Identification and Characterization of Four Autophagy-Related Genes That Are Expressed in Response to Hypoxia in the Brain of the Oriental River Prawn (Macrobrachium nipponense)
Abstract
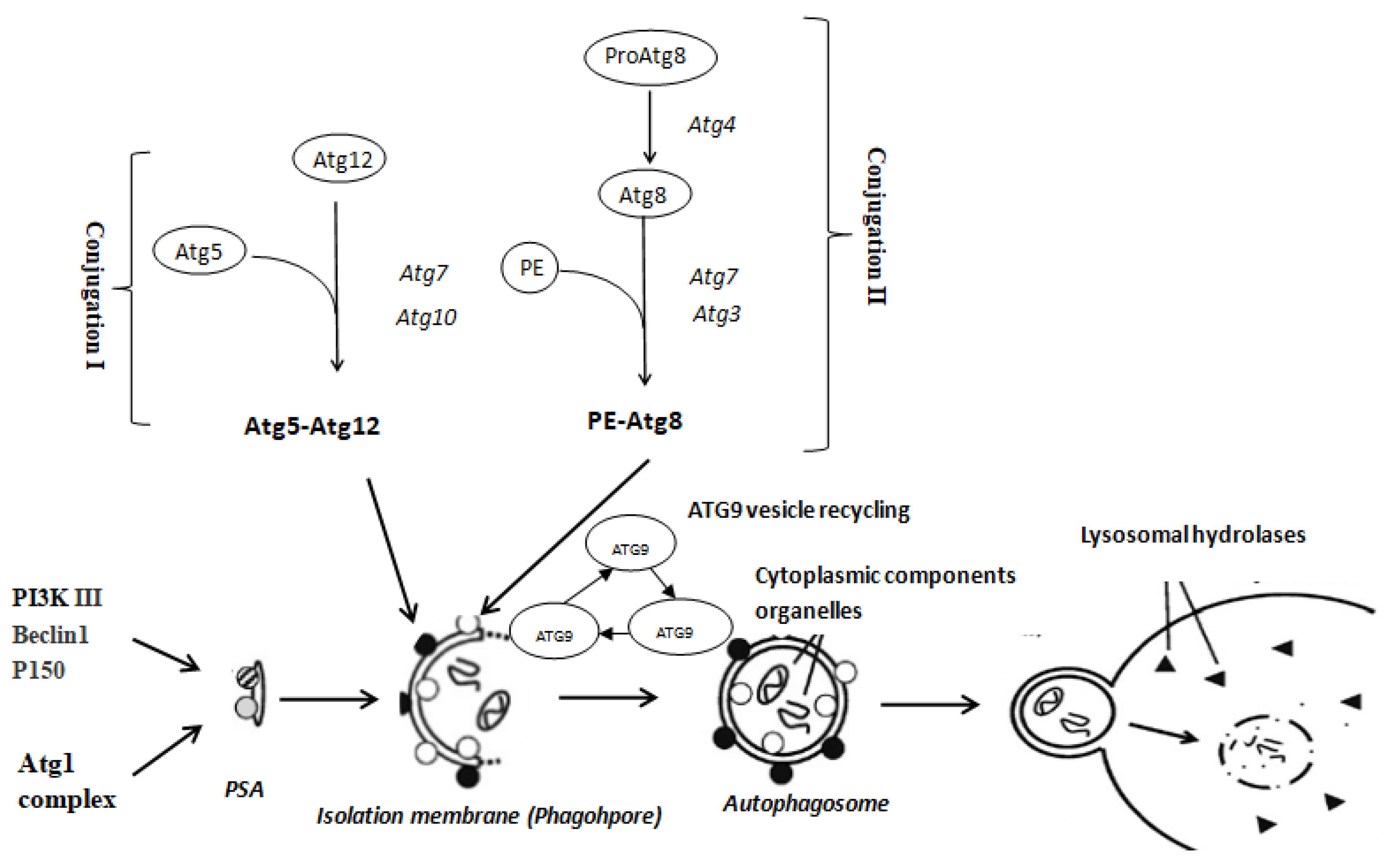
1. Introduction
2. Results
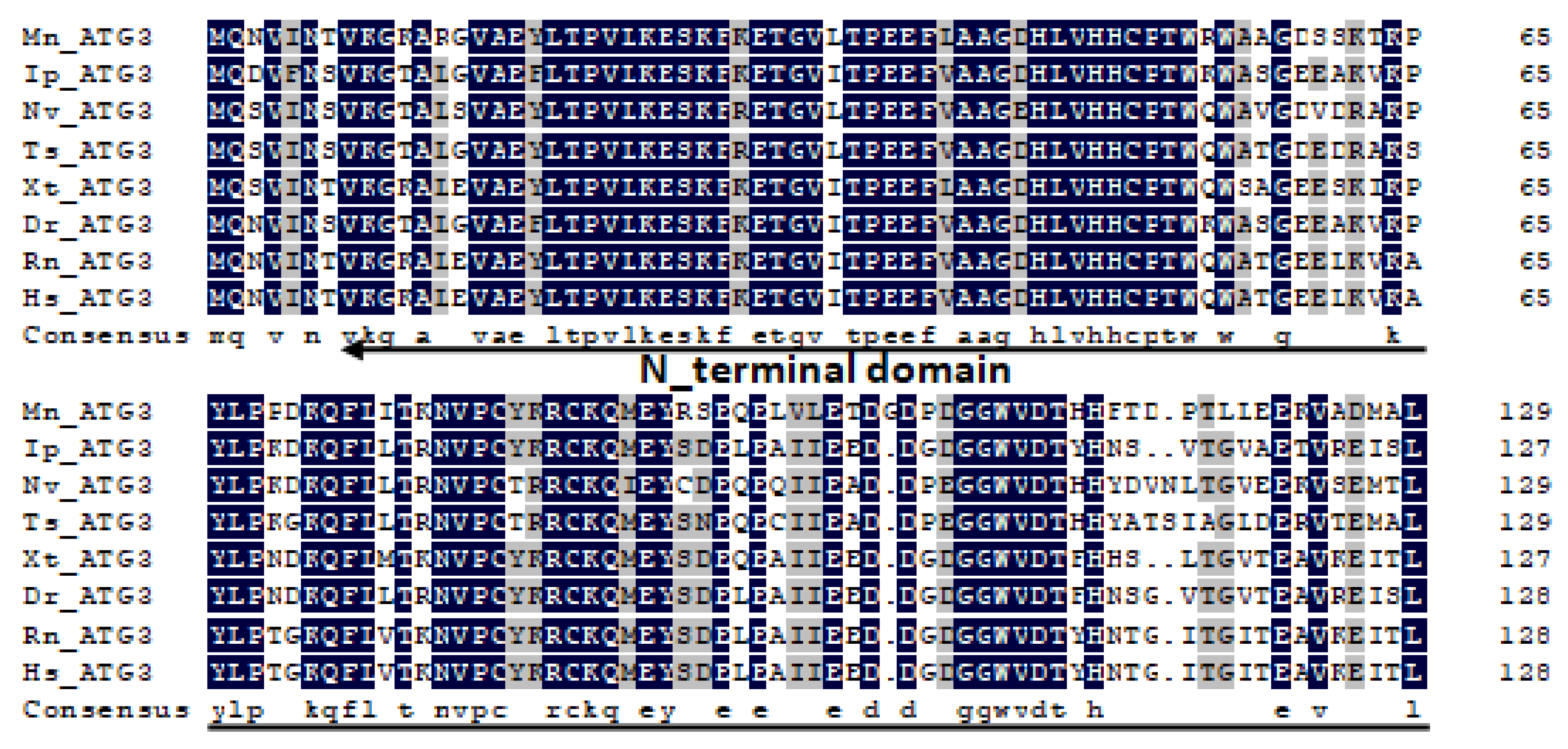
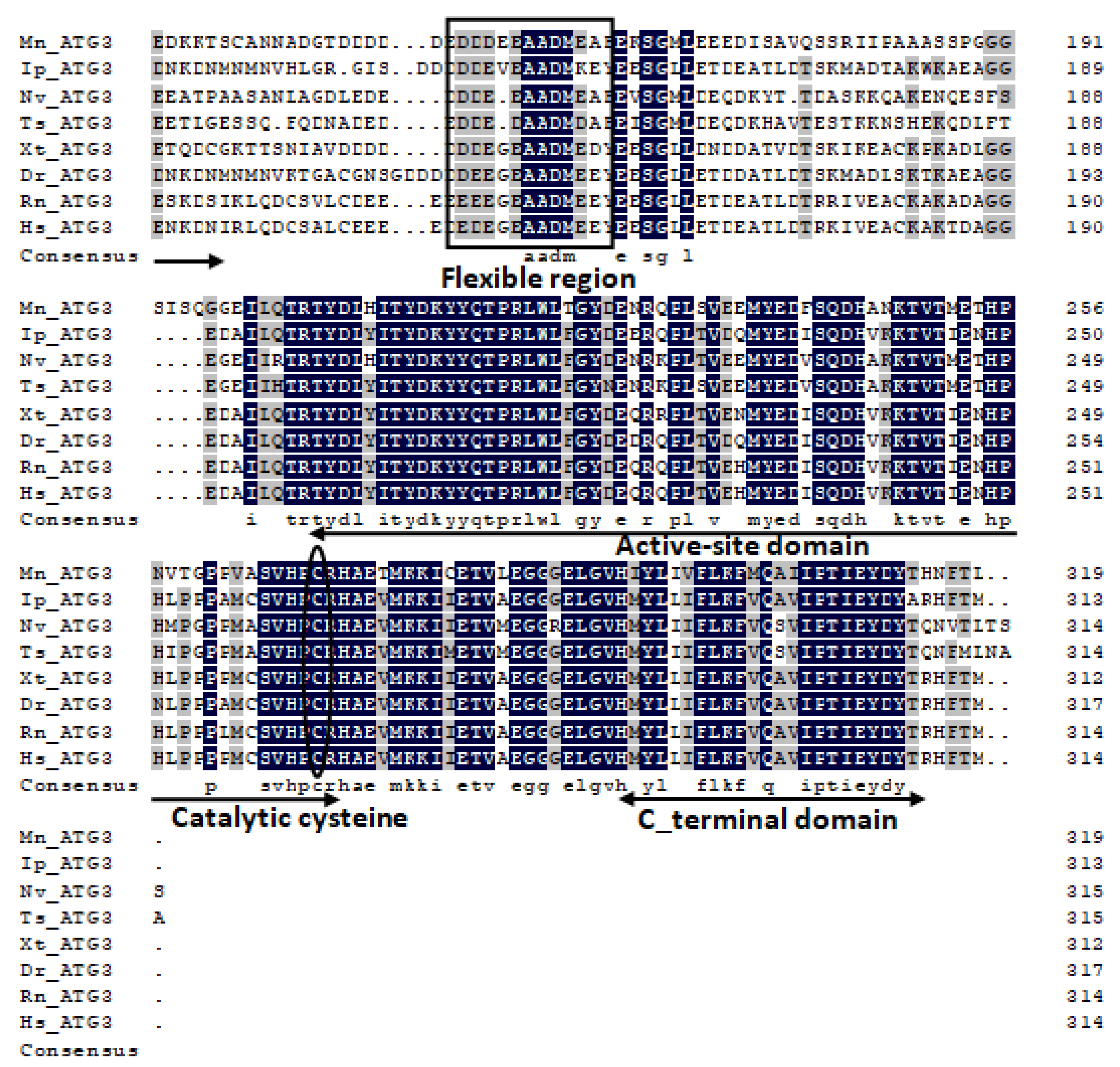
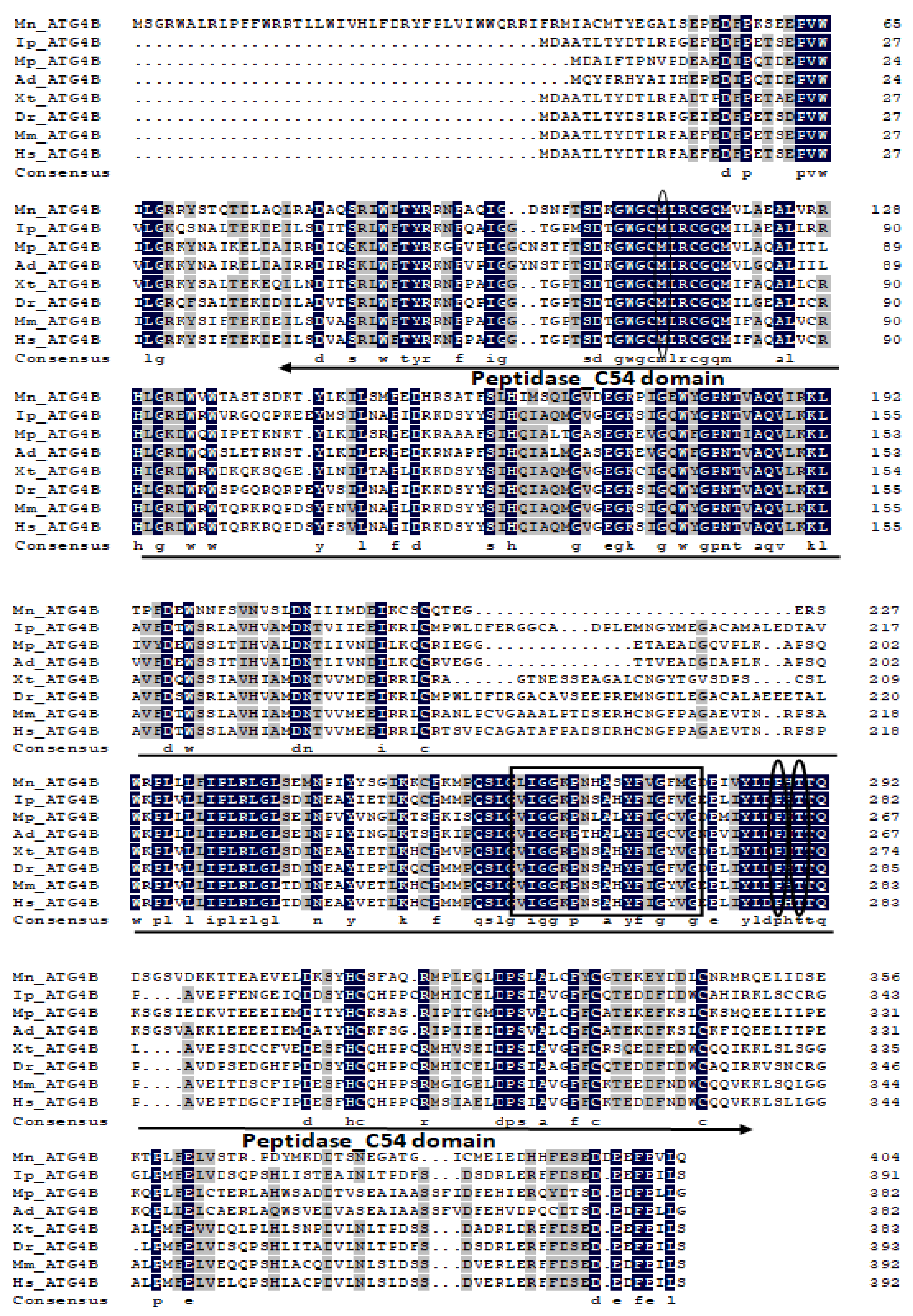
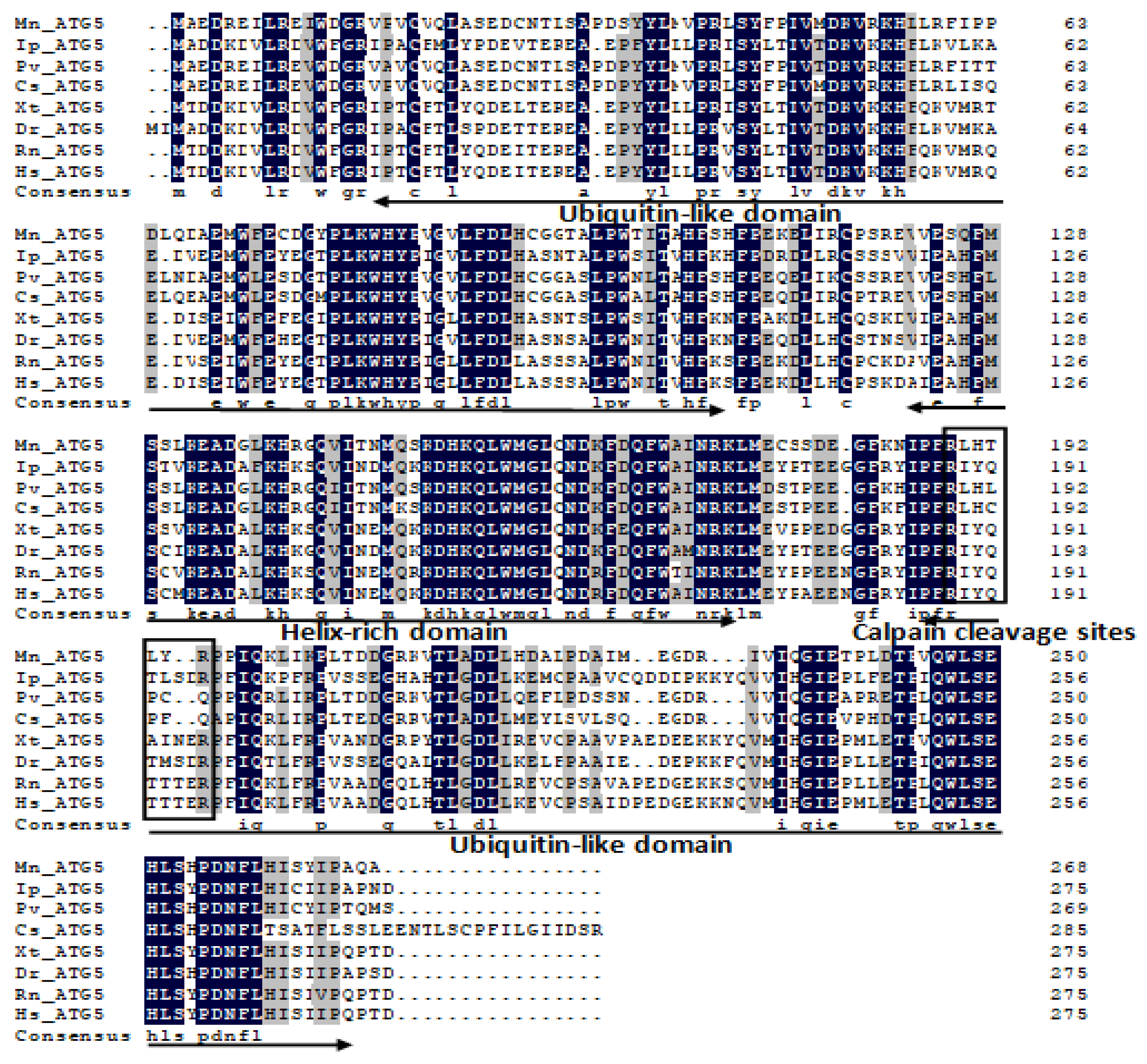
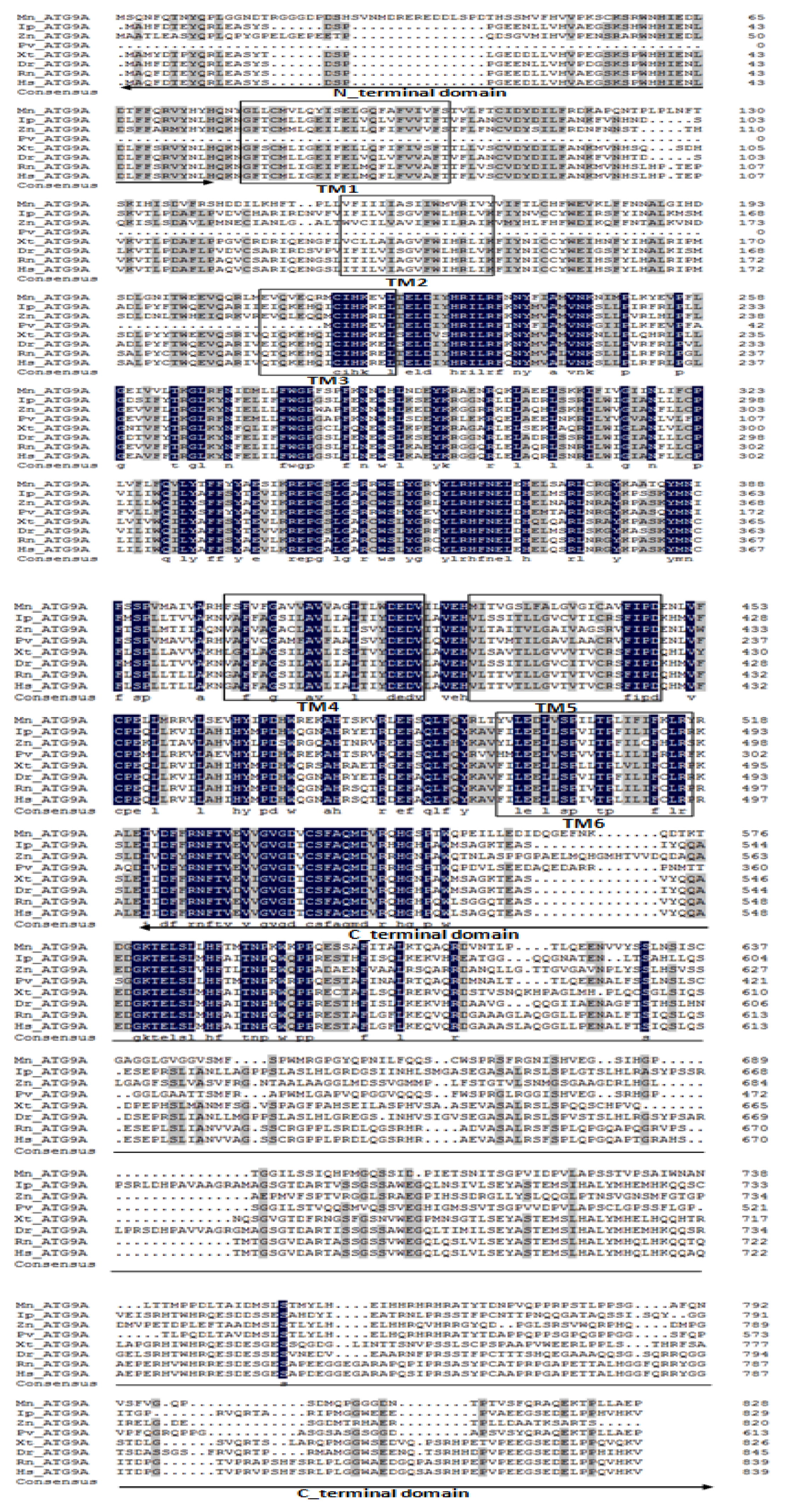
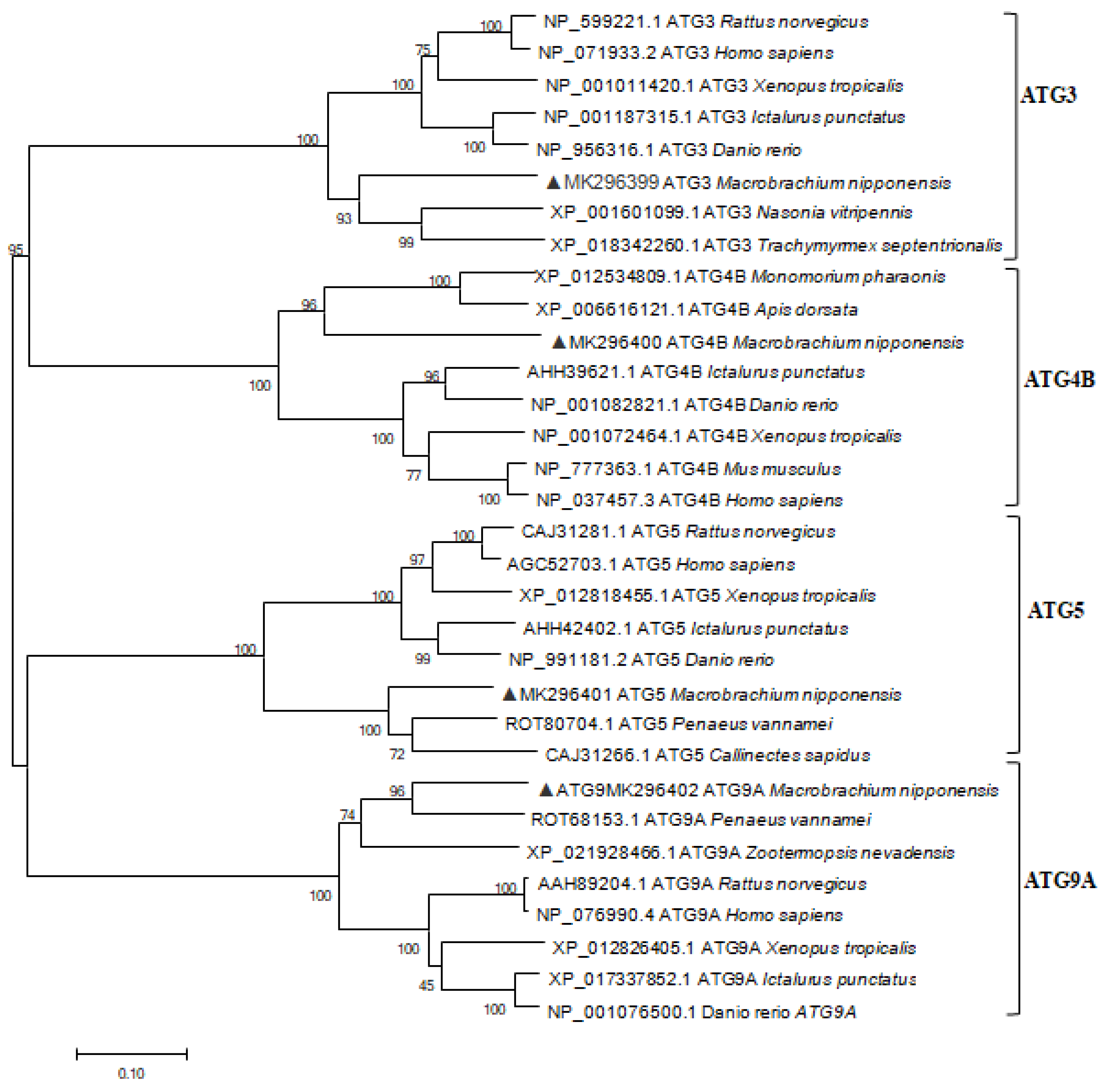
2.1. Characterization and Phylogenetic Analysis of M. nipponense ATG3, ATG4B, ATG5, and ATG9A.
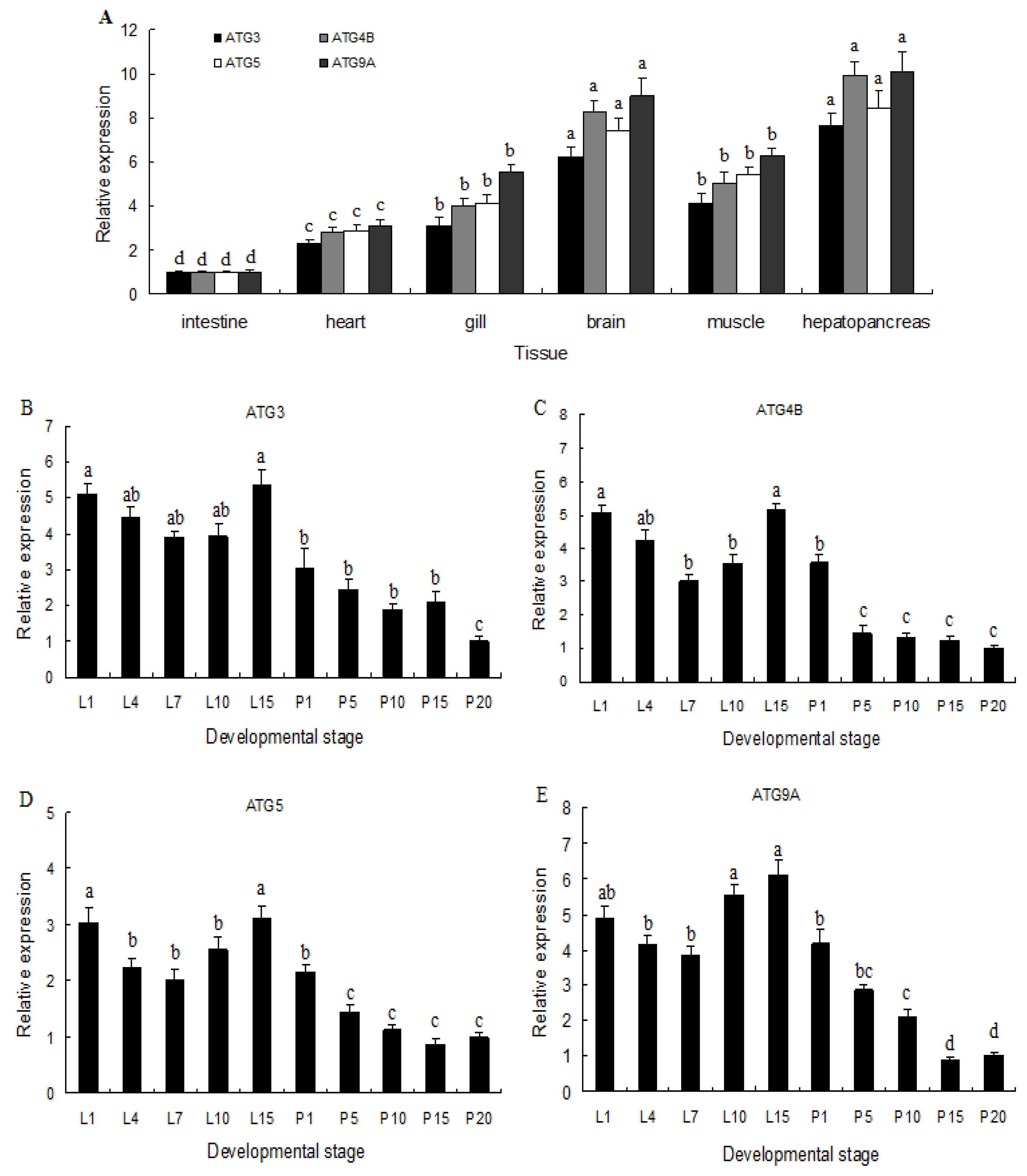
2.2. Expression of ATG3, ATG4B, ATG5, and ATG9A in Different Tissues and Developmental Stages
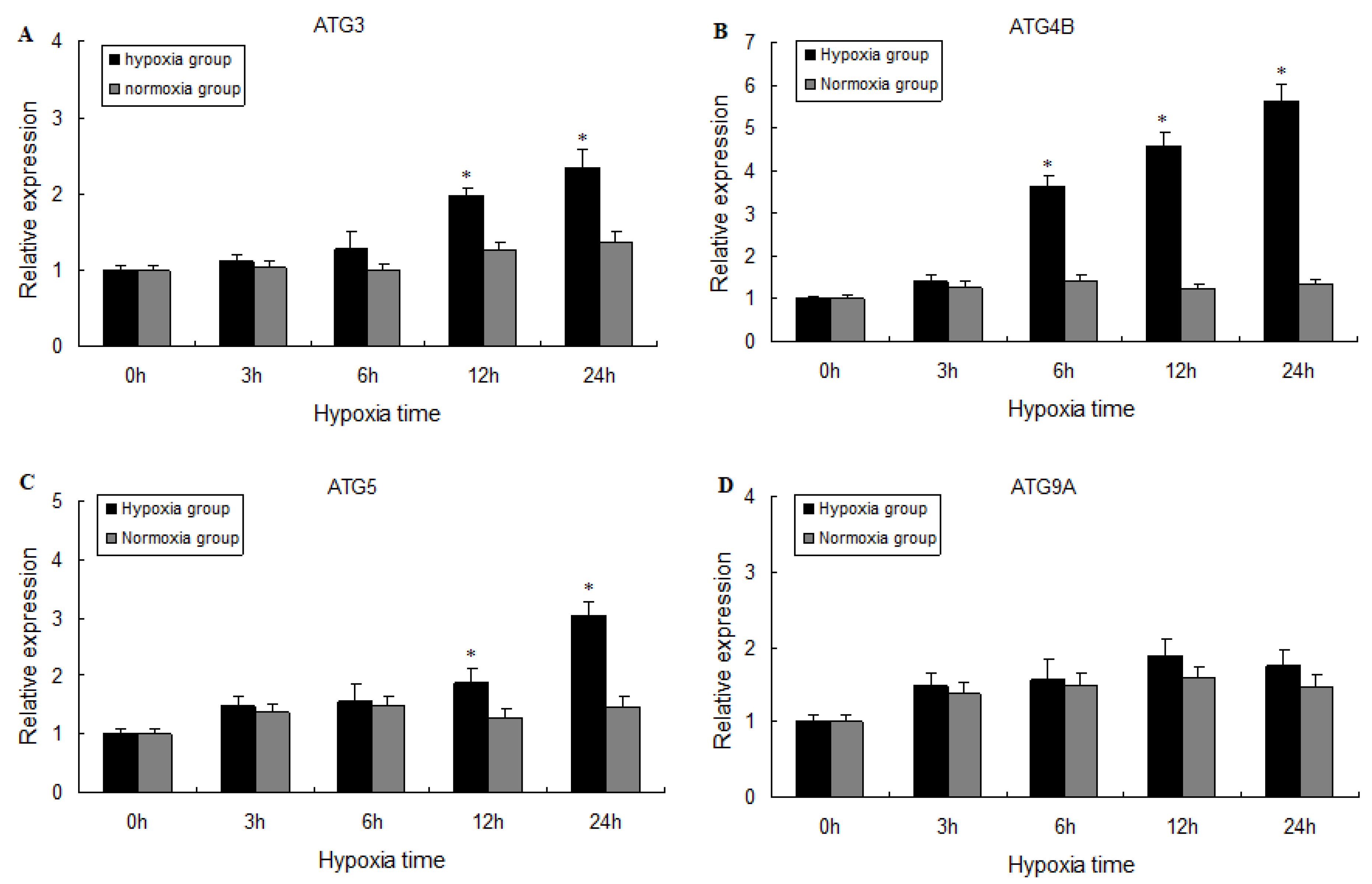
2.3. Expression of ATG3, ATG4B, ATG5, and ATG9A Following Hypoxia
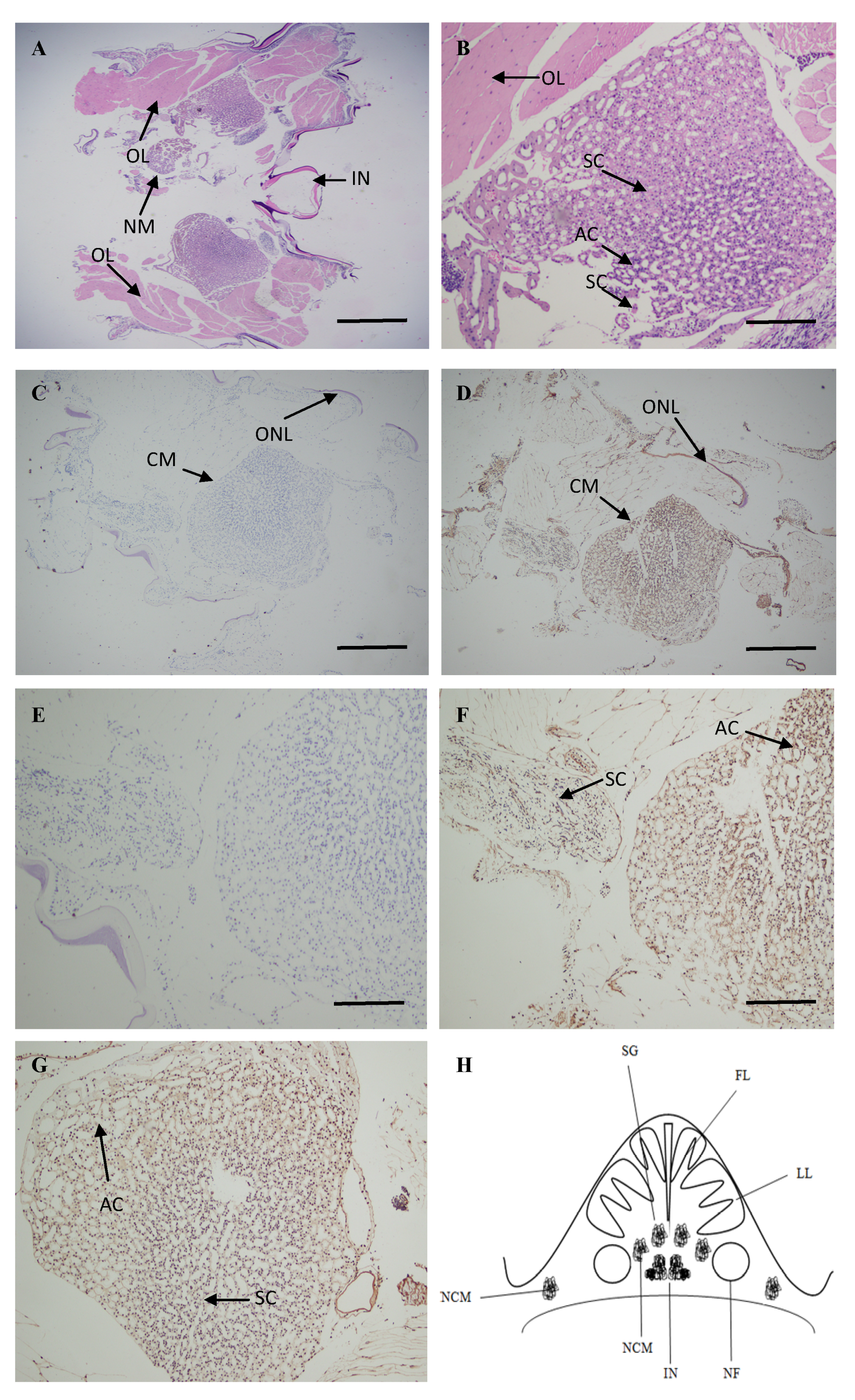
2.4. Localization of ATG4B mRNA in the Brain
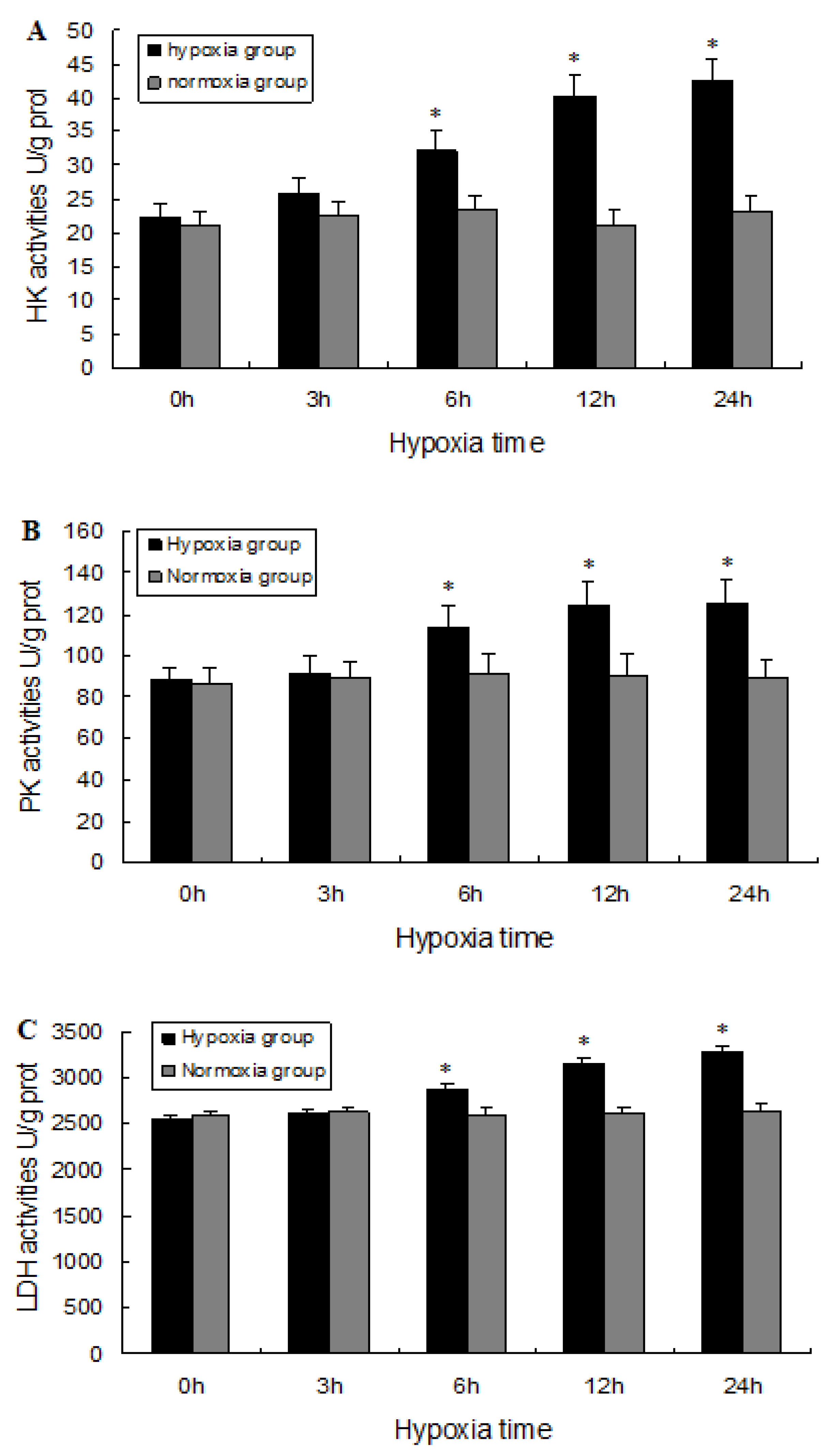
2.5. Biochemical Analysis
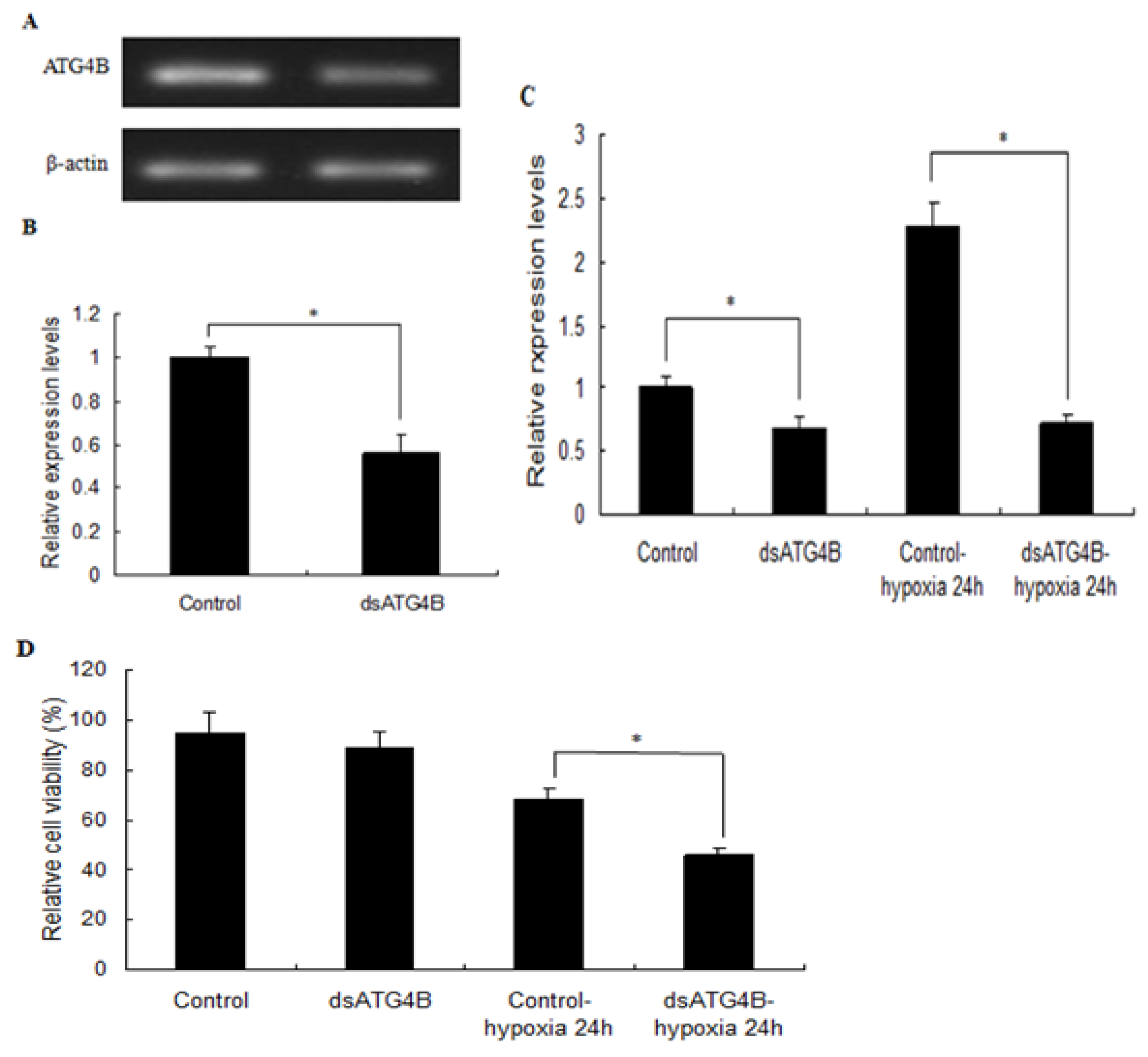
2.6. Effect of ATG4B Gene Silencing on ATG8 Expression and Cell Viability in Brain
2.7. Pull-down Analysis
3. Discussion
4. Materials and Methods
4.1. Experimental Organisms and Hypoxia Treatment
4.2. Full-length cDNA Cloning
4.3. Expression of ATGs in Different Tissues and Developmental Stages, and Under Hypoxia Conditions
4.4. In Situ Hybridization
4.5. Synthesis of Double-Stranded RNA (dsRNA) and Silencing of M. nipponense ATG4B
4.6. Cell Viability Assay and Biochemical Analysis
4.7. Pull-down Analysis
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| qPCR | Quantitative real-time reverse transcription PCR |
| ATG | Autophagy-related genes |
| ORF | Open reading frame |
| ROS | Reactive oxygen species |
| mRNA | Messenger RNA |
| ISH | In situ hybridization |
| dsRNA | Double-stranded RNA |
| cDNA | Complementary DNA |
| FFRC | Freshwater Fisheries Research Center |
| SDS | Sodium dodecyl sulfate |
| PAGE | polyacrylamide gel electrophoresis |
| HK | Hexokinase |
| PK | Pyruvate kinase |
| LDH | Lactate dehydrogenase |
Appendix A
References
- Vaquer-Sunyer, R.; Duarte, C.M. Thresholds of hypoxia for marine biodiversity. Proc. Natl. Acad. Sci. USA 2008, 105, 15452–15457. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Guo, Z.B.; Fu, H.T.; Zhu, J.; Ge, X.P. Integrated metabolomic and transcriptomic analysis of brain energy metabolism in the male Oriental river prawn (Macrobrachium nipponense) in response to hypoxia and reoxygenation. Environ. Pollut. 2018, 243, 1154–1165. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Wu, Y.; Fu, H.T.; Yang, M.; Ge, X.P.; Zhu, J.; Xuan, F.J.; Wu, X.G. Evaluating expression of autophagy-related genes in oriental river prawn Macrobrachium nipponense as potential biomarkers for hypoxia exposure. Ecotox. Environ. Saf. 2019, 171, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Levine, B. Eating oneself and uninvited guests: Autophagy-related pathways in cellular defense. Cell 2005, 120, 159–162. [Google Scholar] [PubMed]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef]
- Hurley, J.H.; Young, L.N. Mechanisms of Autophagy Initiation. Annu. Rev. Biochem. 2017, 86, 225–244. [Google Scholar] [CrossRef]
- Suwansa-ard, S.; Kankuan, W.; Thongbuakaew, T.; Saetan, J.; Kornthong, N.; Kruangkum, T.; Khornchatri, K.; Cummins, S.F.; Isidoro, C.; Sobhon, P. Transcriptomic analysis of the autophagy machinery in crustaceans. BMC Genomics 2016, 17, 587. [Google Scholar] [CrossRef]
- Xie, Z.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell. Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef]
- Gao, D.; Xu, Z.; Kuang, X.D.; Qiao, P.P.; Liu, S.; Zhang, L.; He, P.H.; Jadwiga, W.S.; Wang, Y.N.; Min, W.P. Molecular characterization and expression analysis of the autophagic gene Beclin1 from the purse red common carp (Cyprinus carpio) exposed to cadmium. Comp. Biochem. Physiol. C 2014, 160, 15–22. [Google Scholar]
- Kong, H.J.; Moon, J.Y.; Nam, B.H.; Kim, Y.O.; Kim, W.J.; Lee, J.H.; Kim, K.K.; Kim, B.S.; Yeo, S.Y.; Lee, C.H.; et al. Molecular characterization of the autophagy-related gene Beclin-1 from the olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2011, 31, 189–195. [Google Scholar] [CrossRef]
- Liu, X.H.; Wang, Z.J.; Chen, D.M.; Chen, M.F.; Jin, X.X.; Huang, J.; Zhang, Y.G. Molecular characterization of Beclin1 in rare minnow (Gobiocypris rarus) and its expression after waterborne cadmium exposure. Fish Physiol. Biochem. 2016, 42, 111–123. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, J.; Zhang, Q. Expression pattern and functions of autophagy-related gene atg5 in zebra fish organogenesis. Autophagy 2011, 7, 1514–1527. [Google Scholar] [CrossRef]
- Wei, C.C.; Luo, Z.; Song, Y.F.; Pan, Y.X.; Wu, K.; You, W.J. Identification of autophagy related genes LC3 and ATG4 from yellow catfish Pelteobagrus fulvidraco and their transcriptional responses to waterborne and dietborne zinc exposure. Chemosphere 2017, 175, 228–238. [Google Scholar] [CrossRef]
- Lum, J.J.; Bauer, D.E.; Kong, M.; Harris, M.H.; Li, C.; Lindsten, T.; Thompson, C.B. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005, 120, 237–248. [Google Scholar] [CrossRef]
- Seiliez, I.; Gutierrez, J.; Salmerón, C.; Skiba-Cassy, S.; Chauvin, C.; Dias, K.; Kaushik, S.; Tesseraud, S.; Panserat, S. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. B 2010, 157, 258–266. [Google Scholar] [CrossRef]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Jing, X.G.; Jiang, T.C.; Dai, L.L.; Wang, X.; Cheng, Z. Hypoxia-induced autophagy activation through NF-κB pathway regulates cell proliferation and migration to induce pulmonary vascular remodeling. Exp. Cell Res. 2018, 368, 174–183. [Google Scholar] [CrossRef]
- Marques, A.P.; Rosmaninho-Salgado, J.; Estrada, M.; Cortez, V.; Nobre, R.J.; Cavadas, C. Hypoxia mimetic induces lipid accumulation through mitochondrial dysfunction and stimulates autophagy in murine preadipocyte cell line. Biochimica et Biophysica Acta (BBA)—General Subjects 2017, 1861, 673–682. [Google Scholar] [CrossRef]
- Liu, K.X.; Chen, G.P.; Lin, P.L.; Huang, J.C.; Lin, X.; Qi, J.C.; Lin, C.Q. Detection and analysis of apoptosis- and autophagy-related miRNAs of mouse vascular endothelial cells in chronic intermittent hypoxia model. Life Sci. 2018, 193, 194–199. [Google Scholar] [CrossRef]
- Baskaran, R.; Poornima, P.; Priya, L.B.; Huang, C.Y.; Padma, V.V. Neferine prevents autophagy induced by hypoxia through activation of Akt/mTOR pathway and Nrf2 in muscle cells. Biomed. Pharmacother. 2016, 83, 1407–1413. [Google Scholar] [CrossRef]
- Chaachouay, H.; Fehrenbacher, B.; Toulany, M.; Schaller, M.; Rodemann, H.P. AMPK-independent autophagy promotes radioresistance of human tumor cells under clinical relevant hypoxia in vitro. Radiother. Oncol. 2015, 116, 409–416. [Google Scholar] [CrossRef]
- Sugawara, K.; Suzuki, N.N.; Fujioka, Y.; Mizushima, N.; Ohsumi, Y.; Inagaki, F. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J. Biol. Chem. 2005, 280, 40058–40065. [Google Scholar] [CrossRef]
- Kumanomidou, T.; Mizushima, T.; Komatsu, M.; Suzuki, A.; Tanida, I.; Sou, Y.S.; Ueno, T.; Kominami, E.; Tanaka, K.; Yamane, T. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome forming modifiers. J. Mol. Biol. 2006, 355, 612–618. [Google Scholar] [CrossRef]
- Satoo, K.; Noda, N.N.; Kumeta, H.; Fujioka, Y.; Mizushima, N.; Ohsumi, Y.; Inagaki, F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009, 28, 1341–1350. [Google Scholar] [CrossRef]
- Amar, N.; Lustig, G.; Ichimura, Y.; Ohsumi, Y.; Elazar, Z. Two newly identified sites in the ubiquitin-like protein Atg8 are essential for autophagy. EMBO Rep. 2006, 7, 635–642. [Google Scholar] [CrossRef]
- Nath, S.; Dancourt, J.; Shteyn, V.; Puente, G.; Fong, W.M.; Nag, S.; Bewersdorf, J.; Yamamoto, A.; Antonny, B.; Melia, T.J. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 2014, 16, 415–442. [Google Scholar] [CrossRef]
- Otomo, C.; Metlagel, Z.; Takaesu, G.; Otomo, T. Structure of the human ATG12-ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013, 20, 59–66. [Google Scholar] [CrossRef]
- Rao, Y.; Perna, M.G.; Hofmann, B.; Beier, V.; Wollert, T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat. Commun. 2016, 7, 10338. [Google Scholar] [CrossRef]
- Orsi, A.; Razi, M.; Dooley, H.C.; Robinson, D.; Weston, A.E.; Collinson, L.M.; Tooze, S.A. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 2012, 23, 1860–1873. [Google Scholar] [CrossRef]
- He, H.; Dang, Y.; Dai, F.; Guo, Z.; Wu, J.; She, X.; Pei, Y.; Chen, Y.; Ling, W.; Wu, C.; et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J. Biol. Chem. 2003, 278, 29278–29287. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Dang, Y.; Su, W.; Liu, C.; Ma, H.; Shan, Y.; Pei, Y.; Wan, B.; Guo, J.; Yu, L. Molecular cloning and characterization of rat LC3A and LC3B two novel markers of autophagosome. Biochem. Biophys. Res. Commun. 2006, 339, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Flores, F.J.M.; Gutiérrez, O.A.; Rosario, C.R.; Padilla, C.E.; Alvarez, A.H.; Martínez, V.M. Molecular cloning and characterization of two novel autophagy-related genes belonging to the ATG8 family from the cattle tick Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). Exp. Appl. Acarol. 2014, 64, 533–542. [Google Scholar] [CrossRef]
- Descloux, C.; Ginet, V.; Rummel, C.; Truttmann, A.C.; Puyal, J. Enhanced autophagy contributes to excitotoxic lesions in a rat model of preterm brain injury. Cell Death. Dis. 2018, 9, 853. [Google Scholar] [CrossRef]
- Wilkinson, S.; O’Prey, J.; Fricker, M.; Ryan, K.M. Hypoxia-selective macroautophagy and cell survival signaled by autocrine PDGFR activity. Genes Dev. 2009, 23, 1283–1288. [Google Scholar] [CrossRef]
- Balduini, W.; Carloni, S.; Buonocore, G. Autophagy in hypoxia-ischemia induced brain injury: Evidence and speculations. Autophagy 2009, 5, 221–223. [Google Scholar] [CrossRef]
- Abdul-Rahim, S.A.; Dirkse, A.; Oudin, A.; Schuster, A.; Bohler, J.; Barthelemy, V.; Muller, A.; Vallar, L.; Janji, B.; Golebiewska, A.; et al. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. Br. J. Cancer 2017, 117, 813–825. [Google Scholar] [CrossRef]
- Madeo, F.; Eisenberg, T.; Kroemer, G. Autophagy for the avoidance of neurodegeneration. Genes Dev. 2009, 23, 2253–2259. [Google Scholar] [CrossRef]
- Bai, H.; Qiao, H.; Li, F.J.; Fu, H.T.; Jiang, S.F.; Zhang, W.Y.; Yan, Y.D.; Xiong, Y.W.; Sun, S.M.; Jin, B.; et al. Molecular and functional characterization of the vitellogenin receptor in oriental river prawn, Macrobrachium nipponense. Comp. Biochem. Physiol. A 2016, 194, 45–55. [Google Scholar] [CrossRef]
- Zauner, A.; Daugherty, W.P.; Bullok, M.R.; Warner, D.S. Brain oxygenation and energy metabolism. Neurosurgery 2002, 51, 289–301. [Google Scholar]
- Lim, Y.; Cho, H.C.; Kim, E.K. Brain metabolism as a modulator of autophagy in neurodegeneration. Brain Res. 2016, 1649, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Abreu, S.; Kriegenburg, F.; Gómez-Sánchez, R.; Mari, M.; Sánchez-Wandelmer, J.; Skytte Rasmussen, M.; Soares, G.R.; Zens, B.; Schuschnig, M.; Hardenberg, R.; et al. Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation. EMBO Rep. 2017, 18, 765–780. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.M.; Xuan, F.J.; Ge, X.P.; Fu, H.; Zhu, J.; Zhang, S. Identification of differentially expressed genes in hepatopancreas of oriental river prawn, Macrobrachium nipponense exposed to environmental hypoxia. Gene 2014, 534, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, Q.; Chen, H.; Zhu, X.; Cui, Z.; Qiu, G. The morphological and histological observation of embryonic development in the oriental river prawn Macrobrachium nipponense. J. Shanghai Ocean Univ. 2012, 21, 33–40. [Google Scholar]
- Sun, S.M.; Gu, Z.M.; Fu, H.T.; Zhu, J.; Ge, X.P.; Xuan, F.J. Molecular cloning, characterization, and expression analysis of p53 from the oriental river prawn, Macrobrachium nipponense, in response to hypoxia. Fish Shellfish Immunol. 2016, 54, 68–76. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−△△CtCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, F.; Qiao, H.; Fu, H.T.; Zhang, W.Y.; Jin, S.B.; Jiang, S.F.; Gong, Y.S.; Xiong, Y.W.; Wu, Y.; Hu, Y.N.; et al. Identification and characterization of opsin gene and its role in ovarian maturation in the oriental river prawn Macrobrachium nipponense. Comp. Biochem. Physiol. B 2018, 218, 1–12. [Google Scholar] [CrossRef]
- Sun, S.M.; Xuan, F.J.; Fu, H.T.; Ge, X.P.; Zhu, J.; Qiao, H.; Jin, S.B.; Zhang, W.Y. Molecular characterization and mRNA expression of hypoxia inducible factor-1 and cognate inhibiting factor in Macrobrachium nipponense in response to hypoxia. Comp. Biochem. Physiol. B 2016, 196–197, 48–56. [Google Scholar] [CrossRef]
- Sun, S.M.; Xuan, F.J.; Fu, H.T.; Zhu, J.; Ge, X.P. Molecular cloning and functional characterization of a hexokinase from the oriental river prawn Macrobrachium nipponense in response to hypoxia. Int. J. Mol. Sci. 2017, 18, 1256. [Google Scholar] [CrossRef]
- Duan, Y.F.; Zhang, J.S.; Dong, H.B.; Wang, Y.; Liu, Q.; Li, H. Effect of desiccation and resubmersion on the oxidative stress response of the kuruma shrimp Marsupenaeus japonicas. Fish Shellfish Immunol. 2016, 49, 91–99. [Google Scholar] [CrossRef]
- Bradford, M.M. A refined and sensitive method for the quantification of microgram quantities of protein utilizusing the principle of protein dye-binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, W.; Ren, Q. Function of gC1qR in innate immunity of Chinese mitten crab, Eriocheir sinensis. Dev. Comp. Immunol. 2016, 61, 34–41. [Google Scholar] [CrossRef] [PubMed]
| Primer | Primer Sequence (5′ to 3′) |
|---|---|
| ATG3-R1 (5′ RACE out primer) | TAAAACACCAGTCTCC |
| ATG3-R2 (5′ RACE in primer) | GGACTCCTTGAGGACCGG |
| ATG3-R3 (5′ RACE in primer) | CTACACCTCGTGCTTTGC |
| ATG3-F1 (3′ RACE out primer) | GGAAACCCACCCAAATGTGACAGG |
| ATG3-F2 (3′ RACE in primer) | CTGTGTTAGAGGGTGGTGGTGAGC |
| ATG4B-R1 (5′ RACE out primer) | TCCGACGATAAGTGAG |
| ATG4B-R2 (5′ RACE out primer) | CTGTGCAAGATCTGTCTG |
| ATG4B-R3 (5′ RACE out primer) | TTGAATAACGCCGTCCTA |
| ATG4B-F1 (3′ RACE out primer) | TGCTCTTTTGCTCAACGGATGCCT |
| ATG4B-F2 (3′ RACE in primer) | AACAATTGGACCCCTCTTTGGCAC |
| ATG5-R1 (5′ RACE out primer) | TATGAATCTGGAGCAC |
| ATG5-R2 (5′ RACE out primer) | AGCTGGACACAGACTGGG |
| ATG5-R3 (5′ RACE out primer) | GACCATCCCATATCTCGC |
| ATG5-F1 (3′ RACE out primer) | TAATGGAAGGTGACCGAATTGTGA |
| ATG5-F2 (3′ RACE in primer) | CCCTGGATACACCAGTACAATGGC |
| ATG9A-R1 (5′ RACE out primer) | TAGAGTGCGTGTCGGG |
| ATG9A-R2 (5′ RACE out primer) | TCCTCCCTCTCTCTGTCC |
| ATG9A-R3 (5′ RACE out primer) | CCCTGGTGTCATTCCCAC |
| ATG9A-F1 (3′ RACE out primer) | GCTCCAAGTTCCACGGTGCCATCA |
| ATG9A-F2 (3′ RACE in primer) | GCAAATCTCACAACGATGCCCCCA |
| ATG3-F (Real-Time primer) | ACCCAAATGTGACAGGTCCT |
| ATG3-R (Real-Time primer) | TCACCACCACCCTCTAACAC |
| ATG4B-F (Real-Time primer) | ACTTCAGACAAGGGATGGGG |
| ATG4B-R (Real-Time primer) | AAGCTGTCCATACCCAGTCC |
| ATG5-F (Real-Time primer) | TGGTTCCAAGGCTGTCGTAT |
| ATG5-R (Real-Time primer) | AACCACATTTCTGCGTCCTG |
| ATG9A-F (Real-Time primer) | CCTTGGTGTGGGAATATGCG |
| ATG9A-R (Real-Time primer) | AGCTTTCTCTCGCCAGTGAT |
| β-Actin-F (Real-Time primer) | TATGCACTTCCTCATGCCATC |
| β-Actin-R (Real-Time primer) | AGGAGGCGGCAGTGGTCAT |
| dsATG4B-F (RNAi) | TAATACGACTCACTATAGGGGACAGATGGTGCTTGCAGAA |
| dsATG4B-R (RNAi) | TAATACGACTCACTATAGGGTTCATCCCCCATAAAACCAA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Wu, Y.; Fu, H.; Ge, X.; You, H.; Wu, X. Identification and Characterization of Four Autophagy-Related Genes That Are Expressed in Response to Hypoxia in the Brain of the Oriental River Prawn (Macrobrachium nipponense). Int. J. Mol. Sci. 2019, 20, 1856. https://doi.org/10.3390/ijms20081856
Sun S, Wu Y, Fu H, Ge X, You H, Wu X. Identification and Characterization of Four Autophagy-Related Genes That Are Expressed in Response to Hypoxia in the Brain of the Oriental River Prawn (Macrobrachium nipponense). International Journal of Molecular Sciences. 2019; 20(8):1856. https://doi.org/10.3390/ijms20081856
Chicago/Turabian StyleSun, Shengming, Ying Wu, Hongtuo Fu, Xianping Ge, Hongzheng You, and Xugan Wu. 2019. "Identification and Characterization of Four Autophagy-Related Genes That Are Expressed in Response to Hypoxia in the Brain of the Oriental River Prawn (Macrobrachium nipponense)" International Journal of Molecular Sciences 20, no. 8: 1856. https://doi.org/10.3390/ijms20081856
APA StyleSun, S., Wu, Y., Fu, H., Ge, X., You, H., & Wu, X. (2019). Identification and Characterization of Four Autophagy-Related Genes That Are Expressed in Response to Hypoxia in the Brain of the Oriental River Prawn (Macrobrachium nipponense). International Journal of Molecular Sciences, 20(8), 1856. https://doi.org/10.3390/ijms20081856







