Cohort
Abstract
1. Introduction
2. Results
2.1. Optimization of WNV RNA Recovery from Archived WB and UR Specimens
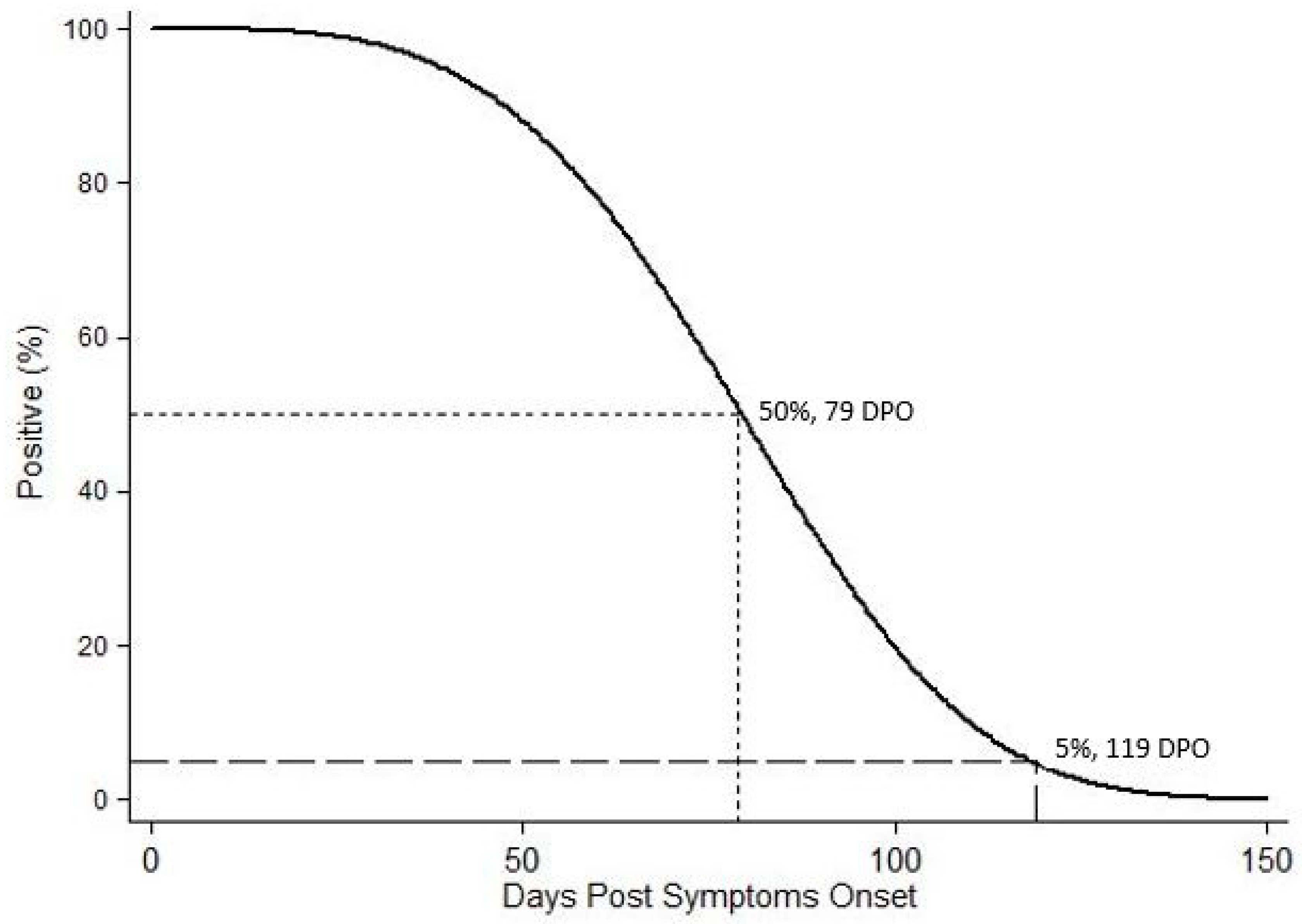
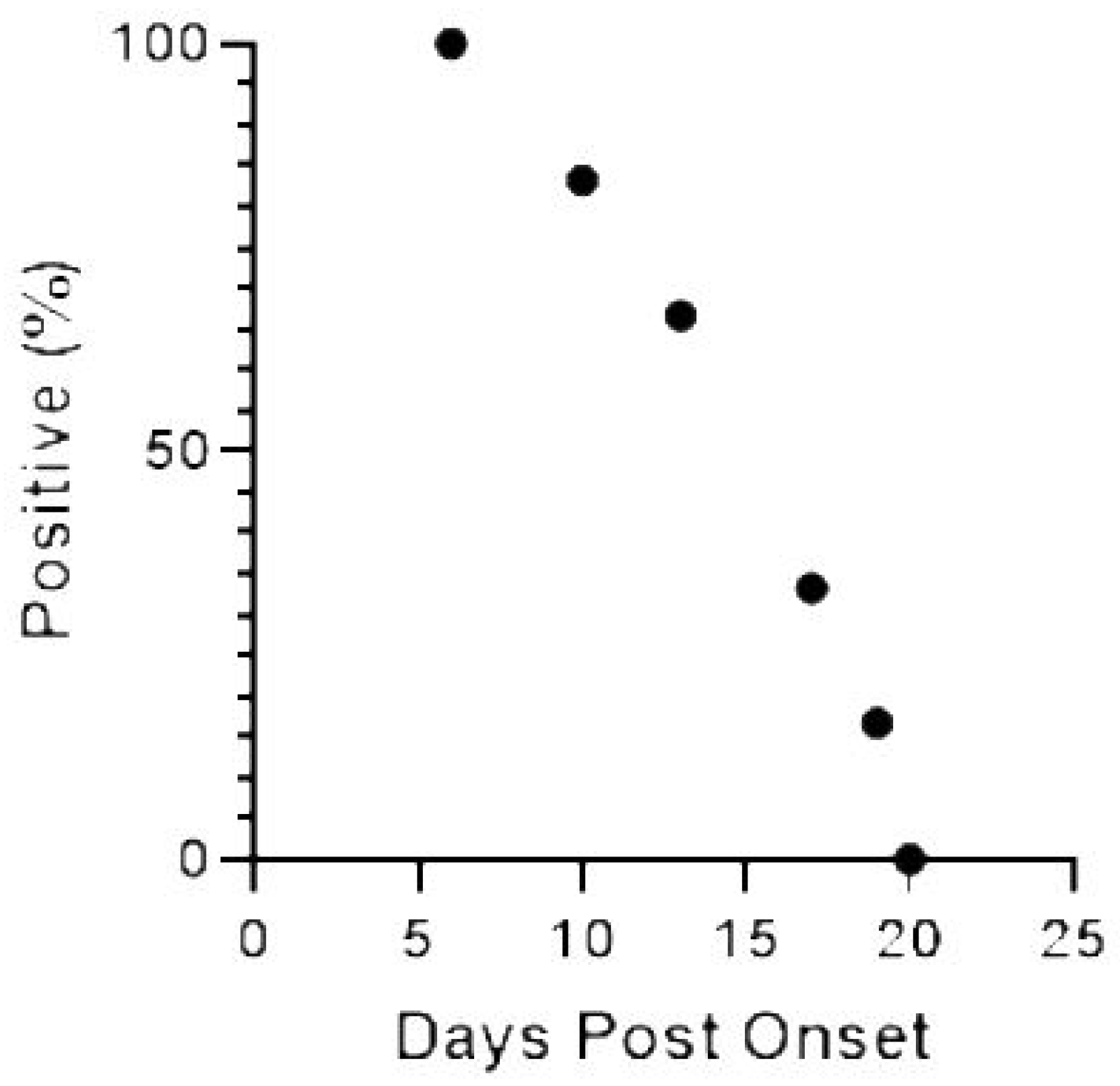
2.2. WNV RNA Recovery from WB from HWNC
2.3. WNV RNA Recovery from UR, Saliva, and Semen from 2018 WNV Cohort Participants
3. Discussion
4. Materials and Methods
4.1. Study Population, Ethics Statement, and Specimen Collection
4.2. Optimization of WNV RNA Extraction from WB and UR Specimens
4.3. Extraction of WNV RNA in Saliva and Semen Specimens
4.4. PCR Detection of WNV RNA in Specimens
4.5. Statistical Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BLQ | Below level of quantification |
| DPO | Days post onset |
| HWNC | Houston West Nile cohort |
| UCB | Urine Conditioning Buffer |
| UR | Urine |
| WB | Whole blood |
| WNV | West Nile virus |
| ZIKV | Zika virus |
References
- Campbell, G.L.; Marfin, A.A.; Lanciotti, R.S.; Gubler, D.J. West Nile virus. Lancet Infect. Dis. 2002, 2, 519–529. [Google Scholar] [CrossRef]
- Granwehr, B.P.; Lillibridge, K.M.; Higgs, S.; Mason, P.W.; Aronson, J.F.; Campbell, G.A.; Barrett, A.D. West Nile virus: Where are we now? Lancet Infect. Dis. 2004, 4, 547–556. [Google Scholar] [CrossRef]
- Murray, K.O.; Walker, C.; Gould, E. The virology, epidemiology, and clinical impact of West Nile virus: A decade of advancements in research since its introduction into the Western Hemisphere. Epidemiol. Infect. 2011, 139, 807–817. [Google Scholar] [CrossRef]
- Krow-Lucal, E.; Lindsey, N.P.; Lehman, J.; Fischer, M.; Staples, J.E. West Nile Virus and Other Nationally Notifiable Arboviral Diseases—United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Barrett, A.D. Economic burden of West Nile virus in the United States. Am. J. Trop. Med. Hyg. 2014, 90, 389–390. [Google Scholar] [CrossRef]
- Weatherhead, J.E.; Miller, V.E.; Garcia, M.N.; Hasbun, R.; Salazar, L.; Dimachkie, M.M.; Murray, K.O. Long-term neurological outcomes in West Nile virus-infected patients: An observational study. Am. J. Trop. Med. Hyg. 2015, 92, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Athar, P.; Hasbun, R.; Nolan, M.S.; Salazar, L.; Woods, S.P.; Sheikh, K.; Murray, K.O. Long-term neuromuscular outcomes of west nile virus infection: A clinical and electromyographic evaluation of patients with a history of infection. Muscle Nerve 2018, 57, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Carson, P.J.; Konewko, P.; Wold, K.S.; Mariani, P.; Goli, S.; Bergloff, P.; Crosby, R.D. Long-term clinical and neuropsychological outcomes of West Nile virus infection. Clin. Infect. Dis. 2006, 43, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.N.; Hause, A.M.; Walker, C.M.; Orange, J.S.; Hasbun, R.; Murray, K.O. Evaluation of prolonged fatigue post-West Nile virus infection and association of fatigue with elevated antiviral and proinflammatory cytokines. Viral Immunol. 2014, 27, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Hasbun, R.; Garcia, M.N.; Kellaway, J.; Baker, L.; Salazar, L.; Woods, S.P.; Murray, K.O. West Nile Virus Retinopathy and Associations with Long Term Neurological and Neurocognitive Sequelae. PLoS ONE 2016, 11, e0148898. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.O.; Garcia, M.N.; Rahbar, M.H.; Martinez, D.; Khuwaja, S.A.; Arafat, R.R.; Rossmann, S. Survival analysis, long-term outcomes, and percentage of recovery up to 8 years post-infection among the Houston West Nile virus cohort. PLoS ONE 2014, 9, e102953. [Google Scholar] [CrossRef]
- Nolan, M.S.; Hause, A.M.; Murray, K.O. Findings of long-term depression up to 8 years post infection from West Nile virus. J. Clin. Psychol. 2012, 68, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Sadek, J.R.; Pergam, S.A.; Harrington, J.A.; Echevarria, L.A.; Davis, L.E.; Goade, D.; Harnar, J.; Nofchissey, R.A.; Sewell, C.M.; Ettestad, P.; et al. Persistent neuropsychological impairment associated with West Nile virus infection. J. Clin. Exp. Neuropsychol. 2010, 32, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Sejvar, J.J. The long-term outcomes of human West Nile virus infection. Clin. Infect. Dis. 2007, 44, 1617–1624. [Google Scholar]
- Baty, S.A.; Gibney, K.B.; Staples, J.E.; Patterson, A.B.; Levy, C.; Lehman, J.; Wadleigh, T.; Feld, J.; Lanciotti, R.; Nugent, C.T.; et al. Evaluation for West Nile Virus (WNV) RNA in urine of patients within 5 months of WNV infection. J. Infect. Dis. 2012, 205, 1476–1477. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.N.; Hasbun, R.; Murray, K.O. Persistence of West Nile virus. Microbes Infect. 2015, 17, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Lanteri, M.C.; Lee, T.H.; Wen, L.; Kaidarova, Z.; Bravo, M.D.; Kiely, N.E.; Kamel, H.T.; Tobler, L.H.; Norris, P.J.; Busch, M.P. West Nile virus nucleic acid persistence in whole blood months after clearance in plasma: Implication for transfusion and transplantation safety. Transfusion 2014, 54, 3232–3241. [Google Scholar] [CrossRef] [PubMed]
- Lustig, Y.; Mannasse, B.; Koren, R.; Katz-Likvornik, S.; Hindiyeh, M.; Mandelboim, M.; Dovrat, S.; Sofer, D.; Mendelson, E. Superiority of West Nile Virus RNA Detection in Whole Blood for Diagnosis of Acute Infection. J. Clin. Microbiol. 2016, 54, 2294–2297. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.; Walker, C.; Herrington, E.; Lewis, J.A.; McCormick, J.; Beasley, D.W.; Tesh, R.B.; Fisher-Hoch, S. Persistent infection with West Nile virus years after initial infection. J. Infect. Dis. 2010, 201, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.O.; Kolodziej, S.; Ronca, S.E.; Gorchakov, R.; Navarro, P.; Nolan, M.S.; Podoll, A.; Finkel, K.; Mandayam, S. Visualization of West Nile Virus in Urine Sediment using Electron Microscopy and Immunogold up to Nine Years Postinfection. Am. J. Trop. Med. Hyg. 2017, 97, 1913–1919. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nagy, A.; Nagy, O.; Ban, E.; Molnar, E.; Muller, Z.; Orban, M.; Kecskes, B.; Harsanyi, E.H.; Kovago, L.; Jobbagy, L.; et al. Detection of West Nile virus in human samples: Follow-up studies during the 2015 seasonal period. Orvosi hetil. 2017, 158, 791–796. [Google Scholar] [CrossRef]
- Lustig, Y.; Lanciotti, R.S.; Hindiyeh, M.; Keller, N.; Milo, R.; Mayan, S.; Mendelson, E. Mutation in West Nile Virus Structural Protein prM during Human Infection. Emerg. Infect. Dis. 2016, 22, 1647–1649. [Google Scholar] [CrossRef]
- Gorchakov, R.; Berry, R.M.; Patel, S.M.; El Sahly, H.M.; Ronca, S.E.; Murray, K.O. Optimizing PCR Detection of Zika Virus from Various Body Fluids. Am. J. Trop. Med. Hyg. 2019, 100, 427–433. [Google Scholar] [CrossRef]
- El Bali, L.; Diman, A.; Bernard, A.; Roosens, N.H.; De Keersmaecker, S.C. Comparative study of seven commercial kits for human DNA extraction from urine samples suitable for DNA biomarker-based public health studies. J. Biomol. Tech. JBT 2014, 25, 96–110. [Google Scholar]
- El-Khoury, V.; Pierson, S.; Kaoma, T.; Bernardin, F.; Berchem, G. Assessing cellular and circulating miRNA recovery: The impact of the RNA isolation method and the quantity of input material. Sci. Rep. 2016, 6, 19529. [Google Scholar] [CrossRef] [PubMed]
- Gibney, K.B.; Lanciotti, R.S.; Sejvar, J.J.; Nugent, C.T.; Linnen, J.M.; Delorey, M.J.; Lehman, J.A.; Boswell, E.N.; Staples, J.E.; Fischer, M. West nile virus RNA not detected in urine of 40 people tested 6 years after acute West Nile virus disease. J. Infect. Dis. 2011, 203, 344–347. [Google Scholar] [CrossRef]
- Tan, S.K.; Sahoo, M.K.; Milligan, S.B.; Taylor, N.; Pinsky, B.A. Stability of Zika virus in urine: Specimen processing considerations and implications for the detection of RNA targets in urine. J. Virol. Methods 2017, 248, 66–70. [Google Scholar] [CrossRef]
- Deng, M.Y.; Wang, H.; Ward, G.B.; Beckham, T.R.; McKenna, T.S. Comparison of six RNA extraction methods for the detection of classical swine fever virus by real-time and conventional reverse transcription-PCR. J. Vet. Diagn. Investig. 2005, 17, 574–578. [Google Scholar] [CrossRef]
- Klungthong, C.; Gibbons, R.V.; Thaisomboonsuk, B.; Nisalak, A.; Kalayanarooj, S.; Thirawuth, V.; Nutkumhang, N.; Mammen, M.P., Jr.; Jarman, R.G. Dengue virus detection using whole blood for reverse transcriptase PCR and virus isolation. J. Clin. Microbiol. 2007, 45, 2480–2485. [Google Scholar] [CrossRef]
- Rios, M.; Daniel, S.; Chancey, C.; Hewlett, I.K.; Stramer, S.L. West Nile virus adheres to human red blood cells in whole blood. Clin. Infect. Dis. 2007, 45, 181–186. [Google Scholar] [CrossRef]
- Lustig, Y.; Mendelson, E.; Paran, N.; Melamed, S.; Schwartz, E. Detection of Zika virus RNA in whole blood of imported Zika virus disease cases up to 2 months after symptom onset, Israel, December 2015 to April 2016. Euro Surveill. 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.O.; Gorchakov, R.; Carlson, A.R.; Berry, R.; Lai, L.; Natrajan, M.; Garcia, M.N.; Correa, A.; Patel, S.M.; Aagaard, K.; et al. Prolonged Detection of Zika Virus in Vaginal Secretions and Whole Blood. Emerg. Infect. Dis. 2017, 23, 99–101. [Google Scholar] [CrossRef]
- Nolan, M.S.; Podoll, A.S.; Hause, A.M.; Akers, K.M.; Finkel, K.W.; Murray, K.O. Prevalence of chronic kidney disease and progression of disease over time among patients enrolled in the Houston West Nile virus cohort. PLoS ONE 2012, 7, e40374. [Google Scholar] [CrossRef] [PubMed]
- Appler, K.K.; Brown, A.N.; Stewart, B.S.; Behr, M.J.; Demarest, V.L.; Wong, S.J.; Bernard, K.A. Persistence of West Nile virus in the central nervous system and periphery of mice. PLoS ONE 2010, 5, e10649. [Google Scholar] [CrossRef] [PubMed]
- Pogodina, V.V.; Frolova, M.P.; Malenko, G.V.; Fokina, G.I.; Koreshkova, G.V.; Kiseleva, L.L.; Bochkova, N.G.; Ralph, N.M. Study on West Nile virus persistence in monkeys. Arch. Virol. 1983, 75, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Saxena, V.; Xie, G.; Li, B.; Farris, T.; Welte, T.; Gong, B.; Boor, P.; Wu, P.; Tang, S.J.; Tesh, R.; et al. A hamster-derived West Nile virus isolate induces persistent renal infection in mice. PLoS Negl. Trop. Dis. 2013, 7, e2275. [Google Scholar] [CrossRef] [PubMed]
- Tesh, R.B.; Siirin, M.; Guzman, H.; Travassos da Rosa, A.P.; Wu, X.; Duan, T.; Lei, H.; Nunes, M.R.; Xiao, S.Y. Persistent West Nile virus infection in the golden hamster: Studies on its mechanism and possible implications for other flavivirus infections. J. Infect. Dis. 2005, 192, 287–295. [Google Scholar] [CrossRef]
- Tonry, J.H.; Xiao, S.Y.; Siirin, M.; Chen, H.; da Rosa, A.P.; Tesh, R.B. Persistent shedding of West Nile virus in urine of experimentally infected hamsters. Am. J. Trop. Med. Hyg. 2005, 72, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lu, L.; Guzman, H.; Tesh, R.B.; Xiao, S.Y. Persistent infection and associated nucleotide changes of West Nile virus serially passaged in hamsters. J. Gen. Virol. 2008, 89 Pt 12, 3073–3079. [Google Scholar] [CrossRef]
- Barzon, L.; Pacenti, M.; Franchin, E.; Pagni, S.; Martello, T.; Cattai, M.; Cusinato, R.; Palu, G. Excretion of West Nile virus in urine during acute infection. J. Infect. Dis. 2013, 208, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Roche, C.; Nhan, T.-X.; Robin, E.; Teissier, A.; Cao-Lormeau, V.-M. Detection of Zika virus in saliva. J. Clin. Virol. 2015, 68, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Barzon, L.; Pacenti, M.; Berto, A.; Sinigaglia, A.; Franchin, E.; Lavezzo, E.; Brugnaro, P.; Palù, G. Isolation of infectious Zika virus from saliva and prolonged viral RNA shedding in a traveller returning from the Dominican Republic to Italy, January 2016. Euro Surveill. 2016, 21, e30159. [Google Scholar] [CrossRef]
- Andries, A.C.; Duong, V.; Ly, S.; Cappelle, J.; Kim, K.S.; Lorn Try, P.; Ros, S.; Ong, S.; Huy, R.; Horwood, P.; et al. Value of routine dengue diagnostic tests in urine and saliva specimens. PLoS Negl. Trop. Dis. 2015, 9, e0004100. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, E.M.; Huuhtamo, E.; Virtala, A.M.; Kantele, A.; Vapalahti, O. Approach to non-incvasive sampling in dengur diagnostics: Exploring virus and NS1 antigen detection in saliva and urine of travelers with dengue. J. Clin. Virol. 2014, 61, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Kurscheidt, F.A.; Mesquita, C.S.; Damke, G.M.; Damke, E.; Analine, R.D.A.; Suehiro, T.T.; Teixeira, J.J.; da Silva, V.R.; Souza, R.P.; Consolaro, M.E. Persistence and clinical relevance of Zika virus in the male genital tract. Nat. Rev. Urol. 2019, 16, 211–230. [Google Scholar] [CrossRef] [PubMed]
- Habu, A.; Murakami, Y.; Ogasa, A.; Fujisaki, Y. Disorder of spermatogenesis and viral discharge into semen in boars infected with Japanese encephalitis virus. Uirusu 1977, 27, 21–26. (In Japanese) [Google Scholar] [CrossRef]
- Santiago, G.A.; Vergne, E.; Quiles, Y.; Cosme, J.; Vazquez, J.; Medina, J.F.; Medina, F.; Colon, C.; Margolis, H.; Munoz-Jordan, J.L. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl. Trop. Dis. 2013, 7, e2311. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorchakov, R.; Gulas-Wroblewski, B.E.; Ronca, S.E.; Ruff, J.C.; Nolan, M.S.; Berry, R.; Alvarado, R.E.; Gunter, S.M.; Murray, K.O. Optimizing PCR Detection of West Nile Virus from Body Fluid Specimens to Delineate Natural History in an Infected Human Cohort. Int. J. Mol. Sci. 2019, 20, 1934. https://doi.org/10.3390/ijms20081934
Gorchakov R, Gulas-Wroblewski BE, Ronca SE, Ruff JC, Nolan MS, Berry R, Alvarado RE, Gunter SM, Murray KO. Optimizing PCR Detection of West Nile Virus from Body Fluid Specimens to Delineate Natural History in an Infected Human Cohort. International Journal of Molecular Sciences. 2019; 20(8):1934. https://doi.org/10.3390/ijms20081934
Chicago/Turabian StyleGorchakov, Rodion, Bonnie E. Gulas-Wroblewski, Shannon E. Ronca, Jeanne C. Ruff, Melissa S. Nolan, Rebecca Berry, R. Elias Alvarado, Sarah M. Gunter, and Kristy O. Murray. 2019. "Optimizing PCR Detection of West Nile Virus from Body Fluid Specimens to Delineate Natural History in an Infected Human Cohort" International Journal of Molecular Sciences 20, no. 8: 1934. https://doi.org/10.3390/ijms20081934
APA StyleGorchakov, R., Gulas-Wroblewski, B. E., Ronca, S. E., Ruff, J. C., Nolan, M. S., Berry, R., Alvarado, R. E., Gunter, S. M., & Murray, K. O. (2019). Optimizing PCR Detection of West Nile Virus from Body Fluid Specimens to Delineate Natural History in an Infected Human Cohort. International Journal of Molecular Sciences, 20(8), 1934. https://doi.org/10.3390/ijms20081934





