Thyroid Hormone Disruptors Interfere with Molecular Pathways of Eye Development and Function in Zebrafish
Abstract
:1. Introduction
2. Results
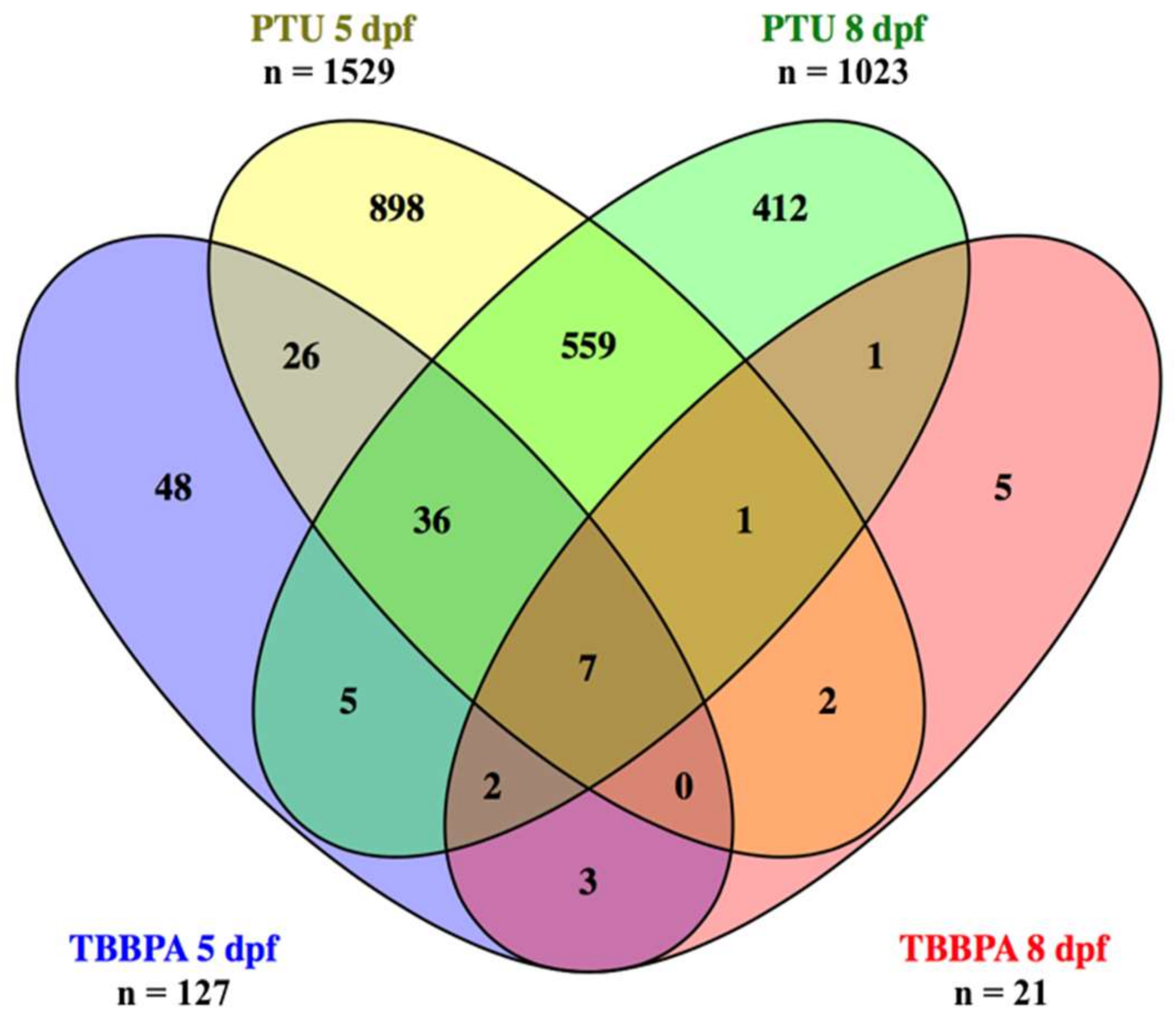
2.1. Overview of Total Numbers of Differentially Expressed Transcripts
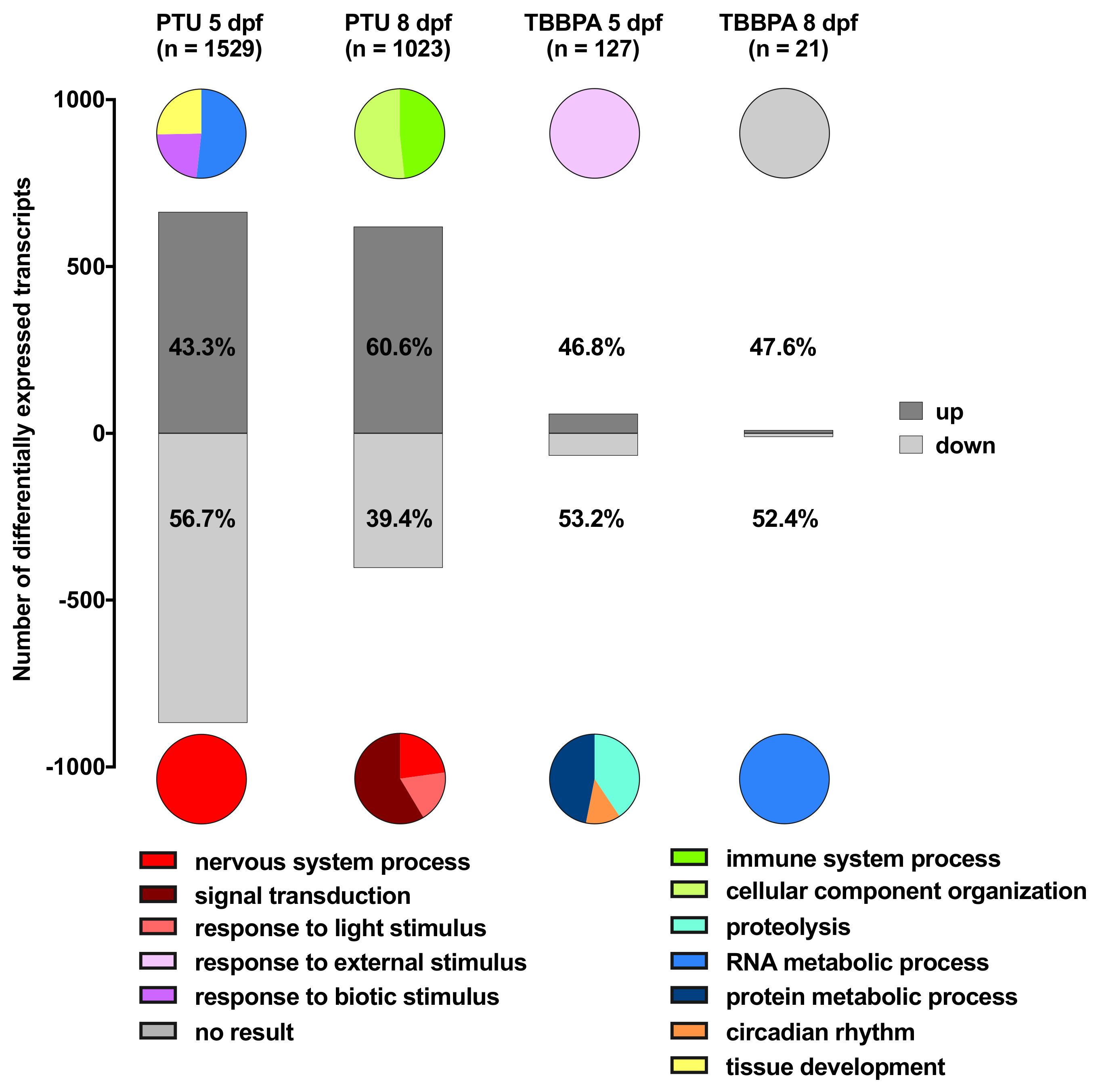
2.2. Descriptive Analysis: Biological Function of Up- or Down-Regulated Transcripts for Each Treatment Group
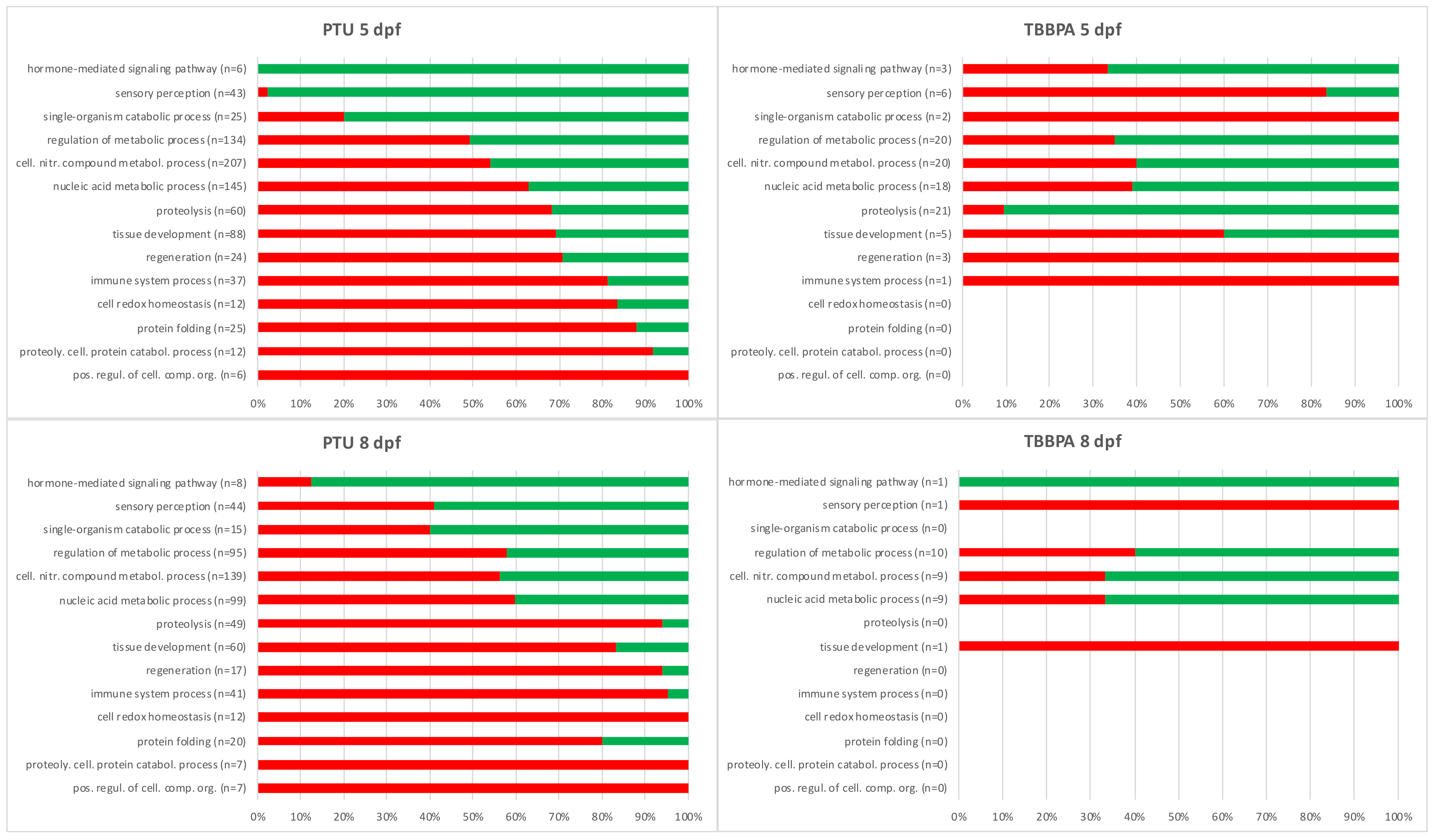
2.3. Comparative Analysis: Biological Function of Transcripts with Distinct Expression Patterns Across Treatments
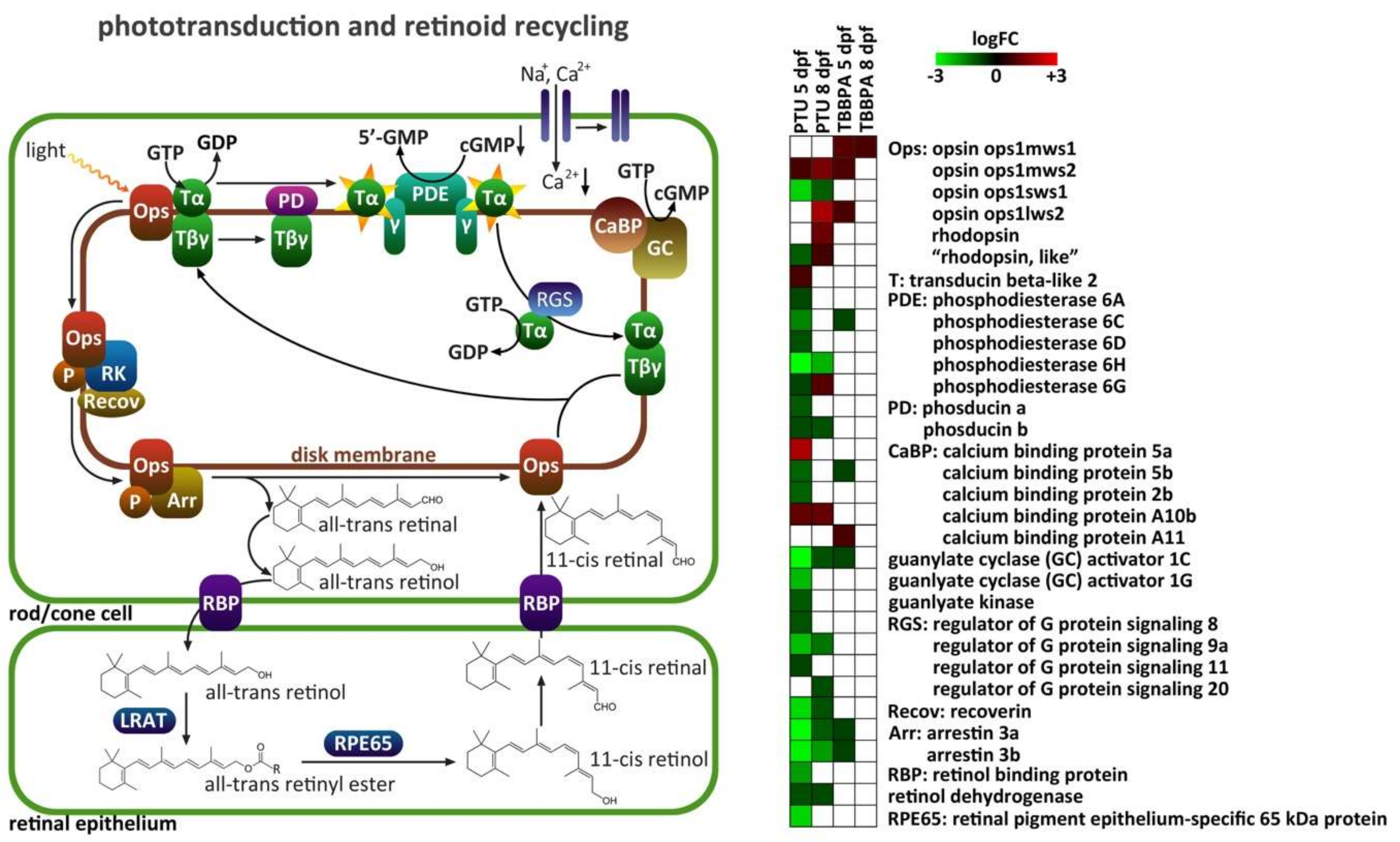
2.4. Specific Analysis: Effects on Vision-Related Transcripts
3. Discussion
3.1. General Response Pattern
3.2. Effects on Vision-Related Transcripts at 5 dpf
3.3. Evidence for Regeneration at 8 dpf
4. Materials and Methods
4.1. Ethics Statement
4.2. Test Chemicals
4.3. Zebrafish Maintenance and Exposure
4.4. Sampling and RNA Isolation
4.5. cRNA Labelling and Hybridization
4.6. Analysis of Microarray Data
4.6.1. Statistical Analysis
4.6.2. Biological Interpretation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| THDC | Thyroid hormone disrupting chemical |
| DPF | Days post fertilization |
| PTU | Propylthiouracil |
| TBBPA | Tetrabromobisphenol-A |
| GO | Gene ontology |
| EDC | Endocrine disrupting chemical |
| TH | Thyroid hormone |
| HPF | Hours post fertilization |
| TR | Thyroid receptor |
References
- Owen, R.; Jobling, S. Environmental science: The hidden costs of flexible fertility. Nature 2012, 485, 441. [Google Scholar] [CrossRef] [PubMed]
- Matthiessen, P.; Wheeler, J.R.; Weltje, L. A review of the evidence for endocrine disrupting effects of current-use chemicals on wildlife populations. Crit. Rev. Toxicol. 2018, 48, 195–216. [Google Scholar] [CrossRef]
- Brown, S.B.; Adams, B.A.; Cyr, D.G.; Eales, J.G. Contaminant effects on the teleost fish thyroid. Environ. Toxicol. Chem. SETAC 2004, 23, 1680–1701. [Google Scholar] [CrossRef]
- Boas, M.; Feldt-Rasmussen, U.; Main, K.M. Thyroid effects of endocrine disrupting chemicals. Mol. Cell. Endocrinol. 2012, 355, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Power, D.M.; Llewellyn, L.; Faustino, M.; Nowell, M.A.; Björnsson, B.T.; Einarsdottir, I.E.; Canario, A.V.M.; Sweeney, G.E. Thyroid hormones in growth and development of fish. Comparat. Biochem. Physiol. Part. C Toxicol. Pharmacol. 2001, 130, 447–459. [Google Scholar]
- Darras, V.M.; Houbrechts, A.M.; Van Herck, S.L.J. Intracellular thyroid hormone metabolism as a local regulator of nuclear thyroid hormone receptor-mediated impact on vertebrate development. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2015, 1849, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, B.L.; Kahana, A. Thyroid hormone and retinoic acid interact to regulate zebrafish craniofacial neural crest development. Dev. Biol. 2013, 373, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.; Hurley, J.B.; Dierks, B.; Srinivas, M.; Saltó, C.; Vennström, B.; Reh, T.A.; Forrest, D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 2001, 27, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Trimarchi, J.M.; Harpavat, S.; Billings, N.A.; Cepko, C.L. Thyroid hormone components are expressed in three sequential waves during development of the chick retina. BMC Dev. Biol. 2008, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, S.; Thisse, B.; Tavares, R.; Sachs, L.; Chaumot, A. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007, 3, e188. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Macaulay, L.J.; Kwok, K.W.; Hinton, D.E.; Ferguson, P.L.; Stapleton, H.M. The PBDE metabolite 6-OH-BDE 47 affects melanin pigmentation and THRβ MRNA expression in the eye of zebrafish embryos. Endocr. Disruptors 2014, 2, e969072. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ptak, D.; Zhang, L.; Walls, E.K.; Zhong, W.; Leung, Y.F. Phenylthiourea specifically reduces zebrafish eye size. PLoS ONE 2012, 7, e40132. [Google Scholar] [CrossRef] [PubMed]
- Reider, M.; Connaughton, V.P. Effects of Low-Dose Embryonic Thyroid Disruption and Rearing Temperature on the Development of the Eye and Retina in Zebrafish. Birth Defects Res. B 2014, 101, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Heijlen, M.; Houbrechts, A.M.; Bagci, E.; Van Herck, S.L.J.; Kersseboom, S.; Esguerra, C.V.; Blust, R.; Visser, T.J.; Knapen, D.; Darras, V.M. Knockdown of Type 3 Iodothyronine Deiodinase Severely Perturbs Both Embryonic and Early Larval Development in Zebrafish. Endocrinology 2014, 155, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Bagci, E.; Heijlen, M.; Vergauwen, L.; Hagenaars, A.; Houbrechts, A.M.; Esguerra, C.V.; Blust, R.; Darras, V.M.; Knapen, D. Deiodinase knockdown during early zebrafish development affects growth, development, energy metabolism, motility and phototransduction. PLoS ONE 2015, 10, e0123285. [Google Scholar] [CrossRef]
- Marelli, F.; Carra, S.; Agostini, M.; Cotelli, F.; Peeters, R.; Chatterjee, K.; Luca Persani, M.D. Patterns of thyroid hormone receptor expression in zebrafish and generation of a novel model of resistance to thyroid hormone action. Mol. Cell. Endocrinol. 2016, 424, 1–45. [Google Scholar] [CrossRef]
- Van der Ven, L.T.M.; van den Brandhof, E.-J.; Vos, J.H.; Power, D.M.; Wester, P.W. Effects of the Antithyroid Agent Propylthiouracil in a Partial Life Cycle Assay with Zebrafish. Environ. Sci. Technol. 2006, 40, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Hamers, T. In Vitro Profiling of the Endocrine-Disrupting Potency of Brominated Flame Retardants. Toxicol. Sci. 2006, 92, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Kotthoff, M.; Rüdel, H.; Jürling, H. Detection of tetrabromobisphenol A and its mono- and dimethyl derivatives in fish, sediment and suspended particulate matter from European freshwaters and estuaries. Anal. Bioanal. Chem. 2017, 409, 3685–3694. [Google Scholar] [CrossRef]
- Harrad, S.; Abdallah, M.A.-E.; Rose, N.L.; Turner, S.D.; Davidson, T.A. Current-use brominated flame retardants in water, sediment, and fish from English lakes. Environ. Sci. Technol. 2009, 43, 9077–9083. [Google Scholar] [CrossRef]
- Morris, S.; Allchin, C.R.; Zegers, B.N.; Haftka, J.J.H.; Boon, J.P.; Belpaire, C.; Leonards, P.E.G.; Van Leeuwen, S.P.J.; de Boer, J. Distribution and fate of HBCD and TBBPA brominated flame retardants in North Sea estuaries and aquatic food webs. Environ. Sci. Technol. 2004, 38, 5497–5504. [Google Scholar] [CrossRef]
- Cariou, R.; Antignac, J.-P.; Zalko, D.; Berrebi, A.; Cravedi, J.-P.; Maume, D.; Marchand, P.; Monteau, F.; Riu, A.; Andre, F.; et al. Exposure assessment of French women and their newborns to tetrabromobisphenol-A: Occurrence measurements in maternal adipose tissue, serum, breast milk and cord serum. Chemosphere 2008, 73, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Baumann, L.; Ros, A.; Rehberger, K.; Neuhauss, S.C.F.; Segner, H. Thyroid disruption in zebrafish (Danio rerio) larvae: Different molecular response patterns lead to impaired eye development and visual functions. Aquat. Toxicol. 2016, 172, 44–55. [Google Scholar] [CrossRef]
- Chan, W.; Chan, K. Disruption of the hypothalamic-pituitary-thyroid axis in zebrafish embryo–larvae following waterborne exposure to BDE-47, TBBPA and BPA. Aquat. Toxicol. 2012, 108, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Parsons, A.; Lange, A.; Hutchinson, T.H.; Miyagawa, S.; Iguchi, T.; Kudoh, T.; Tyler, C.R. Molecular mechanisms and tissue targets of brominated flame retardants, BDE-47 and TBBPA, in embryo-larval life stages of zebrafish (Danio rerio). Aquat. Toxicol. 2019, 209, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Houbrechts, A.M.; Vergauwen, L.; Bagci, E.; Van houcke, J.; Heijlen, M.; Kulemeka, B.; Hyde, D.R.; Knapen, D.; Darras, V.M. Deiodinase knockdown affects zebrafish eye development at the level of gene expression, morphology and function. Mol. Cell. Endocrinol. 2016, 424, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Gestri, G.; Link, B.A.; Neuhauss, S.C.F. The visual system of zebrafish and its use to model human ocular Diseases. Dev. Neurobio. 2012, 72, 302–327. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Sánchez, B.; Clément, A.; Phillips, J.B.; Westerfield, M. Zebrafish Models of Human Eye and Inner Ear Diseases; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; Volume 138, pp. 415–467. [Google Scholar]
- Bhumika, S.; Darras, V.M. Role of thyroid hormones in different aspects of nervous system regeneration in vertebrates. Gen. Comp. Endocrinol. 2014, 203, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.; de Lange, P.; Lombardi, A.; Silvestri, E.; Lanni, A.; Goglia, F. Metabolic effects of thyroid hormone derivatives. Thyroid 2008, 18, 239–253. [Google Scholar] [CrossRef]
- Casals-Casas, C.; Desvergne, B. Endocrine Disruptors: From Endocrine to Metabolic Disruption. Annu. Rev. Physiol. 2011, 73, 135–162. [Google Scholar] [CrossRef]
- Jugan, M.-L.; Levi, Y.; Blondeau, J.-P. Endocrine disruptors and thyroid hormone physiology. Biochem. Pharmacol. 2010, 79, 939–947. [Google Scholar] [CrossRef] [PubMed]
- De Wit, M.; Keil, D.; Remmerie, N.; Ven, K.V.D.; Brandhof, E.-J.V.D.; Knapen, D.; Witters, E.; Coen, W.D. Molecular targets of TBBPA in zebrafish analysed through integration of genomic and proteomic approaches. Chemosphere 2008, 74, 96–105. [Google Scholar] [CrossRef]
- Viets, K.; Eldred, K.C.; Johnston, R.J. Mechanisms of Photoreceptor Patterning in Vertebrates and Invertebrates. Trends Genet. 2016, 32, 1–22. [Google Scholar] [CrossRef]
- Suzuki, S.C.; Bleckert, A.; Williams, P.R.; Takechi, M.; Kawamura, S.; Wong, R.O.L. Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl. Acad. Sci. USA 2013, 110, 15109–15114. [Google Scholar] [CrossRef] [PubMed]
- Orozco, A.; Valverde-R, C. Thyroid hormone deiodination in fish. Thyroid 2005, 15, 799–813. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Wang, M.; Gui, W.; Zhao, Y.; Yu, L.; Zhu, G. Changes in Thyroid Hormone Levels during Zebrafish Development. Zool. Sci. 2012, 29, 181–184. [Google Scholar] [CrossRef]
- Chen, J.; Tanguay, R.L.; Simonich, M.; Nie, S.; Zhao, Y.; Li, L.; Bai, C.; Dong, Q.; Huang, C.; Lin, K. TBBPA chronic exposure produces sex-specific neurobehavioral and social interaction changes in adult zebrafish. Neurotoxicol. Teratol. 2016, 56, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kim, S.-J.; Lee, W.K.; Moon, S.K.; Kwak, S.; Choe, S.-K.; Park, R. Tetrabromobisphenol-A induces apoptotic death of auditory cells and hearing loss. Biochem. Biophys. Res. Commun. 2016, 478, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jarema, K.A.; Hunter, D.L.; Shaffer, R.M.; Behl, M.; Padilla, S. Acute and developmental behavioral effects of flame retardants and related chemicals in zebrafish. Neurotoxicol. Teratol. 2015, 52, 194–209. [Google Scholar] [CrossRef]
- Fraser, T.W.K.; Khezri, A.; Jusdado, J.G.H.; Lewandowska-Sabat, A.M.; Henry, T.; Ropstad, E. Toxicant induced behavioural aberrations in larval zebrafish are dependent on minor methodological alterations. Toxicol. Lett. 2017, 276, 62–68. [Google Scholar] [CrossRef]
- Bouzaffour, M.; Rampon, C.; Ramaugé, M.; Courtin, F.; Vriz, S. Implication of type 3 deiodinase induction in zebrafish fin regeneration. Gen. Comp. Endocrinol. 2010, 168, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Bhumika, S.; Lemmens, K.; Vancamp, P.; Moons, L.; Darras, V.M. Decreased thyroid hormone signaling accelerates the reinnervation of the optic tectum following optic nerve crush in adult zebrafish. Mol. Cell. Neurosci. 2015, 68, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Glatt, C.M.; Ouyang, M.; Welsh, W.; Green, J.W.; Connor, J.O.; Frame, S.R.; Everds, N.E.; Poindexter, G.; Snajdr, S.; Delker, D.A. Molecular Characterization of Thyroid Toxicity: Anchoring Gene Expression Profiles to Biochemical and Pathologic End Points. Environ. Health Perspect. 2005, 113, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, S.; Sun, F.; Zhang, M.; Wu, F.; Xu, F.; Ding, Z. Protective effects of puerarin against tetrabromobisphenol a-induced apoptosis and cardiac developmental toxicity in zebrafish embryo-larvae. Environ. Toxicol. 2014, 30, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Quesada-García, A.; Encinas, P.; Valdehita, A.; Baumann, L.; Segner, H.; Coll, J.M.; Navas, J.M. Thyroid active agents T3 and PTU differentially affect immune gene transcripts in the head kidney of rainbow trout (Oncorynchus mykiss). Aquat. Toxicol. 2016, 174, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Quesada-García, A.; Valdehita, A.; Kropf, C.; Casanova-Nakayama, A.; Segner, H.; Navas, J.M. Thyroid signaling in immune organs and cells of the teleost fish rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2014, 38, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.H.; Sin, Y.M.; Gong, Z.; Lam, T.J. Effects of thyroid hormone on the development of immune system in zebrafish. Gen. Comp. Endocrinol. 2005, 142, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Geven, E.J.W.; Klaren, P.H.M. The teleost head kidney: Integrating thyroid and immune signalling. Dev. Comp. Immunol. 2016, 66, 1–21. [Google Scholar]
- EU Publication Office Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the Protection of Animals used for Scientific Purposes. Text with EEA Relevance. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=celex:32010L0063 (accessed on 25 January 2019).
- OECD. Fish Embryo Acute Toxicity (FET) Test; Test No. 236; OECD: Paris, France, 2013; pp. 1–22. [Google Scholar]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Valentim, A.M.; van Eeden, F.J.; Strähle, U.; Olsson, I.A.S. Euthanizing zebrafish legally in Europe. EMBO Rep. 2016, 17, 1688–1689. [Google Scholar] [CrossRef] [PubMed]
- Kerr, M.K.; Churchill, G.A. Experimental design for gene expression microarrays. Biostatistics 2001, 2, 183–201. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann, A.; Gentleman, R.; Huber, W. Array quality metrics—A bioconductor package for quality assessment of microarray data. Bioinformatics 2009, 25, 415–416. [Google Scholar] [CrossRef] [PubMed]
- Sclep, G.; Allemeersch, J.; Liechti, R.; De Meyer, B.; Beynon, J.; Bhalerao, R.; Moreau, Y.; Nietfeld, W.; Renou, J.-P.; Reymond, P.; et al. CATMA, a comprehensive genome-scale resource for silencing and transcript profiling of Arabidopsis genes. BMC Bioinform. 2007, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Silver, J.; Oshlack, A.; Holmes, M.; Diyagama, D.; Holloway, A.; Smyth, G.K. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007, 23, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G. Statistical Applications in Genetics and Molecular Biology; Berkeley Electronic Press: Berkeley, CA, USA, 2004; Volume 3. [Google Scholar]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Vergauwen, L.; Benoot, D.; Blust, R.; Knapen, D. Long-term warm or cold acclimation elicits a specific transcriptional response and affects energy metabolism in zebrafish. Comp. Biochem. Physiol. Part A 2010, 157, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. JSTOR 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
| 0 dpf | 1 dpf | 2 dpf | 3 dpf | 4 dpf | 5 dpf | 6 dpf | 7 dpf | 8 dpf | |
|---|---|---|---|---|---|---|---|---|---|
| Control | W | W | W | W | W | X | |||
| Control recovery | W | W | W | W | W | W | W | W | X |
| PTU (350 mg/L) | C | C | C | C | C | X | |||
| PTU recovery | C | C | C | C | C | W | W | W | X |
| TBBPA (200 µg/L) | C | C | C | C | C | X | |||
| TBBPA recovery | C | C | C | C | C | W | W | W | X |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baumann, L.; Segner, H.; Ros, A.; Knapen, D.; Vergauwen, L. Thyroid Hormone Disruptors Interfere with Molecular Pathways of Eye Development and Function in Zebrafish. Int. J. Mol. Sci. 2019, 20, 1543. https://doi.org/10.3390/ijms20071543
Baumann L, Segner H, Ros A, Knapen D, Vergauwen L. Thyroid Hormone Disruptors Interfere with Molecular Pathways of Eye Development and Function in Zebrafish. International Journal of Molecular Sciences. 2019; 20(7):1543. https://doi.org/10.3390/ijms20071543
Chicago/Turabian StyleBaumann, Lisa, Helmut Segner, Albert Ros, Dries Knapen, and Lucia Vergauwen. 2019. "Thyroid Hormone Disruptors Interfere with Molecular Pathways of Eye Development and Function in Zebrafish" International Journal of Molecular Sciences 20, no. 7: 1543. https://doi.org/10.3390/ijms20071543
APA StyleBaumann, L., Segner, H., Ros, A., Knapen, D., & Vergauwen, L. (2019). Thyroid Hormone Disruptors Interfere with Molecular Pathways of Eye Development and Function in Zebrafish. International Journal of Molecular Sciences, 20(7), 1543. https://doi.org/10.3390/ijms20071543







