Synthetic miR-143 Inhibits Growth of HER2-Positive Gastric Cancer Cells by Suppressing KRAS Networks Including DDX6 RNA Helicase
Abstract
1. Introduction
2. Results
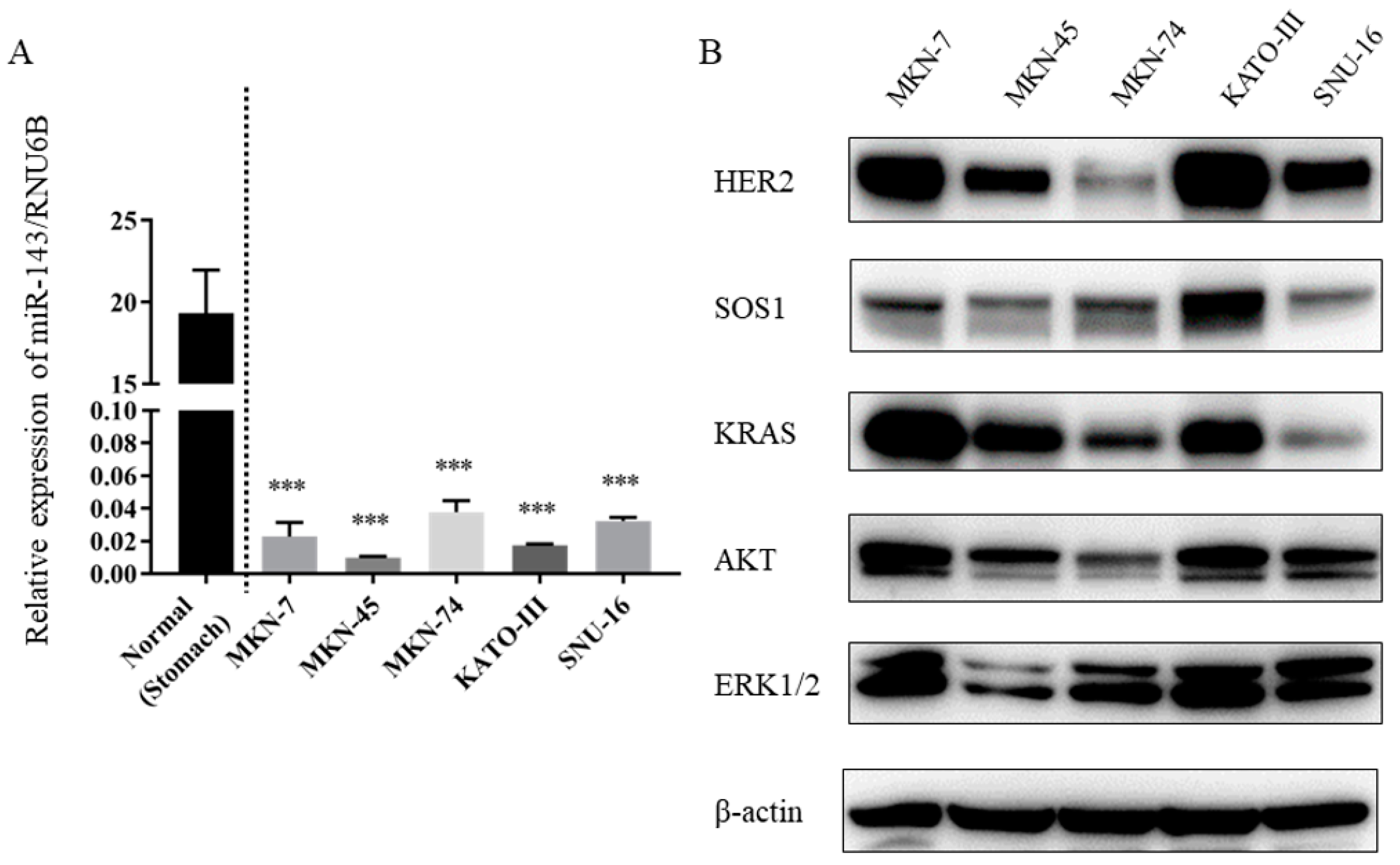
2.1. Expression Levels of miR-143 Were Down-Regulated in Gastric Cancer Cell Lines
2.2. Expression Levels of KRAS and Downstream Molecules Were Up-Regulated in HER2-Positive Gastric Cancer Cell Lines
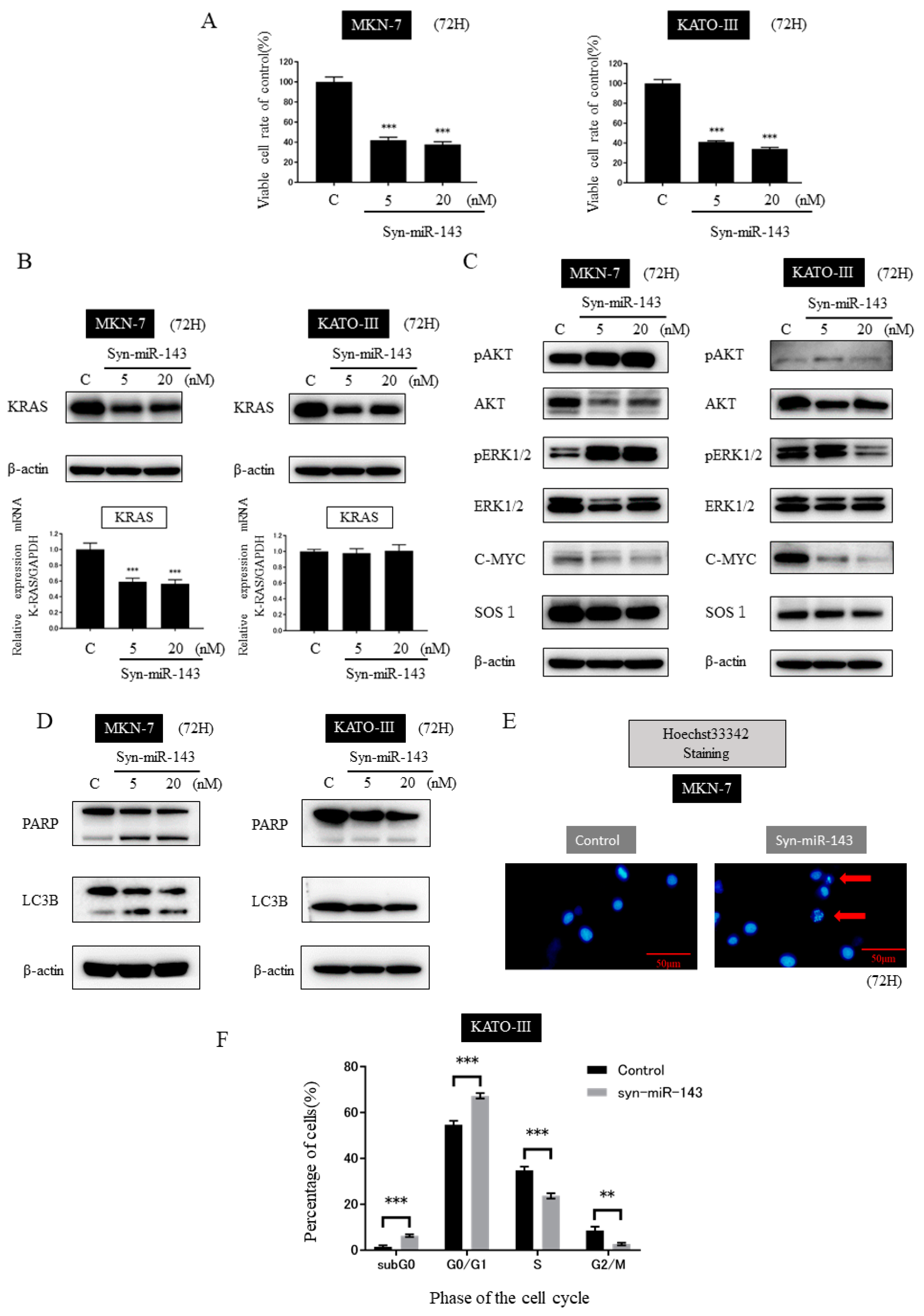
2.3. Ectopic Expression of miR-143 Inhibited the Growth of MKN-7 and KATO-III Cells by Targeting KRAS and Its Related Signaling Molecules
2.4. Transfection of syn-miR-143 Induced Apoptosis in MKN-7 Cells and Cell-Cycle Arrest in KATO-III Cells
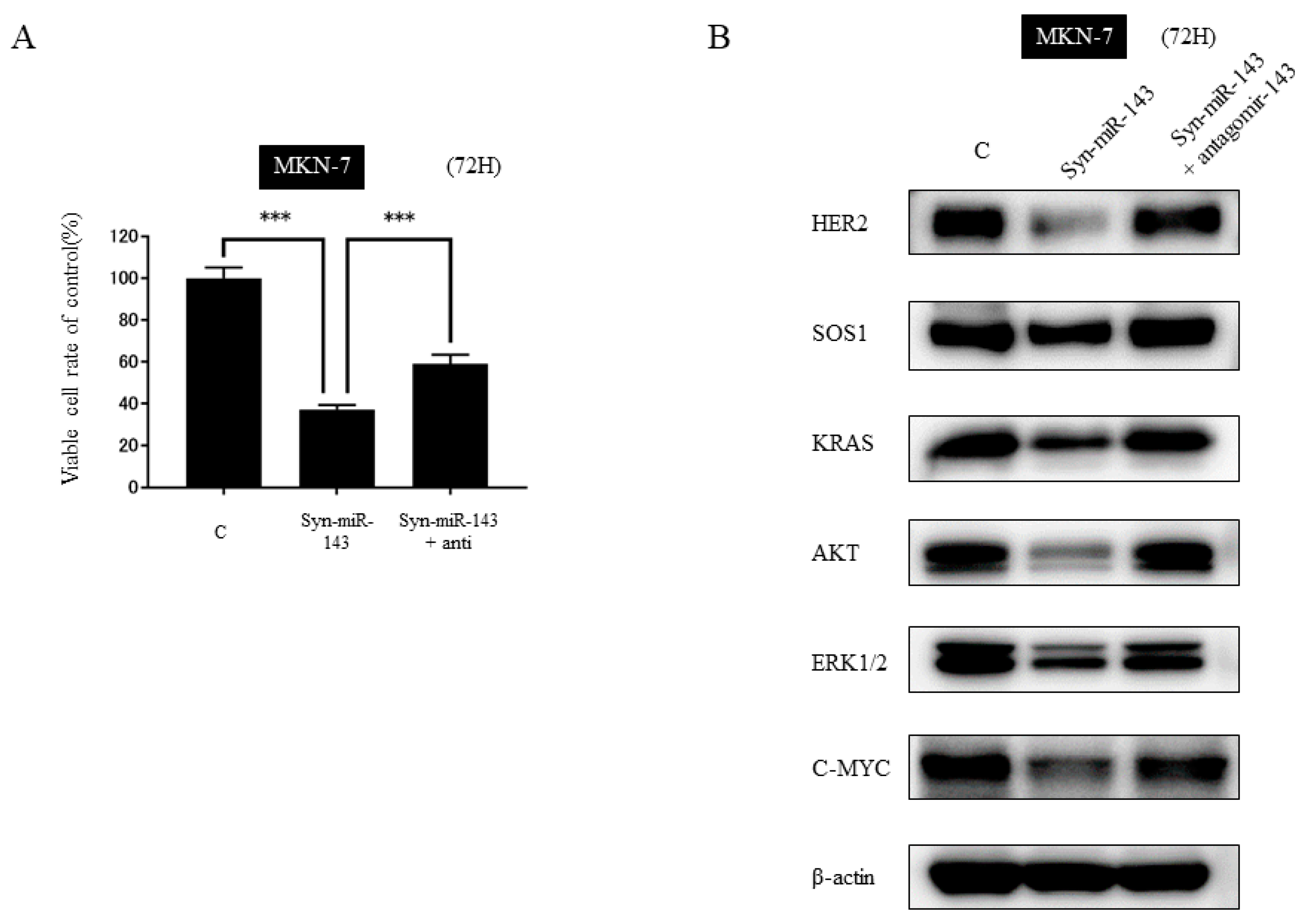
2.5. KRAS and Its Related Molecules are Targets of syn-miR-143
2.6. DDX6 Is a Direct Target Gene of syn-miR-143 and syn-miR-143 Down-Regulated the Expression Level of HER2 through Targeting DDX6
2.7. Knockdown of KRAS Alone Was Not Sufficient to Suppress the Cell Growth and the Effector Signals of KRAS in HER2-Positive Gastric Cancer Cell Lines
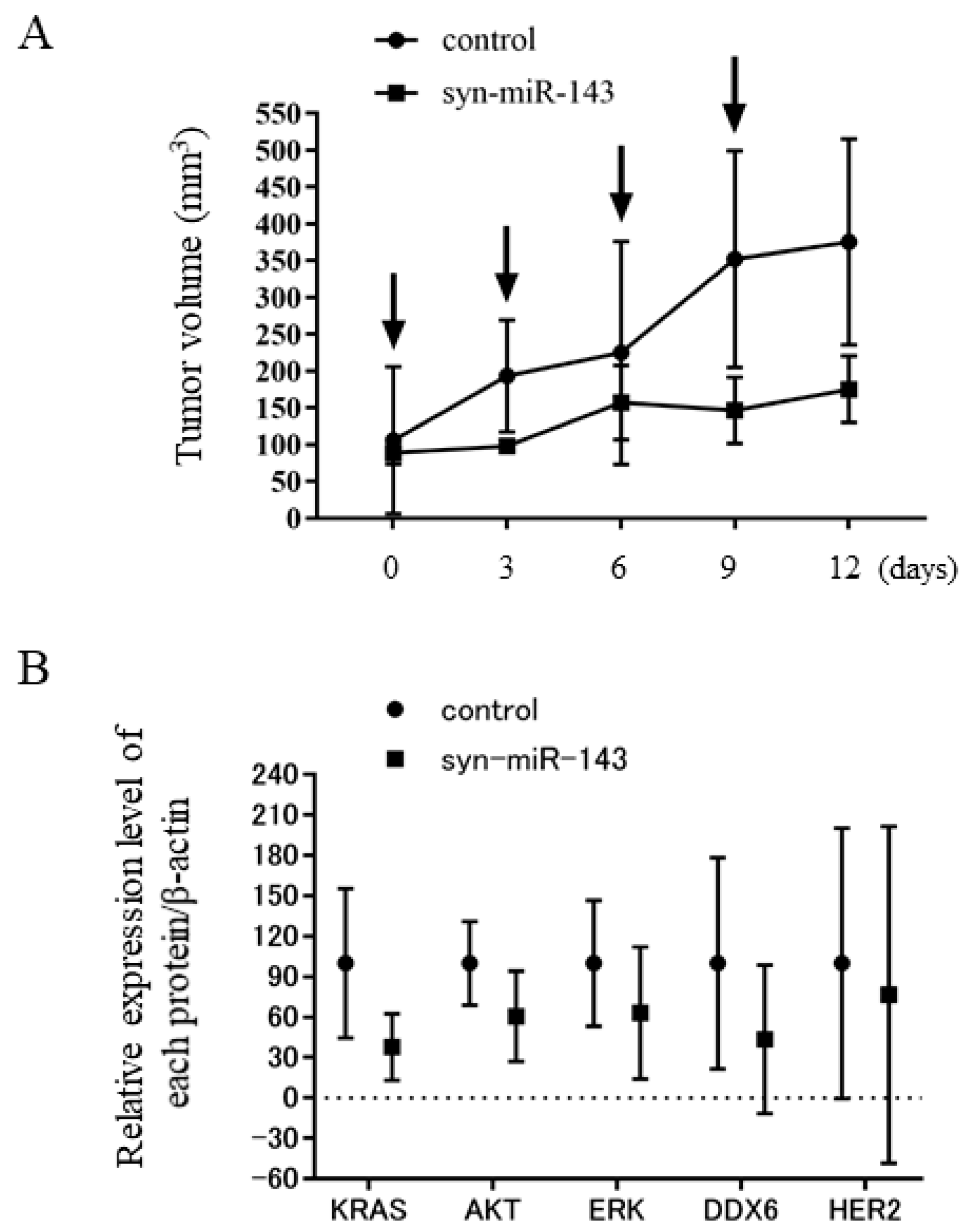
2.8. Syn-miR-143 Exhibited an Anti-Tumor Effect on Xenografted Tumors in Nude Mice
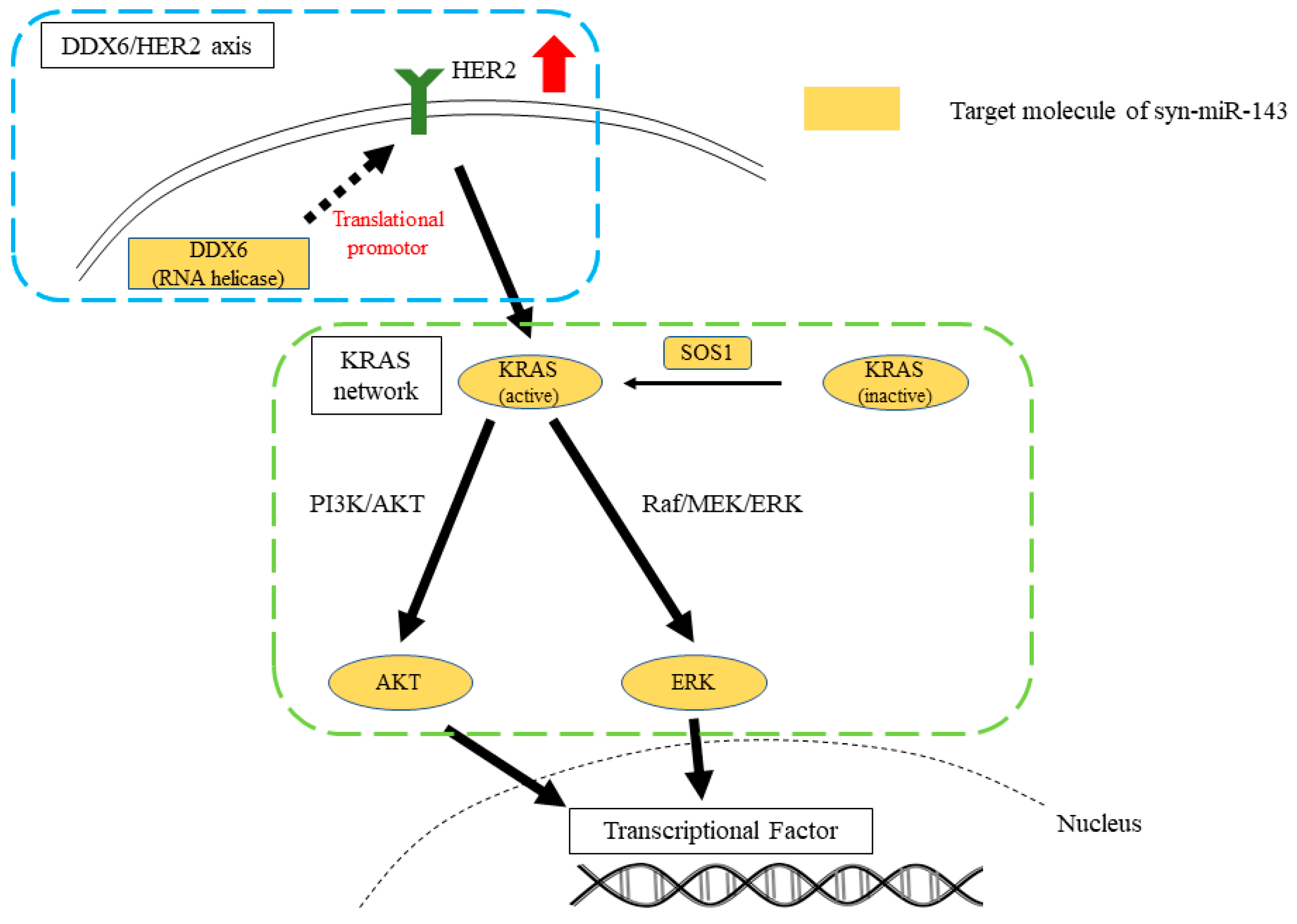
3. Discussion
4. Material and Methods
4.1. Cell Culture and Cell Viability
4.2. Cell Transfection Experiment with miRNA or siRNA
4.3. Western Blotting Analysis
4.4. Quantitative RT-PCR
4.5. Dual luciferase Reporter Assay
4.6. Hoechst 33352 Staining
4.7. Cell-Cycle Analysis
4.8. Human Tumor Xenograft Model
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DDX6 | DEAD/H-box RNA helicase 6 |
| Syn-miR-143 | synthetic miR-143 |
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Ferro, A.; Peleteiro, B.; Malvezzi, M.; Bosetti, C.; Bertuccio, P.; Levi, F.; Negri, E.; La Vecchia, C.; Lunet, N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur. J. Cancer 2014, 50, 1330–1344. [Google Scholar] [CrossRef] [PubMed]
- Sant, M.; Allemani, C.; Santaquilani, M.; Knijn, A.; Marchesi, F.; Capocaccia, R.; Group, E.W. EUROCARE-4. Survival of cancer patients diagnosed in 1995–1999. Results and commentary. Eur. J. Cancer 2009, 45, 931–991. [Google Scholar] [CrossRef]
- Boku, N. HER2-positive gastric cancer. Gastric. Cancer 2014, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gravalos, C.; Jimeno, A. HER2 in gastric cancer: A new prognostic factor and a novel therapeutic target. Ann. Oncol. 2008, 19, 1523–1529. [Google Scholar] [CrossRef]
- Pazo Cid, R.A.; Anton, A. Advanced HER2-positive gastric cancer: Current and future targeted therapies. Crit. Rev. Oncol. Hematol. 2013, 85, 350–362. [Google Scholar] [CrossRef]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Sukawa, Y.; Yamamoto, H.; Nosho, K.; Ito, M.; Igarashi, H.; Naito, T.; Mitsuhashi, K.; Matsunaga, Y.; Takahashi, T.; Mikami, M.; et al. HER2 expression and PI3K-Akt pathway alterations in gastric cancer. Digestion 2014, 89, 12–17. [Google Scholar] [CrossRef]
- Hou, F.; Shi, D.B.; Chen, Y.Q.; Gao, P. Human Epidermal Growth Factor Receptor-2 Promotes Invasion and Metastasis in Gastric Cancer by Activating Mitogen-activated Protein Kinase Signaling. Appl. Immunohistochem. Mol. Morphol. AIMM 2018. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Noguchi, S.; Yasui, Y.; Iwasaki, J.; Kumazaki, M.; Yamada, N.; Naito, S.; Akao, Y. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013, 328, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Chen, X.; Liang, H.; Deng, T.; Chen, W.; Zhang, S.; Liu, M.; Gao, X.; Liu, Y.; Zhao, C.; et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol. Cancer 2014, 13, 220. [Google Scholar] [CrossRef] [PubMed]
- Borralho, P.M.; Kren, B.T.; Castro, R.E.; da Silva, I.B.; Steer, C.J.; Rodrigues, C.M. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009, 276, 6689–6700. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.L.; Cheng, B.; Li, P.Y.; Huang, H.J.; Zhao, Q.; Dan, Z.L.; Tian, D.A.; Zhang, P. MicroRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J. Gastroenterol. 2013, 19, 7758–7765. [Google Scholar] [CrossRef] [PubMed]
- Akao, Y.; Kumazaki, M.; Shinohara, H.; Sugito, N.; Kuranaga, Y.; Tsujino, T.; Yoshikawa, Y.; Kitade, Y. Impairment of K-Ras signaling networks and increased efficacy of epidermal growth factor receptor inhibitors by a novel synthetic miR-143. Cancer Sci. 2018. [Google Scholar] [CrossRef]
- Tajirika, T.; Tokumaru, Y.; Taniguchi, K.; Sugito, N.; Matsuhashi, N.; Futamura, M.; Yanagihara, K.; Akao, Y.; Yoshida, K. DEAD-Box Protein RNA-Helicase DDX6 Regulates the Expression of HER2 and FGFR2 at the Post-Transcriptional Step in Gastric Cancer Cells. Int. J. Mol. Sci. 2018, 19, 2005. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Li, J.; Liu, L.; Hou, N.; Chang, D.; Zhao, L.; Li, Z.; Song, T.; Huang, C. Dysregulation of miRNAs and their potential as biomarkers for the diagnosis of gastric cancer. Biomedical. Rep. 2013, 1, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, H.; Kimura, M.; Takeyama, H. Role of microRNAs in gastric cancer. World J. Gastroenterol. 2014, 20, 5694–5699. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Iio, A.; Nakagawa, Y.; Naoe, T.; Tanigawa, N.; Akao, Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology 2009, 77, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, A.; Lerma, E.; Escuin, D.; Tibau, A.; Munoz, J.; Ojeda, B.; Barnadas, A.; Adrover, E.; Sanchez-Tejada, L.; Giner, D.; et al. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br. J. Cancer 2012, 106, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- Betts, G.; Valentine, H.; Pritchard, S.; Swindell, R.; Williams, V.; Morgan, S.; Griffiths, E.A.; Welch, I.; West, C.; Womack, C. FGFR2, HER2 and cMet in gastric adenocarcinoma: Detection, prognostic significance and assessment of downstream pathway activation. Virchows Arch. Int. J. Pathol. 2014, 464, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Akao, Y.; Marukawa, O.; Morikawa, H.; Nakao, K.; Kamei, M.; Hachiya, T.; Tsujimoto, Y. The rck/p54 candidate proto-oncogene product is a 54-kilodalton D-E-A-D box protein differentially expressed in human and mouse tissues. Cancer Res. 1995, 55, 3444–3449. [Google Scholar] [PubMed]
- Nakagawa, Y.; Morikawa, H.; Hirata, I.; Shiozaki, M.; Matsumoto, A.; Maemura, K.; Nishikawa, T.; Niki, M.; Tanigawa, N.; Ikegami, M.; et al. Overexpression of rck/p54, a DEAD box protein, in human colorectal tumours. Br. J. Cancer 1999, 80, 914–917. [Google Scholar] [CrossRef]
- Taniguchi, K.; Iwatsuki, A.; Sugito, N.; Shinohara, H.; Kuranaga, Y.; Oshikawa, Y.; Tajirika, T.; Futamura, M.; Yoshida, K.; Uchiyama, K.; et al. Oncogene RNA helicase DDX6 promotes the process of c-Myc expression in gastric cancer cells. Mol. Carcinog. 2018, 57, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.R.; Bang, Y.J.; Qin, S.K.; Chung, H.C.; Xu, J.M.; Park, J.O.; Jeziorski, K.; Shparyk, Y.; Hoff, P.M.; Sobrero, A.; et al. Lapatinib in Combination With Capecitabine Plus Oxaliplatin in Human Epidermal Growth Factor Receptor 2-Positive Advanced or Metastatic Gastric, Esophageal, or Gastroesophageal Adenocarcinoma: TRIO-013/LOGiC--A Randomized Phase III Trial. J. Clin. Oncol. 2016, 34, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Xu, R.H.; Chung, H.C.; Sun, G.P.; Doi, T.; Xu, J.M.; Tsuji, A.; Omuro, Y.; Li, J.; Wang, J.W.; et al. Lapatinib plus paclitaxel versus paclitaxel alone in the second-line treatment of HER2-amplified advanced gastric cancer in Asian populations: TyTAN—A randomized, phase III study. J. Clin. Oncol. 2014, 32, 2039–2049. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokumaru, Y.; Tajirika, T.; Sugito, N.; Kuranaga, Y.; Shinohara, H.; Tsujino, T.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K. Synthetic miR-143 Inhibits Growth of HER2-Positive Gastric Cancer Cells by Suppressing KRAS Networks Including DDX6 RNA Helicase. Int. J. Mol. Sci. 2019, 20, 1697. https://doi.org/10.3390/ijms20071697
Tokumaru Y, Tajirika T, Sugito N, Kuranaga Y, Shinohara H, Tsujino T, Matsuhashi N, Futamura M, Akao Y, Yoshida K. Synthetic miR-143 Inhibits Growth of HER2-Positive Gastric Cancer Cells by Suppressing KRAS Networks Including DDX6 RNA Helicase. International Journal of Molecular Sciences. 2019; 20(7):1697. https://doi.org/10.3390/ijms20071697
Chicago/Turabian StyleTokumaru, Yoshihisa, Toshihiro Tajirika, Nobuhiko Sugito, Yuki Kuranaga, Haruka Shinohara, Takuya Tsujino, Nobuhisa Matsuhashi, Manabu Futamura, Yukihiro Akao, and Kazuhiro Yoshida. 2019. "Synthetic miR-143 Inhibits Growth of HER2-Positive Gastric Cancer Cells by Suppressing KRAS Networks Including DDX6 RNA Helicase" International Journal of Molecular Sciences 20, no. 7: 1697. https://doi.org/10.3390/ijms20071697
APA StyleTokumaru, Y., Tajirika, T., Sugito, N., Kuranaga, Y., Shinohara, H., Tsujino, T., Matsuhashi, N., Futamura, M., Akao, Y., & Yoshida, K. (2019). Synthetic miR-143 Inhibits Growth of HER2-Positive Gastric Cancer Cells by Suppressing KRAS Networks Including DDX6 RNA Helicase. International Journal of Molecular Sciences, 20(7), 1697. https://doi.org/10.3390/ijms20071697






