Abstract
Breast cancer is the most common malignancy in women worldwide and can be categorized into several subtypes according to histopathological parameters or genomic signatures. Such heterogeneity of breast cancer can arise from the reactivation of mammary stem cells in situ during tumorigenesis. Moreover, different breast cancer subtypes exhibit varieties of cancer incidence, therapeutic response, and patient prognosis, suggesting that a specific therapeutic protocol is required for each breast cancer subtype. Recent studies using molecular and cellular assays identified a link between specific genetic/epigenetic alterations and distinct cells of origin of breast cancer subtypes. These alterations include oncogenes, tumor suppressor genes, and cell-lineage determinants, which can induce cell reprogramming (dedifferentiation and transdifferentiation) among two lineage-committed mammary epithelial cells, namely basal and luminal cells. The interconversion of cell states through cell reprogramming into the intermediates of mammary stem cells can give rise to heterogeneous breast cancers that complicate effective therapies of breast cancer. A better understanding of mechanisms underlying cell reprogramming in breast cancer can help in not only elucidating tumorigenesis but also developing therapeutics for breast cancer. This review introduces recent findings on cancer gene-mediated cell reprogramming in breast cancer and discusses the therapeutic potential of targeting cell reprogramming.
1. Introduction
Embryonic stem cells (ESCs) are pluripotent stem cells that can differentiate into three germ layers: endoderm, mesoderm, and ectoderm. After the differentiation of ESCs into different germ layers, the resulting daughter stem/progenitor cells and terminally differentiated cells can be distinguished based on the differential expression of lineage- and tissue-specific markers. For example, GATA4, FOXA2 (HNF-3β), HNF-4A, SOX17, and alpha fetoprotein are specifically expressed in the endoderm; NES (nestin), SOX1, MAP2, and GFAP are expressed in the ectoderm; and TBXT (Brachyury), KDR (FLK1), VIM (vimentin), and FN1 (fibronectin) are expressed in the mesoderm. Many of these marker genes encode transcription factors (TFs) that are critical for cell fate specification. After lineage commitment, stem/progenitor cells usually undergo downward, lineage-specific differentiation and cannot go back to the stem-cell state. However, Takahashi and Yamanaka [1] introduced a cell reprogramming method that utilizes a combination of four TFs, namely OCT4, SOX2, KLF4, and MYC (OSKM), to convert differentiated fibroblasts back to an ESC-like state; the resulting cells are called induced pluripotent stem cells (iPSCs). This cell reprogramming method was proven to be successful in numerous cell types with various differentiation statuses and was applied in many research fields, including cancer research. For example, Corominas-Faja et al. [2] used OSKM to reprogram the MCF-7 human breast cancer cells into SOX2-overexpressing cancer stem cell (CSC)-like cells that exhibit activated mammalian target of rapamycin (mTOR) kinase activity. In addition, OSKM could reprogram MCF-10A cells, a non-tumorigenic human mammary epithelial cell line, into CSC-like cells, which express the stem-cell marker CD44 and feature enhanced malignancy [3].
In addition to the OSKM-mediated cell reprogramming of differentiated cells into iPSCs, many studies used single or a few lineage-specific factors, usually TFs, to directly convert one cell type into another. Such lineage switch is a process of direct reprogramming (DR) or transdifferentiation [4]. For example, Tani et al. [5] reported that a combination of three cardiac-specific TFs (GATA4, MEF2C, and TBX5) can directly reprogram fibroblasts into cardiomyocytes and that additional factors, such as microRNAs (miRNAs), cytokines, and epigenetic factors, can modulate this cardiac DR. In the cases of mammary and breast cancer cells, overexpression of GATA3 or NOTCH1 in mammary basal cells (BCs) can convert BCs to luminal cells (LCs) [6,7]. By contrast, forced expression of TP63 reprograms LCs to BCs [8,9]. Such interconversion between mammary BCs and LCs demonstrates the cell plasticity of both epithelial lineages in the mammary gland. Because the normal development process and tumorigenesis of the mammary gland epithelium share similar signal pathways [10,11,12,13], study of mechanisms underlying lineage conversion or DR can not only illustrate the control of mammary gland development but also elucidate the tumorigenesis of breast cancer. Lineage interconversion may contribute to tumor heterogeneity and increase the number of breast cancer subtypes under oxidative and therapeutic stresses, which can complicate the curative therapy of advanced cancer [4,12,13]. Thus, a better understanding of cell reprogramming mechanisms in breast cancer can be helpful to unveil the potential therapeutic strategy to target different subtypes of breast cancer.
2. Epithelial Cell Lineages in the Mammary Gland and Subtypes of Breast Cancer
In mouse models, multipotent mammary stem cells (MaSCs) that express both basal (e.g., Trp63, cytokeratin 5 (Krt5), and Krt14) and luminal (e.g., Notch1 and Krt8) signature genes can be found during embryonic development until day 14.5 (E14.5). After E17, only unipotent luminal and basal progenitors (LPs and BPs, respectively) are present in the mammary epithelium, which support tissue homeostasis throughout adulthood [7,8,14]. The mammary gland epithelium contains two major cell types, namely LCs (ductal and secretory alveolar cells) and BCs (myoepithelial cells); both these cell types are generated and maintained in adult mammary tissues by their own long-lived and unipotent LPs and BPs, respectively [15]. Another study found that, in adult mice, a small fraction of BPs can differentiate into both basal and luminal lineages, suggesting that a few BPs are bipotent MaSCs [16]. However, studies showed that, at least in postnatal mice, the homeostasis of the mammary gland epithelium is maintained by lineage-restricted unipotent LPs and BPs [17,18]. Nevertheless, both differentiated basal- and luminal-lineage mammary cells preserve the capacity to dedifferentiate into an intermediate bipotent MaSC state [19] and then develop into BCs or LCs [7,8] (Figure 1).
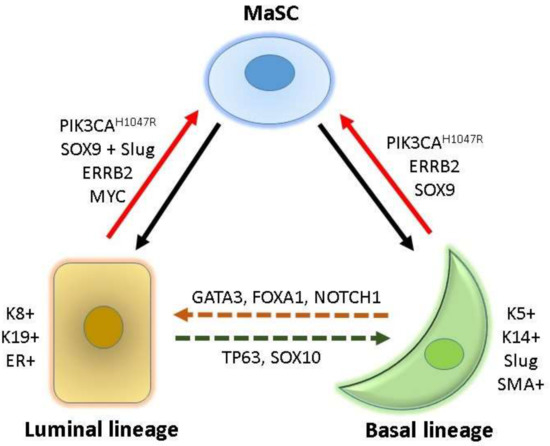
Figure 1.
Cell hierarchy and cell reprogramming in the mammary tissue. Bipotent mammary stem cells (MaSC) can differentiate into luminal and basal lineages (indicated by solid back lines). Cytokeratin (K) 8, K19, and estrogen receptor (ER) are luminal lineage-specific markers, and K5, K14, Slug, and smooth muscle actin (SMA) are basal lineage-specific markers. Overexpression of lineage-specific factors may induce interconversion between luminal and basal lineages (dotted lines). Oncogene activation can reprogram lineage-committed basal and luminal cells into MaSC (red lines), which can give rise to heterogeneous breast cancers.
TFs are critical fate-determinants for the mammary development of basal and luminal lineages [20,21,22]. For the luminal differentiation from bipotent MaSCs, NOTCH1, ELF5, and EHF contribute to the development of LPs [7,9,22,23] and GATA3, FOXA1, and ESR1 (estrogen receptor alpha) are critical for further differentiation into mature LCs [6,22,24]. TP63, NF1, and SNAI2 (Slug) are required for the differentiation of the basal lineage [9,22]. In addition to TFs, other cell surface markers and lineage-specific molecules are useful for the identification and purification of various lineage-restricted cells from mammary tissues. For example, smooth muscle actin, KRT5, KRT14, and vimentin are specifically expressed in the basal lineage, and ESR1, progesterone receptor (PR), E-cadherin (CDH1), EPCAM, KRT8, KRT18, and KRT19 are predominant in the luminal lineage [20,21]. These lineage-specific makers and TFs are commonly used to classify and trace the cell of origin of various mammary epithelial and breast cancer cells (Figure 1).
Breast cancers are organized and constituted of heterogeneous mammary cell types in a hierarchy pattern. According to the histological expression of ERα, PR, and HER2/ERRB2, breast cancer patients are divided into three therapeutic groups: ER-positive, HER2, and triple-negative breast cancer (TNBC) who receive hormone therapy, anti-HER2 target therapy, and chemotherapy, respectively. According to their intrinsic transcriptional profiles, breast cancers can also be classified into five subtypes: luminal A, luminal B, HER2, TNBC/basal-like, and normal like [25,26,27,28]. Additional subtypes, such as claudin low, were identified through further analyses with the aid of more breast cancer cohorts [25,26,27,28]. These heterogeneous breast cancers exhibit different incidences, therapeutic responses, and prognoses [29]. However, the connection between breast cancer subtypes and the cell of origin during tumorigenesis is not fully elucidated. Several recent studies revealed that heterogeneous breast cancers can arise from different mammary cells through lineage interconversion and cell reprogramming; some examples and their therapeutic implications are given below.
3. Cancer Gene-Mediated Cell Reprogramming in Breast Cancer
Recent studies demonstrated that either the activation of an oncogene or the inactivation of a tumor suppressor gene plays a critical role in cell reprogramming of lineage-restricted BCs or LCs into bipotent MaSCs (Figure 1), which may serve as tumor-initiating cells (TICs) or CSCs and confer heterogeneous breast cancers with diverse cells of origin (Table 1) [12,13]. Understanding the actions of these cell reprogramming factors in different mammary epithelial cells can facilitate the elucidation of mechanisms underlying the tumorigenesis of various breast cancer subtypes and the development of specific therapeutics for heterogeneous breast cancers.

Table 1.
Cell reprogramming factors in the tumorigenesis of breast cancer.
3.1. PIK3CA
PIK3CA is one of the most frequently mutated genes in human breast cancer with distinct subtypes [30], implying the contribution of the PIK3CA mutation to breast cancer heterogeneity. The most recurrent activating mutation of PIK3CA in breast cancers is H1047R, which causes constitutive activation of the phosphatidylinositol 3-kinase (PI3K) pathway and proliferation of heterogeneous mammary epithelial cells [31,32,33]. In 2015, two independent studies used lineage tracing mouse models to show that the oncogenic expression of Pik3caH1047R in lineage-committed BCs or LCs can reprogram these differentiated cells into MaSCs and give rise to heterogeneous types of breast cancers [34,35]. The expression of Pik3caH1047R can also interconvert BCs into LCs and LCs into BCs. Transcriptional profiling revealed that Pik3caH1047R clearly rewires gene expression signatures in these cells, by which the switches of cell-identity signatures are correlated with the cells of origin and tumor types developed. These results demonstrate that the PIK3CAH1047R mutation can initiate heterogeneous breast cancers by triggering the MaSC genetic program in differentiated BCs and LCs, thus providing new insights into the development of therapeutic strategies for PI3K-related breast cancers.
3.2. MYC
MYC is a member of OSKM acting on cell reprogramming [1] and is also a frequently deregulated oncogene in breast cancers, particularly in the basal-like subtype [30,36,37]. Poli et al. [38] reported that MYC acts as an oncogenic reprogramming factor to convert TERT-immortalized human mammary epithelial cells and luminal breast cancer cells into the basal/stem cell-like state and gives rise to TICs favoring the onset of mammary tumorigenesis. Overexpression of MYC induces alternative transcriptional and epigenetic programs, through which luminal lineage-specific enhancers and TFs, such as GATA3 and ESR1, are suppressed. By contrast, MYC promotes the activation of de novo enhancers, which support the activation of oncogenic pathways (e.g., WNT and EGFR) and the onset of a stem cell-like state, represented by enhanced mammosphere formation [38]. Therefore, the luminal differentiation program is suppressed and mammary tumors emerge. These data suggest that MYC induces mammary tumorigenesis through cell reprogramming, which can be attributed to MYC-mediated activation of oncogenic enhancers and, meanwhile, repression of luminal fate-specific enhancers. Moreover, a mouse model with overexpression of Myc together with Pik3caH1047R endows long-term tumorigenicity and enhanced metastatic potential. Furthermore, metadata analyses indicate that oncogenic pathways activated by MYC-regulated enhancers are associated with a poor prognosis of patients with basal-like breast cancers [38]. Therefore, targeting MYC and MYC-regulated enhancers or oncogenic pathways represents potential therapeutic strategies for basal-like breast cancers.
3.3. ERBB2 and Polyomavirus Middle T
Basal-like breast cancers can arise from LCs with the oncogenic activation of ERBB2 signaling or the expression of the polyomavirus middle T (PyMT) antigen [39]. Hein and colleagues [39] used the LC-restricted avian retroviral vector and in vivo tracing experiments to show that the lineage-specific expression of ERBB2 or PyMT in committed LCs can induce these cells to generate mammary tumors derived from additional cellular lineages, revealing that either ERBB2 or PyMT induces the plasticity of LCs during tumorigenesis. These results support the notion that the origin of breast cancer heterogeneity can arise from the activation of oncogenic signaling in the committed luminal lineage in addition to from multipotent stem cells.
3.4. TP53 and BRCA1
Similar to gain of function in oncogenes, the loss of function in tumor-suppressor genes may modulate the consequence of cell reprogramming in breast cancer. TP53 is a roadblock for OSKM-induced cell reprogramming, and inactivation of TP53 significantly increases the reprogramming efficiency of differentiated cells into iPSCs [40,41,42,43]. Inactivation of the TP53 pathway is frequently observed in breast cancer, especially the basal-like TNBC [30], and is correlated with gene expression signatures of ESCs and MaSCs [44]. Zhang et al. [45] used a murine model to identify a Lin−CD29HCD24H subset of TICs that can generate heterogeneous breast cancers in the absence of Trp53. The Lin−CD29HCD24H subpopulation may have arisen from a bipotent MaSC [45]. A recent study reported that MYC is a target of TP53, whose inactivation leads to MYC activation and increases mammary cell plasticity [46]. These studies demonstrate the important role of TP53 in guarding the identity of mammary cells and preventing the reprogramming of differentiated mammary cells into MaSCs and subsequent tumorigenesis.
BRCA1 is a tumor-suppressor gene involved in transcriptional regulation and DNA repair [47]. Breast cancer patients with hereditary BRCA1 mutations usually develop basal-like tumors. Deleting the Brca1 gene in mouse luminal lineage cells generates tumors that are similar to human BRCA1 breast cancer and sporadic basal-like breast cancers according to histological and transcriptional profiling analyses. By contrast, tumors derived from Brca1-defective BCs are histologically unlike human BRCA1 or sporadic basal-like breast cancers [48]. These results indicate that basal-like breast cancer can arise from the luminal lineage instead of the basal lineage. Another study showed that BRCA1- and FANCD2-mediated DNA inter-strand crosslink repair controls the epithelial-to-mesenchymal transition (EMT) and the dedifferentiation of mammary epithelial cells [49]. These results suggest that BRCA1-mediated functions play a crucial role in modulating luminal–basal fate transition, EMT, and cell reprogramming.
3.5. SOX10
Based on the findings of the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) and transcriptional profiling, the TF Sox10 was determined to be a critical fate determinant for cell state interconversion during mammary development [50]. SOX10 is expressed in mouse and human breast cancers where tumor cells highly expressing SOX10 exhibit characteristics of MaSCs, EMT, and neural crest cells. Overexpression of SOX10 can reprogram fibroblasts into a neural crest cell-like state [51], supporting the role of SOX10 in regulating cell state plasticity. The results of RNA-sequencing (RNA-seq) and chromatin immunoprecipitation-sequencing (ChIP-seq) performed in breast cancer cells revealed that SOX10 contributes to the expression of genes related to EMT, stem/progenitor cells, and neural crest cells, and suppresses genes related to epithelial cell differentiation and apoptosis. A single-allele deletion in the Sox10 gene could significantly delay tumor development in a mouse model. In addition, overexpression of Sox10 induces the EMT of mammary epithelial cells [52]. These data demonstrate the critical role of SOX10 in regulating cell state plasticity and mammary tumorigenesis.
Taken together, these findings demonstrate that the activation of oncogenes and the inactivation of tumor-suppressor genes in lineage-restricted mammary epithelial cells can reprogram these cells into different lineages and the MaSC-like state, supporting the development of breast cancers and the evolution of tumor heterogeneity [12].
4. Targeting Cell Reprogramming for Breast Cancer Therapy
Through identifying critical cell reprogramming factors that contribute to tumorigenesis and tumor heterogeneity in breast cancer, researchers can develop therapeutic strategies to specifically target these cell reprogramming factors, which may improve breast cancer treatment (Table 2).

Table 2.
Targeting cell reprogramming for breast cancer treatment.
Recently, Wang et al. described an engineered MYC-fusion protein drug containing a dominant-negative MYC peptide (OmoMYC) and a functional penetrating “Phylomer” peptide (FPPa), with an improved drug delivery efficiency [53]. FPPa-OmoMYC triggers apoptosis in TNBC but not in non-tumorigenic cells. Gene expression signatures induced by FPPa-OmoMYC are different from those of MYC-induced cell reprogramming, which include transcriptional, metabolic, and apoptotic processes. FPPa-OmoMYC also synergistically enhances the efficacy of chemotherapeutic agents, including docetaxel, doxorubicin, and cetuximab, in the killing of breast cancer cells. Implementation of FPPa-OmoMYC and its derivatives in future clinical trials is expected for the treatment of MYC-associated breast cancer, such as basal-like TNBC.
Because PI3K signaling is activated at a high frequency in breast cancer [30] and contributes to cell reprogramming as described above [34,35], many therapeutics were developed to inhibit PI3K signaling [54]. For example, ER-positive breast cancers induced by estrogen in Pik3caH1047R mice are sensitive to the PI3K inhibitor BYL719 (alpelisib) in combination with BH3 mimetics, which inhibit BCL2 family members [55]. However, either BYL719 or BH3 mimetic alone does not efficiently suppress estrogen-induced Pik3caH1047R breast cancer. These results indicate that the anti-apoptosis function of BCL2 family members is important for cell survival during PIK3CAH1047R-induced cell reprogramming toward ER-positive breast cancer, consistent with apoptosis being a barrier for cell reprogramming [56,57]. Therefore, the combined inhibition of anti-apoptosis and cell reprogramming factors may improve the efficacy of anticancer therapy.
In addition to directly targeting cell reprogramming factors, manipulation of other fate-determinant TFs or lineage-committed molecules may also inhibit oncogenic cell reprogramming and tumorigenicity, because forced expression of such lineage-specific factors can induce differentiation of oncogenic MaSCs or CSCs, thus reducing tumorigenicity. For example, KRT19 is dominantly expressed in the mammary epithelium of the luminal lineage. Overexpression of KRT19 in KU-CSLC cells, a breast cancer stem-like cell line, significantly reduces CSC properties, whereas knockdown of KRT19 in MDA-MB-231 TNBC cells increases tumor growth, sphere formation, cell migration, and drug resistance to doxorubicin [58]. These results suggest that overexpression of luminal-specific KRT19 can reprogram basal-like TNBC cells and mammary CSCs into less aggressive and drug-sensitive states.
GATA3 is a master TF that induces mammary luminal differentiation [6,24] and is usually not expressed in basal-like breast cancers. Several studies showed that overexpression of GATA3 reduces the tumorigenicity, EMT, and metastasis of basal TNBC cell lines, such as MDA-MB-231 and Hs578T [59,60,61,62]. In a mouse model of PyMT-induced breast cancer, overexpression of Gata3 in late-stage tumors induces tumor differentiation and suppresses tumor dissemination [63]. GATA3 inhibits the expression of the metastasis-promoting genes ID1/-3, KRTHB1, LY6E, and RARRES3 but upregulates the inhibitors of metastasis including the deleted in liver cancer 1 (DLC1) and progestagen-associated endometrial protein (PAEP) [59]. In addition, the expression of mesenchymal genes (vimentin, N-cadherin, and MMP9) is repressed, and the epithelial gene E-cadherin is activated by GATA3 [60]. GATA3 also inhibits the expression of lysyl oxidase, a metastasis-promoting and matrix-remodeling protein, through promoter methylation, and reprograms basal TNBC cells into a less aggressive phenotype [61]. Therefore, GATA3 increment in tumor cells may be a helpful method to treat basal-like TNBC.
FOXA1 plays a crucial role in the regulation of luminal differentiation and is not expressed in the basal subtype of breast cancer. Kong and colleagues [64] showed that overexpression of FOXA1, together with GATA3 and ESR1 in the TNBC cell lines MDA-MB-231 and BT-549, induces cell reprogramming of these cells into estrogen-responsive luminal-like cells, which are susceptible to hormone therapy. By contrast, inhibition of the expression of FOXA1 in luminal breast cancer cell lines, such as MCF7 and T47D, and SKBR3 not only leads to silencing of luminal genes but also causes the induction of basal genes and enhanced cell migration and invasion, which represent the basal phenotype [65]. Therefore, although the targeting of FOXA1 is proposed as a strategy to treat luminal types of breast cancers, this approach can result in cells reprogramming into more aggressive cancers [66,67]. There is still a need to determine strategies to precisely control the pros and cons of the utility of cell reprogramming and cell fate-determinant factors in cancer therapy.
In addition to PI3K inhibitors, Yuan et al. [68] screened a kinase inhibitor library and demonstrated that treatment with Rho-associated protein kinase (ROCK) and mTOR kinase inhibitors could reprogram TNBC cells into NANOG-expressing stem/progenitor cells. This result is supported by the finding of previous studies that ROCK inhibition promotes ESC survival [69] and mTOR inhibition prevents epithelial stem cell senescence [70]. ROCK and mTOR inhibitor-induced stem/progenitor cells can be differentiated into terminal adipogenic cells. These fat-like cells exhibit a gene expression profile similar to that of normal adipocytes and show reduced tumorigenicity in in vitro and in vivo assays. These results support the notion that induction of cancer cells undergoing differentiation can be a therapeutic strategy to treat breast cancer [71]. However, there are still obstacles for applying this reprogramming and differentiation approach in cancer patients, because we cannot ensure whether reprogramming-derived stem/progenitor cells will show a normal or less aggressive phenotype; in addition, it is difficult to know the right time window to induce the differentiation of these stem/progenitor cells in vivo [4]. Nevertheless, the induction of cancer cell reprogramming and differentiation is worthy of development in the future.
5. Epigenetic Perspectives on Breast Cancer Cell Reprogramming and Therapy
Although all cells derived from a fertilized egg have the same genetic information, descendent cells, including ESCs, and all differentiated cells in different lineages have distinct chromatin configurations and epigenetic states, which can be represented by specific posttranslational modifications (PTMs) of histones or histone codes. These differences define a unique phenotypic characteristic for each cell type and become a barrier to prevent switching between different cell lineages and states. Similarly, cancer cells and their normal counterparts share similar genetics (except for tumorigenic alterations) but exhibit considerably different epigenetic features. Thus, cell reprogramming and tumorigenesis, both of which change cell states and chromatin configurations, require overcoming epigenetic barriers present among the hierarchies of stem/progenitor cells and various cell lineages, as well as between normal and cancer cells [72]. It is important to better understand mechanisms underlying epigenetic reprogramming and chromatin remodeling between MaSCs/progenitors and basal/luminal lineages, which can be helpful in developing favorable therapeutic strategies for treating aggressive breast cancers [73].
During the cell reprogramming of differentiated cells into stem cells, lineage-restricted genes are gradually silenced and chromatin regions containing these genes are packaged into a more compact, closed configuration and cannot be accessed by TFs, RNA polymerase, and other cellular factors. By contrast, reprogramming factors, such as OCT4, SOX2, and KLF4, which are pioneer TFs, can access their target DNA sequences and enhancers and release the highly-packaged chromatin into an open state, allowing gene expression [73]. Chromatin regions spanning active enhancers usually feature monomethylation of histone H3 at lysine 4 (H3K4me1) and acetylation of histone H3 at lysine 27 (H3K27ac), which can be examined using ChIP assays. Therefore, changes in chromatin states or chromatin remodeling during cell reprogramming are correlated with changes in histone PTMs and gene expression profiles, through which one can distinguish between different cell identities accordingly. These changes can be genome-wide and comprehensively examined using recent advanced DNA sequencing technologies, such as RNA-seq, ChIP-seq, and ATAC-seq [73]. Integral data from RNA-seq, ChIP-seq, and ATAC-seq can generate high-resolution results to show genetic and epigenetic changes throughout the genome, allowing us to decipher more precisely the controlling mechanism for the cell reprogramming of breast cancers.
For example, using the aforementioned three techniques, Poli and colleagues [38] demonstrated that the MYC-induced cell reprogramming of mammary cells is linked to de novo activation of enhancers, which are enriched for the pioneer TFs of FOX- and SOX-family members and can derive stem/progenitor cell transcriptional programs and stem/progenitor cell phenotypes. Meanwhile, MYC induces a downregulation of lineage-specific TFs, such as GATA3 and ESR1, which leads to decommissioning of luminal-specific enhancers. Similarly, Tu et al. [74] used ChIP-seq and RNA-seq approaches to further show that MYC represses the expression of tumor-suppressor genes, such as CDKN1A, GADD45A, and HMOX1, through interacting with G9A (EHMT2), a histone methyltransferase that causes H3K9 methylation and gene repression in breast cancer cells. Importantly, inhibition of G9A unleashes MYC-mediated suppression of these tumor-suppressor genes and reduces tumorigenicity, particularly in basal-like breast cancers. These results not only unveil molecular mechanisms underlying selective gene activation or repression by MYC but also identify a drug target for MYC-driven basal breast cancers. In addition to the MYC–G9A interaction, many cross-talks between cell reprogramming factors and chromatin remodelers were identified in breast cancer [50,73,75,76,77]; however, there might be many additional such networks that require further investigation.
6. Conclusions
Taking the advantages of these comprehensive and high-resolution genome-wide analyses including single-cell sequencing [78,79], we can uncover an increasing number of relationships between the cell reprogramming of lineage-committed mammary epithelial cells and the tumorigenesis of heterogeneous breast cancers [22,50,73,80,81]. In addition, by performing these analyses, many candidate therapeutic targets can be found and examined for improving the treatment of breast cancer patients. In conclusion, recent works using new technologies to decipher cell reprogramming mechanisms in breast cancer can extend our knowledge on the formation of breast cancer heterogeneity and speed up the development of therapeutic strategies.
Author Contributions
P.Y.C., M.F.H., J.C.L., L.F.C., and C.S.L. wrote the paper.
Funding
This research was funded by the Ministry of Science and Technology (MOST), Taiwan, grant numbers MOST106-2314-B-442-001-MY3 (P.Y.C.) and MOST106-2320-B-037-001-MY3 (C.S.L.).
Acknowledgments
We thank to the members in the laboratory of C.S.L. for valuable discussion and contributions. This manuscript was edited by Wallace Academic Editing.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Corominas-Faja, B.; Cufi, S.; Oliveras-Ferraros, C.; Cuyas, E.; Lopez-Bonet, E.; Lupu, R.; Alarcon, T.; Vellon, L.; Iglesias, J.M.; Leis, O.; et al. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell Cycle 2013, 12, 3109–3124. [Google Scholar] [CrossRef] [PubMed]
- Nishi, M.; Sakai, Y.; Akutsu, H.; Nagashima, Y.; Quinn, G.; Masui, S.; Kimura, H.; Perrem, K.; Umezawa, A.; Yamamoto, N.; et al. Induction of cells with cancer stem cell properties from nontumorigenic human mammary epithelial cells by defined reprogramming factors. Oncogene 2014, 33, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Lin, Y.C.; Nakamura, Y.; Eckner, R.; Wuputra, K.; Kuo, K.K.; Lin, C.S.; Yokoyama, K.K. Potential application of cell reprogramming techniques for cancer research. Cell Mol. Life Sci. 2019, 76, 45–65. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Sadahiro, T.; Ieda, M. Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration. Int. J. Mol. Sci. 2018, 19, 2629. [Google Scholar] [CrossRef] [PubMed]
- Kouros-Mehr, H.; Slorach, E.M.; Sternlicht, M.D.; Werb, Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 2006, 127, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Lilja, A.M.; Rodilla, V.; Huyghe, M.; Hannezo, E.; Landragin, C.; Renaud, O.; Leroy, O.; Rulands, S.; Simons, B.D.; Fre, S. Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat. Cell Biol. 2018, 20, 677–687. [Google Scholar] [CrossRef]
- Wuidart, A.; Sifrim, A.; Fioramonti, M.; Matsumura, S.; Brisebarre, A.; Brown, D.; Centonze, A.; Dannau, A.; Dubois, C.; Van Keymeulen, A.; et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat. Cell Biol. 2018, 20, 666–676. [Google Scholar] [CrossRef]
- Yalcin-Ozuysal, O.; Fiche, M.; Guitierrez, M.; Wagner, K.U.; Raffoul, W.; Brisken, C. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 2010, 17, 1600–1612. [Google Scholar] [CrossRef]
- Spike, B.T.; Engle, D.D.; Lin, J.C.; Cheung, S.K.; La, J.; Wahl, G.M. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell 2012, 10, 183–197. [Google Scholar] [CrossRef]
- Zvelebil, M.; Oliemuller, E.; Gao, Q.; Wansbury, O.; Mackay, A.; Kendrick, H.; Smalley, M.J.; Reis-Filho, J.S.; Howard, B.A. Embryonic mammary signature subsets are activated in Brca1-/- and basal-like breast cancers. Breast Cancer Res. 2013, 15, R25. [Google Scholar] [CrossRef]
- Zhang, M.; Lee, A.V.; Rosen, J.M. The Cellular Origin and Evolution of Breast Cancer. Cold Spring Harbor Perspect. Med. 2017, 7, a027128. [Google Scholar] [CrossRef]
- Zhang, M.; Rosen, J.M. Stem cells in the etiology and treatment of cancer. Curr. Opin. Genet. Dev. 2006, 16, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Bland, P.; Howard, B.A. Mammary lineage restriction in development. Nat. Cell Biol. 2018, 20, 637–639. [Google Scholar] [CrossRef]
- Van Keymeulen, A.; Rocha, A.S.; Ousset, M.; Beck, B.; Bouvencourt, G.; Rock, J.; Sharma, N.; Dekoninck, S.; Blanpain, C. Distinct stem cells contribute to mammary gland development and maintenance. Nature 2011, 479, 189–193. [Google Scholar] [CrossRef]
- Rios, A.C.; Fu, N.Y.; Lindeman, G.J.; Visvader, J.E. In situ identification of bipotent stem cells in the mammary gland. Nature 2014, 506, 322–327. [Google Scholar] [CrossRef]
- Wuidart, A.; Ousset, M.; Rulands, S.; Simons, B.D.; van Keymeulen, A.; Blanpain, C. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016, 30, 1261–1277. [Google Scholar] [CrossRef]
- Davis, F.M.; Lloyd-Lewis, B.; Harris, O.B.; Kozar, S.; Winton, D.J.; Muresan, L.; Watson, C.J. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat. Commun. 2016, 7, 13053. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Keckesova, Z.; Donaher, J.L.; Shibue, T.; Tischler, V.; Reinhardt, F.; Itzkovitz, S.; Noske, A.; Zurrer-Hardi, U.; Bell, G.; et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012, 148, 1015–1028. [Google Scholar] [CrossRef]
- Inman, J.L.; Robertson, C.; Mott, J.D.; Bissell, M.J. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development 2015, 142, 1028–1042. [Google Scholar] [CrossRef]
- Visvader, J.E.; Stingl, J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes Dev. 2014, 28, 1143–1158. [Google Scholar] [CrossRef]
- Pellacani, D.; Bilenky, M.; Kannan, N.; Heravi-Moussavi, A.; Knapp, D.; Gakkhar, S.; Moksa, M.; Carles, A.; Moore, R.; Mungall, A.J.; et al. Analysis of Normal Human Mammary Epigenomes Reveals Cell-Specific Active Enhancer States and Associated Transcription Factor Networks. Cell Rep. 2016, 17, 2060–2074. [Google Scholar] [CrossRef] [PubMed]
- Bouras, T.; Pal, B.; Vaillant, F.; Harburg, G.; Asselin-Labat, M.L.; Oakes, S.R.; Lindeman, G.J.; Visvader, J.E. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 2008, 3, 429–441. [Google Scholar] [CrossRef]
- Asselin-Labat, M.L.; Sutherland, K.D.; Barker, H.; Thomas, R.; Shackleton, M.; Forrest, N.C.; Hartley, L.; Robb, L.; Grosveld, F.G.; van der Wees, J.; et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 2007, 9, 201–209. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Herschkowitz, J.I.; Simin, K.; Weigman, V.J.; Mikaelian, I.; Usary, J.; Hu, Z.; Rasmussen, K.E.; Jones, L.P.; Assefnia, S.; Chandrasekharan, S.; et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007, 8, R76. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Network, T.C.G.A. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Koren, S.; Bentires-Alj, M. Mouse models of PIK3CA mutations: One mutation initiates heterogeneous mammary tumors. FEBS J. 2013, 280, 2758–2765. [Google Scholar] [CrossRef]
- Meyer, D.S.; Brinkhaus, H.; Muller, U.; Muller, M.; Cardiff, R.D.; Bentires-Alj, M. Luminal expression of PIK3CA mutant H1047R in the mammary gland induces heterogeneous tumors. Cancer Res. 2011, 71, 4344–4351. [Google Scholar] [CrossRef]
- Liu, P.; Cheng, H.; Santiago, S.; Raeder, M.; Zhang, F.; Isabella, A.; Yang, J.; Semaan, D.J.; Chen, C.; Fox, E.A.; et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat. Med. 2011, 17, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Reavie, L.; Couto, J.P.; de Silva, D.; Stadler, M.B.; Roloff, T.; Britschgi, A.; Eichlisberger, T.; Kohler, H.; Aina, O.; et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature 2015, 525, 114–118. [Google Scholar] [CrossRef]
- Van Keymeulen, A.; Lee, M.Y.; Ousset, M.; Brohee, S.; Rorive, S.; Giraddi, R.R.; Wuidart, A.; Bouvencourt, G.; Dubois, C.; Salmon, I.; et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 2015, 525, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Chandriani, S.; Frengen, E.; Cowling, V.H.; Pendergrass, S.A.; Perou, C.M.; Whitfield, M.L.; Cole, M.D. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS ONE 2009, 4, e6693. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Poli, V.; Fagnocchi, L.; Fasciani, A.; Cherubini, A.; Mazzoleni, S.; Ferrillo, S.; Miluzio, A.; Gaudioso, G.; Vaira, V.; Turdo, A.; et al. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat. Commun. 2018, 9, 1024. [Google Scholar] [CrossRef]
- Hein, S.M.; Haricharan, S.; Johnston, A.N.; Toneff, M.J.; Reddy, J.P.; Dong, J.; Bu, W.; Li, Y. Luminal epithelial cells within the mammary gland can produce basal cells upon oncogenic stress. Oncogene 2016, 35, 1461–1467. [Google Scholar] [CrossRef]
- Hong, H.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Kanagawa, O.; Nakagawa, M.; Okita, K.; Yamanaka, S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009, 460, 1132–1135. [Google Scholar] [CrossRef]
- Kawamura, T.; Suzuki, J.; Wang, Y.V.; Menendez, S.; Morera, L.B.; Raya, A.; Wahl, G.M.; Izpisua Belmonte, J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009, 460, 1140–1144. [Google Scholar] [CrossRef] [PubMed]
- Marion, R.M.; Strati, K.; Li, H.; Murga, M.; Blanco, R.; Ortega, S.; Fernandez-Capetillo, O.; Serrano, M.; Blasco, M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009, 460, 1149–1153. [Google Scholar] [CrossRef] [PubMed]
- Utikal, J.; Polo, J.M.; Stadtfeld, M.; Maherali, N.; Kulalert, W.; Walsh, R.M.; Khalil, A.; Rheinwald, J.G.; Hochedlinger, K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009, 460, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Spike, B.T.; Wahl, G.M.; Levine, A.J. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc. Natl. Acad. Sci. USA 2010, 107, 22745–22750. [Google Scholar] [CrossRef]
- Zhang, M.; Behbod, F.; Atkinson, R.L.; Landis, M.D.; Kittrell, F.; Edwards, D.; Medina, D.; Tsimelzon, A.; Hilsenbeck, S.; Green, J.E.; et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008, 68, 4674–4682. [Google Scholar] [CrossRef]
- Santoro, A.; Vlachou, T.; Luzi, L.; Melloni, G.; Mazzarella, L.; D’Elia, E.; Aobuli, X.; Pasi, C.E.; Reavie, L.; Bonetti, P.; et al. p53 Loss in Breast Cancer Leads to Myc Activation, Increased Cell Plasticity, and Expression of a Mitotic Signature with Prognostic Value. Cell Rep. 2019, 26, 624–638.e8. [Google Scholar] [CrossRef]
- Gorski, J.J.; James, C.R.; Quinn, J.E.; Stewart, G.E.; Staunton, K.C.; Buckley, N.E.; McDyer, F.A.; Kennedy, R.D.; Wilson, R.H.; Mullan, P.B.; et al. BRCA1 transcriptionally regulates genes associated with the basal-like phenotype in breast cancer. Breast Cancer Res. Treat. 2010, 122, 721–731. [Google Scholar] [CrossRef]
- Molyneux, G.; Geyer, F.C.; Magnay, F.A.; McCarthy, A.; Kendrick, H.; Natrajan, R.; Mackay, A.; Grigoriadis, A.; Tutt, A.; Ashworth, A.; et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 2010, 7, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bierie, B.; Li, A.G.; Pathania, S.; Toomire, K.; Dimitrov, S.D.; Liu, B.; Gelman, R.; Giobbie-Hurder, A.; Feunteun, J.; et al. BRCA1/FANCD2/BRG1-Driven DNA Repair Stabilizes the Differentiation State of Human Mammary Epithelial Cells. Mol. Cell 2016, 63, 277–292. [Google Scholar] [CrossRef]
- Dravis, C.; Chung, C.Y.; Lytle, N.K.; Herrera-Valdez, J.; Luna, G.; Trejo, C.L.; Reya, T.; Wahl, G.M. Epigenetic and Transcriptomic Profiling of Mammary Gland Development and Tumor Models Disclose Regulators of Cell State Plasticity. Cancer Cell 2018, 34, 466–482.e6. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lim, H.; Li, Z.; Oh, Y.; Kovlyagina, I.; Choi, I.Y.; Dong, X.; Lee, G. Generation of multipotent induced neural crest by direct reprogramming of human postnatal fibroblasts with a single transcription factor. Cell Stem Cell 2014, 15, 497–506. [Google Scholar] [CrossRef]
- Dravis, C.; Spike, B.T.; Harrell, J.C.; Johns, C.; Trejo, C.L.; Southard-Smith, E.M.; Perou, C.M.; Wahl, G.M. Sox10 Regulates Stem/Progenitor and Mesenchymal Cell States in Mammary Epithelial Cells. Cell Rep. 2015, 12, 2035–2048. [Google Scholar] [CrossRef]
- Wang, E.; Sorolla, A.; Cunningham, P.T.; Bogdawa, H.M.; Beck, S.; Golden, E.; Dewhurst, R.E.; Florez, L.; Cruickshank, M.N.; Hoffmann, K.; et al. Tumor penetrating peptides inhibiting MYC as a potent targeted therapeutic strategy for triple-negative breast cancers. Oncogene 2019, 38, 140–150. [Google Scholar] [CrossRef]
- Paplomata, E.; O’Regan, R. The PI3K/AKT/mTOR pathway in breast cancer: Targets, trials and biomarkers. Ther. Adv. Med. Oncol. 2014, 6, 154–166. [Google Scholar] [CrossRef]
- Stratikopoulos, E.E.; Kiess, N.; Szabolcs, M.; Pegno, S.; Kakit, C.; Wu, X.; Poulikakos, P.I.; Cheung, P.; Schmidt, H.; Parsons, R. Mouse ER+/PIK3CA(H1047R) breast cancers caused by exogenous estrogen are heterogeneously dependent on estrogen and undergo BIM-dependent apoptosis with BH3 and PI3K agents. Oncogene 2019, 38, 47–59. [Google Scholar] [CrossRef]
- Kim, E.J.Y.; Anko, M.L.; Flensberg, C.; Majewski, I.J.; Geng, F.S.; Firas, J.; Huang, D.C.S.; van Delft, M.F.; Heath, J.K. BAK/BAX-Mediated Apoptosis Is a Myc-Induced Roadblock to Reprogramming. Stem Cell Rep. 2018, 10, 331–338. [Google Scholar] [CrossRef]
- Li, Y.; Feng, H.; Gu, H.; Lewis, D.W.; Yuan, Y.; Zhang, L.; Yu, H.; Zhang, P.; Cheng, H.; Miao, W.; et al. The p53-PUMA axis suppresses iPSC generation. Nat. Commun. 2013, 4, 2174. [Google Scholar] [CrossRef]
- Saha, S.K.; Kim, K.; Yang, G.M.; Choi, H.Y.; Cho, S.G. Cytokeratin 19 (KRT19) has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells. Int. J. Mol. Sci. 2018, 19, 1423. [Google Scholar] [CrossRef]
- Dydensborg, A.B.; Rose, A.A.; Wilson, B.J.; Grote, D.; Paquet, M.; Giguere, V.; Siegel, P.M.; Bouchard, M. GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene 2009, 28, 2634–2642. [Google Scholar] [CrossRef]
- Yan, W.; Cao, Q.J.; Arenas, R.B.; Bentley, B.; Shao, R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J. Biol. Chem. 2010, 285, 14042–14051. [Google Scholar] [CrossRef]
- Chu, I.M.; Michalowski, A.M.; Hoenerhoff, M.; Szauter, K.M.; Luger, D.; Sato, M.; Flanders, K.; Oshima, A.; Csiszar, K.; Green, J.E. GATA3 inhibits lysyl oxidase-mediated metastases of human basal triple-negative breast cancer cells. Oncogene 2012, 31, 2017–2027. [Google Scholar] [CrossRef] [PubMed]
- Chu, I.M.; Lai, W.C.; Aprelikova, O.; El Touny, L.H.; Kouros-Mehr, H.; Green, J.E. Expression of GATA3 in MDA-MB-231 triple-negative breast cancer cells induces a growth inhibitory response to TGFss. PLoS ONE 2013, 8, e61125. [Google Scholar] [CrossRef] [PubMed]
- Kouros-Mehr, H.; Bechis, S.K.; Slorach, E.M.; Littlepage, L.E.; Egeblad, M.; Ewald, A.J.; Pai, S.Y.; Ho, I.C.; Werb, Z. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell 2008, 13, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.L.; Li, G.; Loh, S.L.; Sung, W.K.; Liu, E.T. Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Mol. Syst. Biol. 2011, 7, 526. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, G.M.; Bebek, G.; Ginther, C.L.; Sizemore, S.T.; Lozada, K.L.; Miedler, J.D.; Anderson, L.A.; Godwin, A.K.; Abdul-Karim, F.W.; Slamon, D.J.; et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene 2013, 32, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Qin, L.; Sun, T.; Wu, H.; He, T.; Yang, Z.; Mo, Q.; Liao, L.; Xu, J. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene 2017, 36, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Nakayama, Y. Down-regulation of Forkhead box protein A1 (FOXA1) leads to cancer stem cell-like properties in tamoxifen-resistant breast cancer cells through induction of interleukin-6. J. Biol. Chem. 2017, 292, 8136–8148. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, F.; You, M.; Yang, Q. Identification of protein kinase inhibitors to reprogram breast cancer cells. Cell Death Dis. 2018, 9, 915. [Google Scholar] [CrossRef]
- Watanabe, K.; Ueno, M.; Kamiya, D.; Nishiyama, A.; Matsumura, M.; Wataya, T.; Takahashi, J.B.; Nishikawa, S.; Muguruma, K.; Sasai, Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007, 25, 681–686. [Google Scholar] [CrossRef]
- Iglesias-Bartolome, R.; Patel, V.; Cotrim, A.; Leelahavanichkul, K.; Molinolo, A.A.; Mitchell, J.B.; Gutkind, J.S. mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell 2012, 11, 401–414. [Google Scholar] [CrossRef]
- Centritto, F.; Paroni, G.; Bolis, M.; Garattini, S.K.; Kurosaki, M.; Barzago, M.M.; Zanetti, A.; Fisher, J.N.; Scott, M.F.; Pattini, L.; et al. Cellular and molecular determinants of all-trans retinoic acid sensitivity in breast cancer: Luminal phenotype and RARalpha expression. EMBO Mol. Med. 2015, 7, 950–972. [Google Scholar] [CrossRef]
- Apostolou, E.; Hochedlinger, K. Chromatin dynamics during cellular reprogramming. Nature 2013, 502, 462–471. [Google Scholar] [CrossRef]
- Swinstead, E.E.; Paakinaho, V.; Hager, G.L. Chromatin reprogramming in breast cancer. Endocr. Relat. Cancer 2018, 25, R385–R404. [Google Scholar] [CrossRef]
- Tu, W.B.; Shiah, Y.J.; Lourenco, C.; Mullen, P.J.; Dingar, D.; Redel, C.; Tamachi, A.; Ba-Alawi, W.; Aman, A.; Al-Awar, R.; et al. MYC Interacts with the G9a Histone Methyltransferase to Drive Transcriptional Repression and Tumorigenesis. Cancer Cell 2018, 34, 579–595. [Google Scholar] [CrossRef]
- Breindel, J.L.; Skibinski, A.; Sedic, M.; Wronski-Campos, A.; Zhou, W.; Keller, P.J.; Mills, J.; Bradner, J.; Onder, T.; Kuperwasser, C. Epigenetic Reprogramming of Lineage-Committed Human Mammary Epithelial Cells Requires DNMT3A and Loss of DOT1L. Stem Cell Rep. 2017, 9, 943–955. [Google Scholar] [CrossRef]
- Swinstead, E.E.; Miranda, T.B.; Paakinaho, V.; Baek, S.; Goldstein, I.; Hawkins, M.; Karpova, T.S.; Ball, D.; Mazza, D.; Lavis, L.D.; et al. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell 2016, 165, 593–605. [Google Scholar] [CrossRef]
- Takaku, M.; Grimm, S.A.; Shimbo, T.; Perera, L.; Menafra, R.; Stunnenberg, H.G.; Archer, T.K.; Machida, S.; Kurumizaka, H.; Wade, P.A. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 2016, 17, 36. [Google Scholar] [CrossRef]
- Colacino, J.A.; Azizi, E.; Brooks, M.D.; Harouaka, R.; Fouladdel, S.; McDermott, S.P.; Lee, M.; Hill, D.; Madden, J.; Boerner, J.; et al. Heterogeneity of Human Breast Stem and Progenitor Cells as Revealed by Transcriptional Profiling. Stem Cell Rep. 2018, 10, 1596–1609. [Google Scholar] [CrossRef]
- Giraddi, R.R.; Chung, C.Y.; Heinz, R.E.; Balcioglu, O.; Novotny, M.; Trejo, C.L.; Dravis, C.; Hagos, B.M.; Mehrabad, E.M.; Rodewald, L.W.; et al. Single-Cell Transcriptomes Distinguish Stem Cell State Changes and Lineage Specification Programs in Early Mammary Gland Development. Cell Rep. 2018, 24, 1653–1666. [Google Scholar] [CrossRef]
- Rodilla, V.; Fre, S. Cellular Plasticity of Mammary Epithelial Cells Underlies Heterogeneity of Breast Cancer. Biomedicines 2018, 6, 103. [Google Scholar] [CrossRef]
- Wahl, G.M.; Spike, B.T. Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. NPJ Breast Cancer 2017, 3, 14. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).