Effects of Pressure and Temperature on the Atomic Fluctuations of Dihydrofolate Reductase from a Psychropiezophile and a Mesophile
Abstract
1. Introduction
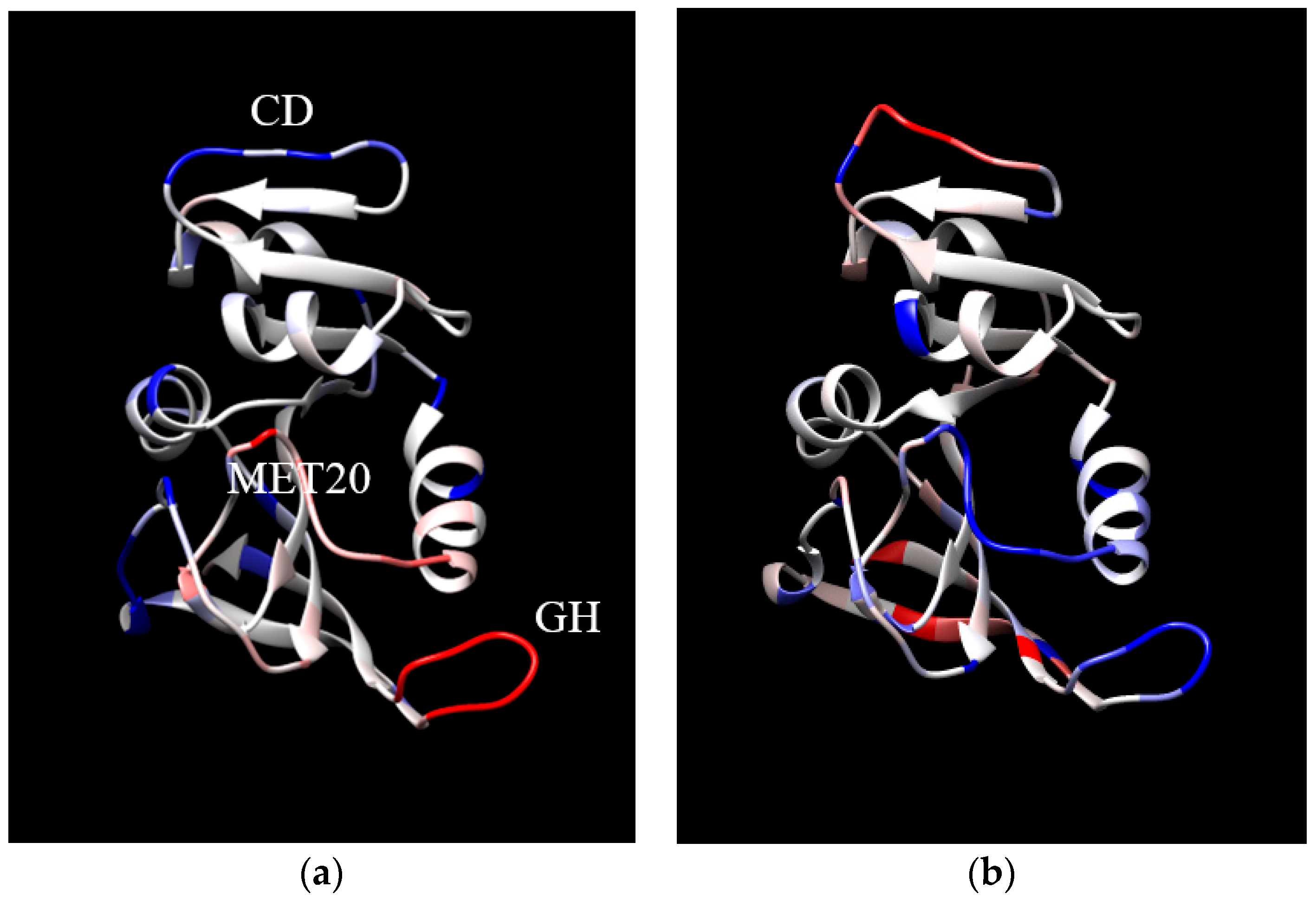
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Seelig, J.; Schonfeld, H.-J. Thermal protein unfolding by differential scanning calorimetry and circular dichroism spectroscopy Two-state model versus sequential unfolding. Q. Rev. Biophys. 2016, 49, e9. [Google Scholar] [CrossRef]
- Winter, R. High pressure effects in molecular bioscience. In Chemistry at Extreme Conditions; Manaa, M.R., Ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2005; pp. 29–82. [Google Scholar]
- Meersman, F.; Daniel, I.; Bartlett, D.H.; Winter, R.; Hazael, R.; McMillan, P.F. High-pressure biochemistry and biophysics. In Carbon in Earth; Hazen, R.M., Jones, A.P., Baross, J.A., Eds.; Mineralogical Society of America: Washington, DC, USA, 2013; Volume 75, pp. 607–648. [Google Scholar]
- Royer, C.A. Review: Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta 2002, 1595, 201–209. [Google Scholar] [CrossRef]
- Hummer, G.; Garde, S.; Garcia, A.E.; Paulaitis, M.E.; Pratt, L.R. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 1552–1555. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.L.; Weber, G. Pressure stability of proteins. Ann. Rev. Phys. Chem. 1993, 44, 89–113. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Caro, J.A.; Noberto, D.R.; Barthe, P.; Roumestand, C.; Schlessman, J.L.; Garcia, A.E.; Garcia-Moreno, B.E.; Royer, C.A. Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 6945–6950. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Hummer, G.; Quillin, M.L.; Matthews, B.W.; Gruner, S.M. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc. Natl. Acad. Sci. USA 2005, 46, 16668–16671. [Google Scholar] [CrossRef] [PubMed]
- Nagae, T.; Kawamura, T.; Chavas, L.M.G.; Niwa, K.; Hasegawa, M.; Kato, C.; Watanabe, N. High-pressure-induced water penetration into 3-isopropylmalate dehydrogenase. Acta Crysta 2012, D68, 300–309. [Google Scholar] [CrossRef]
- Nagae, T.; Yamada, H.; Watanabe, N. High-pressure protein crystal structure analysis of Escherichia coli dihydrofolate reductase complexed with folate and NADP+. Acta Crysta 2018, D74, 895–905. [Google Scholar] [CrossRef]
- Panick, G.; Malessa, R.; Winter, R.; Rapp, G.; Frye, K.J.; Royer, C.A. Structural characterization of the pressure-denatured state and unfolding/refolding kinetics of staphylococcal nuclease by synchrotron small-angle X-ray scattering and Fourier-transform infrared spectroscopy. J. Mol. Biol. 1998, 275, 389–402. [Google Scholar] [CrossRef]
- Vidugiris, G.J.; Markley, J.L.; Royer, C.A. Evidence for a molten globule-like transition state in protein folding from determination of activation volumes. Biochemistry 1995, 34, 4909–4912. [Google Scholar] [CrossRef]
- McCammon, J.A.; Gelin, B.R.; Karplus, M. Dynamics of folded proteins. Nature 1977, 267, 585–590. [Google Scholar] [CrossRef] [PubMed]
- McCammon, J.A.; Harvey, S.C. Dynamics of Proteins and Nucleic Acids; Cambridge University Press: New York, NY, USA, 1987. [Google Scholar]
- Frauenfelder, H.; Petkso, G.A.; Tsernoglou, D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature 1979, 280, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Yayanos, A.A. Microbiology to 10,500 meters in the deep sea. Ann. Rev. Microbiol. 1995, 49, 777–805. [Google Scholar] [CrossRef] [PubMed]
- Kato, C. Microbiology of piezophiles in deep-sea environments. In Extremophiles: Microbiology and Biotechnology; Anitori, R.P., Ed.; Caister Academic Press: Poole, UK, 2012; pp. 233–263. [Google Scholar]
- Gross, M.; Jaenicke, R. Review: Proteins under pressure: The influence of high hydrostatic pressure on structure, function and assembly of proteins and protein complexes. Eur. J. Biochem. 1994, 221, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Nagae, T.; Kato, C.; Watanabe, N. Structural analysis of 3-isopropylmalate dehydrogenase from the obligate piezophile Shewanella benthica DB21MT-2 and the nonpiezophile Shewanella oneidensis MR-1. Acta Crysta 2012, F68, 265–268. [Google Scholar]
- Ohmae, E.; Murakami, C.; Tate, S.-I.; Gekko, K.; Hata, K.; Akasaka, K.; Kato, C. Pressure dependence of activity and stability of dihydrofolate reductases of the deep-sea bacterium Moritella profunda and Escherichia coli. Biochim. Biophys. Acta 2012, 1824, 511–519. [Google Scholar] [CrossRef]
- Boonyaratanakornkit, B.B.; Park, C.B.; Clark, D.S. Review: Pressure effects on intra- and intermolecular interactions within proteins. Biochim. Biophys. Acta 2002, 1595, 235–249. [Google Scholar] [CrossRef]
- Tanaka, N.; Ikeda, C.; Kanaori, K.; Hiraga, K.; Konno, T.; Kuniga, S. Pressure effect on the conformational fluctuation of apomyoglobin in the native state. Biochemistry 2000, 39, 12063–12068. [Google Scholar] [CrossRef]
- Somero, G.N. Proteins and temperature. Ann. Rev. Physiol. 1995, 57, 453–468. [Google Scholar] [CrossRef]
- Jaenicke, R.; Závodszky, P. Proteins under extreme physical conditions. FEBS Lett. 1990, 268, 344–349. [Google Scholar] [CrossRef]
- D’Amico, S.; Marx, J.-C.; Gerday, C.; Feller, G. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 2003, 278, 7891–7896. [Google Scholar] [CrossRef]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef]
- Xu, Y.; Nogi, Y.; Kato, C.; Liang, Z.; Rüger, H.-J.; De Kegel, D.; Glansdorff, N. Moritella profunda sp. nov. and Moritella abyssi sp. nov., two psychropiezophilic organisms isolated from deep Atlantic sediments. Int. J. Sys. Evol. Microbiol. 2003, 53, 533–538. [Google Scholar] [CrossRef]
- Evans, R.M.; Behiry, E.M.; Tey, L.-H.; Guo, J.; Loveridge, E.J.; Allemann, R.K. Catalysis by dihydrofolate reductase from the psychropiezophile Moritella profunda. ChemBioChem 2010, 11, 2010–2017. [Google Scholar] [CrossRef]
- Swanwick, R.S.; Shrimpton, P.J.; Allemann, R.K. Pivotal role of gly 121 in dihydrofolate reductase from Escherichia coli: The altered structure of a mutant enzyme may form the basis of its diminished catalytic performance. Biochemistry 2004, 43, 4119–4127. [Google Scholar] [CrossRef]
- Ohmae, E.; Miyashita, Y.; Kato, C. Thermodynamic and functional characteristics of deep-sea enzymes revealed by pressure effects. Extremophiles 2013, 17, 701–709. [Google Scholar] [CrossRef]
- Huang, Q.; Rodgers, J.M.; Hemley, R.J.; Ichiye, T. Extreme biophysics: Enzymes under pressure. J. Comput. Chem. 2017, 38, 1174–1182. [Google Scholar] [CrossRef]
- Rodgers, J.M.; Hemley, R.J.; Ichiye, T. Quasiharmonic analysis of protein energy landscapes from pressure-temperature molecular dynamics simulations. J. Chem. Phys. 2017, 147, 125103. [Google Scholar] [CrossRef]
- Ringe, D.; Petsko, G.A. The glass transition in protein dynamics: What it is, why it occurs, and how to exploit it. Biophys. Chem. 2003, 105, 667–680. [Google Scholar] [CrossRef]
- Huang, Q.; Rodgers, J.M.; Hemley, R.J.; Ichiye, T. Quasi-harmonic analysis of the energy landscapes of dihydrofolate reductase from piezophiles and mesophiles. J. Phys. Chem. B 2018, 122, 5527–5533. [Google Scholar] [CrossRef]
- Akasaka, K. Probing conformational fluctuation of proteins by pressure perturbation. Chem. Rev. 2006, 108, 1814–1835. [Google Scholar] [CrossRef]
- Huang, Q.; Tran, K.N.; Rodgers, J.M.; Bartlett, D.H.; Hemley, R.J.; Ichiye, T. A molecular perspective on the limits of life: Enzymes under pressure. Cond. Matter Phys. 2016, 19, 1–16. [Google Scholar] [CrossRef]
- Meinhold, L.; Smith, J.C.; Kitao, A.; Zewail, A.H. Picosecond fluctuating protein energy landscape mapped by pressure-temperature molecular dynamics simulation. Proc. Natl. Acad. Sci. USA 2007, 104, 17261–17265. [Google Scholar] [CrossRef]
- Harris, K.R.; Newitt, P.J. Self-Diffusion of Water at Low Temperatures and High Pressure. J. Chem. Eng. Data 1997, 42, 346–348. [Google Scholar] [CrossRef]
- Woolf, L.A. Tracer diffusion of tritiated-water (THO) in ordinary water (H2O) under pressure. J. Chem. Soc. 1975, 71, 784–796. [Google Scholar] [CrossRef]
- Krynicki, K.; Green, C.D.; Sawyer, D.W. Pressure and temperature dependence of self-diffusion in water. Faraday Disc. Chem. Soc. 1978, 66, 199–208. [Google Scholar] [CrossRef]
- Harris, K.R.; Woolf, L.A. Pressure and temperature dependence of the self diffusion coefficient of water and oxygen-18 water. J. Chem. Soc. 1980, 76, 377–385. [Google Scholar] [CrossRef]
- Paci, E. High pressure simulations of biomolecules. Biochim. Biophys. Acta 2002, 1595, 185–200. [Google Scholar] [CrossRef]
- Marchi, M.; Akasaka, K. Simulation of hydrated BPTI at high pressure: Changes in hydrogen bonding and its relation with NMR experiments. J. Phys. Chem. B 2001, 105, 711–714. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.; Mittal, J.; Feig, M.; MacKerell, A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- MacKerell, A.D., Jr.; Bashford, D.; Bellot, M.; Dunbrack, R.L., Jr.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef]
- Horn, H.W.; Swope, W.C.; Pitera, J.W.; Madura, J.D.; Dick, T.J.; Hura, G.L.; Head-Gordon, T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J. Chem. Phys. 2004, 120, 9665–9678. [Google Scholar] [CrossRef]
- Tran, K.N.; Tan, M.-L.; Ichiye, T. A single-site multipole model for liquid water. J. Chem. Phys. 2016, 145, 034501. [Google Scholar] [CrossRef]
- Huang, Q.; Rodgers, J.M.; Hemley, R.J.; Ichiye, T. Adaptations for pressure and temperature effects on loop motion in Escherichia coli and Moritella profunda dihydrofolate reductase. High Pressure Res. 2019. accepted. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Hata, K.; Tanaka, T.; Murakami, C.; Ohmae, E.; Gekko, K.; Shiro, Y.; Akasaka, K. Moritella profunda Dihydrofolate reductase complex with NADP+ and Folate. PDB 2010, unpublished. [Google Scholar]
- Sawaya, M.R.; Kraut, J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: Crystallographic evidence. Biochemistry 1997, 36, 586–603. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., III; MacKerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Miller, B.T.; Singh, R.P.; Klauda, J.B.; Hodoscek, M.; Brooks, B.R.; Woodcock, H.L. CHARMMing: A new, flexible web portal for CHARMM. J. Chem. Inf. Model. 2008, 48, 1920–1929. [Google Scholar] [CrossRef]
- CGenFF. Available online: https://cgenff.paramchem.org/ (accessed on 20 May 2016).
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- York, D.M.; Pedersen, L.G.; Darden, T.A. The effect of long-range electrostatic interactions in simulations of macromolecular crystals: A comparison of the Ewald and trucated list methods. J. Chem. Phys. 1993, 99, 8345–8348. [Google Scholar] [CrossRef]
- Rapaport, D.C. Hydrogen-bonds in water network organization and lifetimes. Mol. Phys. 1983, 50, 1151–1162. [Google Scholar] [CrossRef]
- Luzar, A.; Chandler, D. Effect of environment on hydrogen bond dynamics in liquid water. Phys. Rev. Lett. 1996, 76, 928–931. [Google Scholar] [CrossRef]
- Teng, X.; Ichiye, T. Molecular dynamics study of the effects of TMAO on aqueous solutions of urea. J. Phys. Chem. B 2019, 123, 1108–1115. [Google Scholar] [CrossRef]
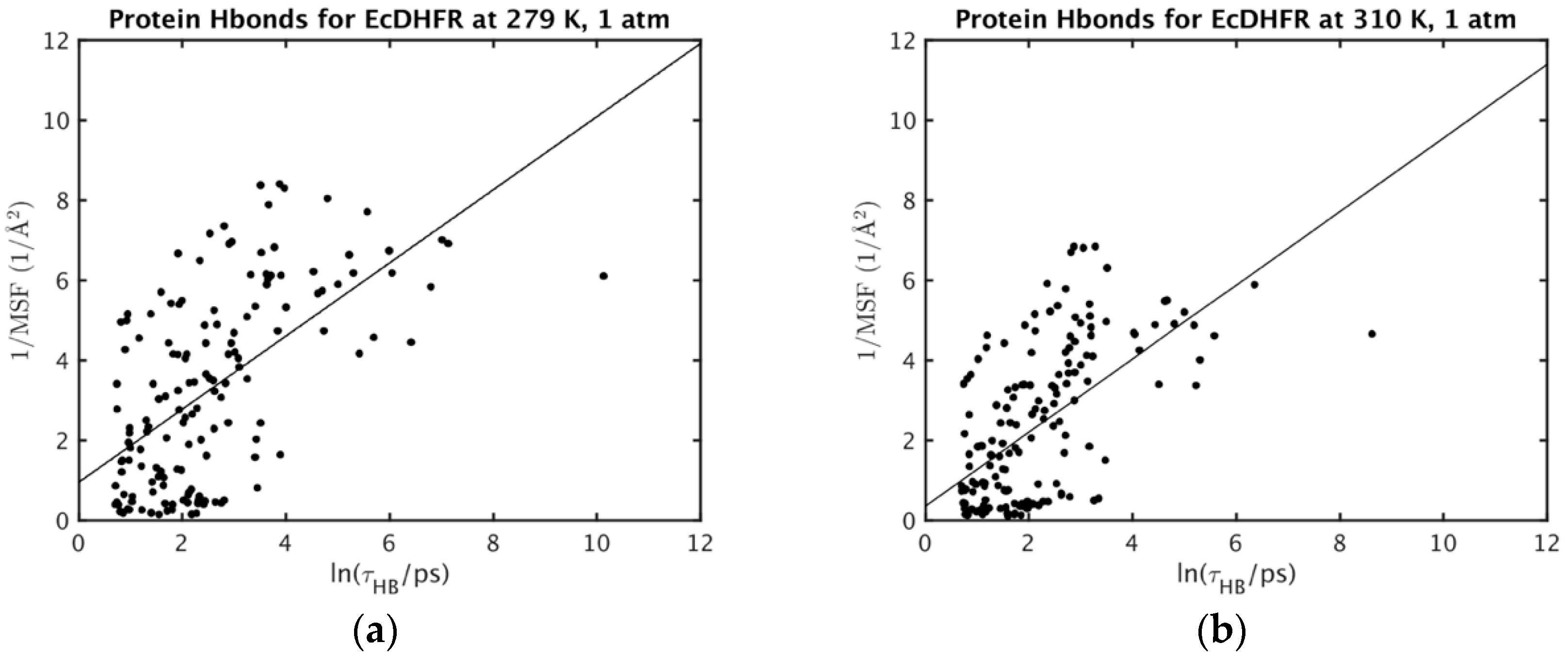
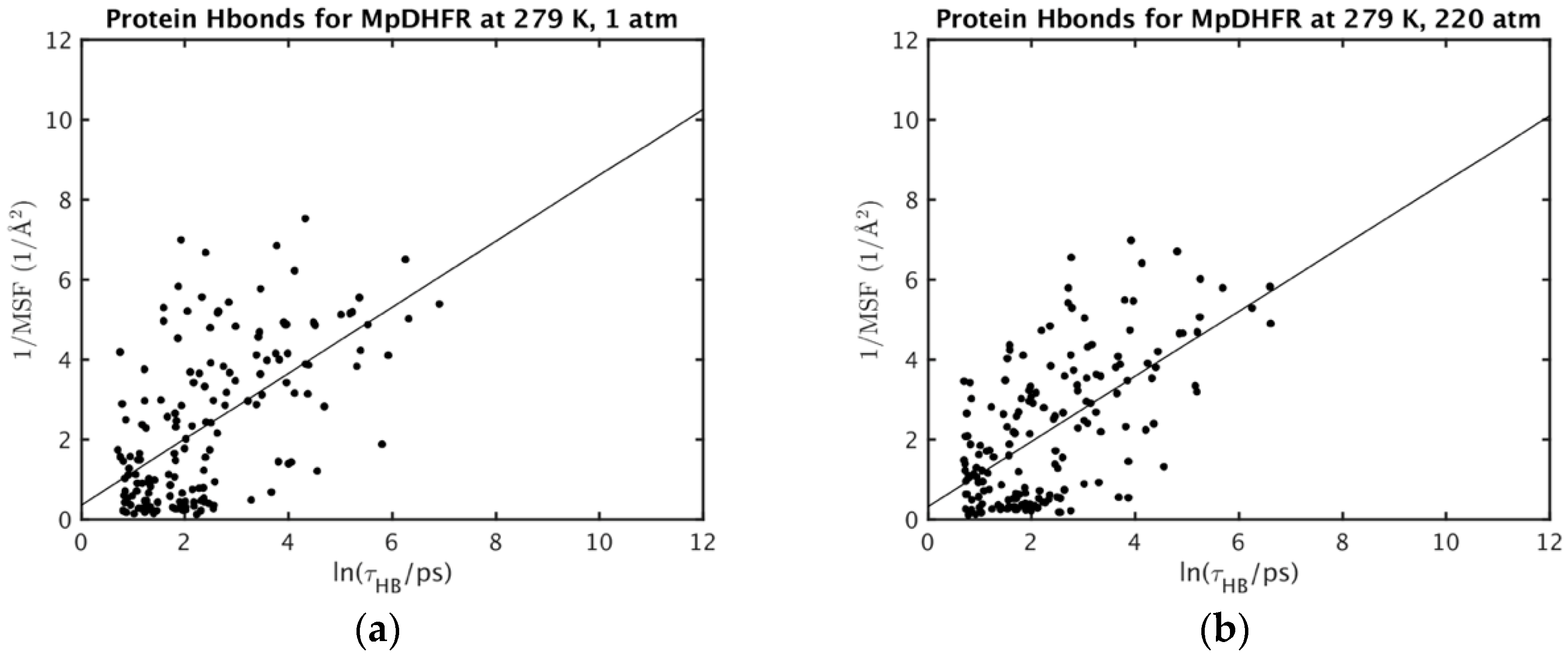
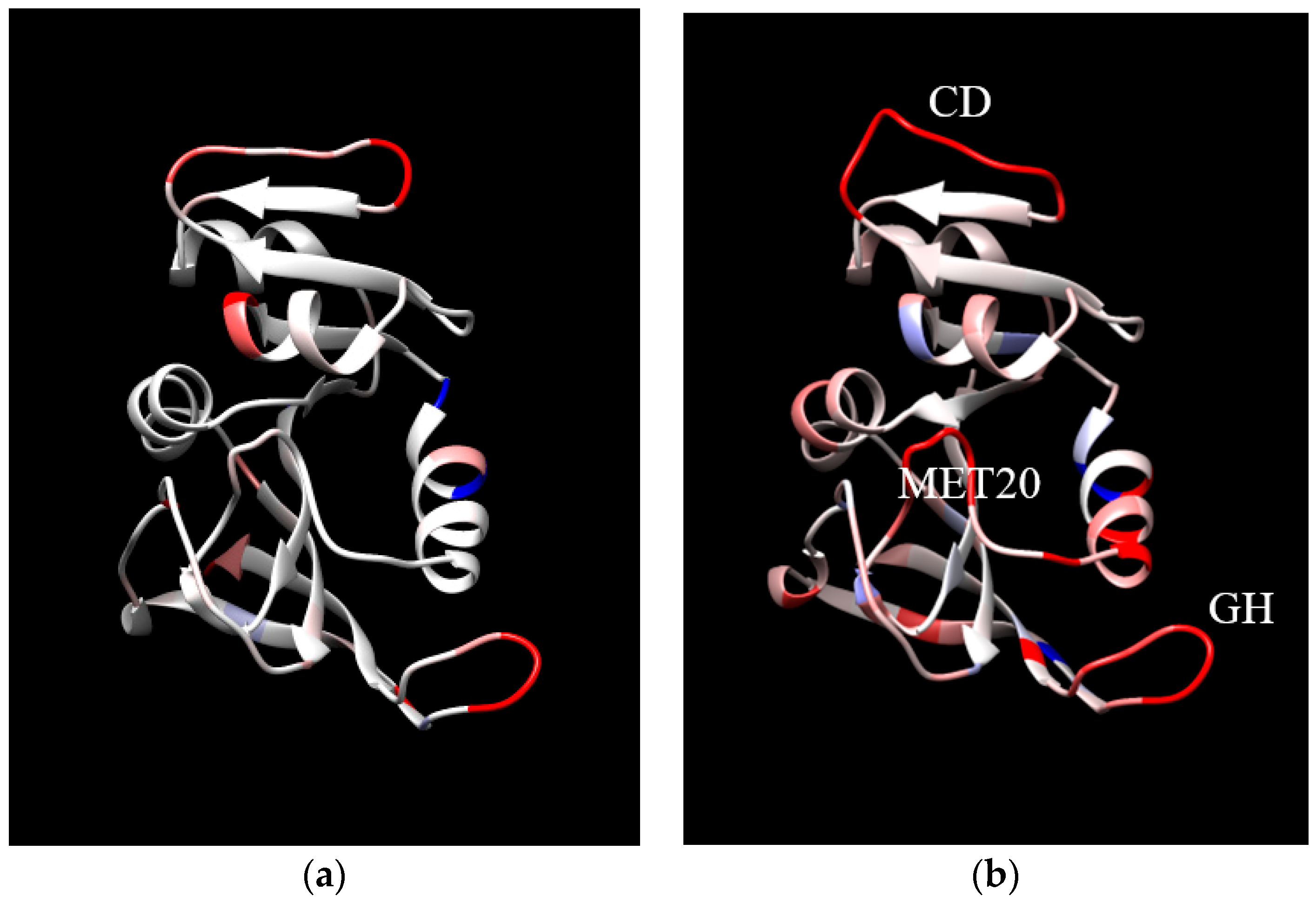
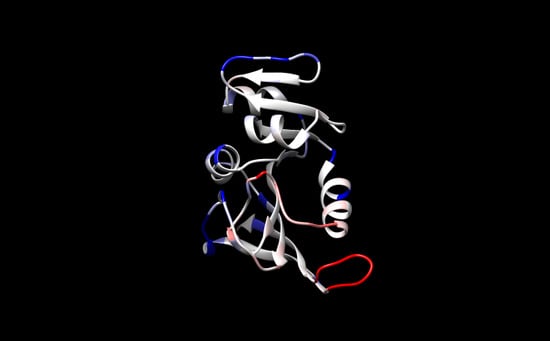
| Protein (T, K; P, bar) | MSF (Å2) * | Vapp/NHA (Å3) * | Nw,in | NHB | τHB (ps) |
|---|---|---|---|---|---|
| EcDHFR (279; 1) | 0.65 ± 0.02 | 17.38 ± 0.02 | 1.96 ± 0.31 | 107 ± 2 | 27.7 ± 3.4 |
| MpDHFR (279; 1) | 0.76 ± 0.08 | 17.73 ± 0.04 | 3.25 ± 0.30 | 103 ± 2 | 30.6 ± 3.7 |
| EcDHFR (279; 220) | 0.71 ± 0.11 | 17.55 ± 0.01 | 2.08 ± 0.31 | 105 ± 1 | 25.5 ± 2.7 |
| MpDHFR (279; 220) | 0.83 ± 0.04 | 17.77 ± 0.03 | 3.28 ± 0.40 | 104 ± 1 | 22.5 ± 1.8 |
| EcDHFR (310; 1) | 0.93 ± 0.07 | 17.87 ± 0.02 | 2.36 ± 0.37 | 106 ± 1 | 11.6 ± 1.2 |
| MpDHFR (310; 1) | 1.39 ± 0.15 | 18.26 ± 0.02 | 4.21 ± 0.32 | 101 ± 2 | 11.2 ± 0.6 |
| EcDHFR (310; 220) | 1.04 ± 0.14 | 17.82 ± 0.01 | 2.47 ± 0.36 | 104 ± 1 | 11.2 ± 1.1 |
| MpDHFR (310; 220) | 1.49 ± 0.16 | 18.25 ± 0.01 | 3.26 ± 0.42 | 101 ± 2 | 11.1 ± 1.1 |
| Protein (P, bar) | ΔT%MSF * | ΔT%Vapp/NHA * | ΔTNw,in | ΔTNHB | ΔT%τHB |
|---|---|---|---|---|---|
| EcDHFR (1) | 43 | 3 | 0.40 | −1 | −58 |
| EcDHFR (220) | 46 | 2 | 0.39 | −1 | −56 |
| MpDHFR (1) | 83 | 3 | 0.96 | −2 | −63 |
| MpDHFR (220) | 80 | 3 | −0.02 | −3 | −51 |
| Protein (T, K) | ΔP%MSF * | ΔP%Vapp/NHA* | ΔPNw,in | ΔPNHB | ΔP%τHB |
|---|---|---|---|---|---|
| EcDHFR (279) | 9 | 1 | 0.12 | −2 | −8 |
| EcDHFR (310) | 12 | 0 | 0.11 | −2 | −3 |
| MpDHFR (279) | 9 | 0 | 0.03 | 1 | −26 |
| MpDHFR (310) | 7 | 0 | −0.95 | 0 | −1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Q.; Rodgers, J.M.; Hemley, R.J.; Ichiye, T. Effects of Pressure and Temperature on the Atomic Fluctuations of Dihydrofolate Reductase from a Psychropiezophile and a Mesophile. Int. J. Mol. Sci. 2019, 20, 1452. https://doi.org/10.3390/ijms20061452
Huang Q, Rodgers JM, Hemley RJ, Ichiye T. Effects of Pressure and Temperature on the Atomic Fluctuations of Dihydrofolate Reductase from a Psychropiezophile and a Mesophile. International Journal of Molecular Sciences. 2019; 20(6):1452. https://doi.org/10.3390/ijms20061452
Chicago/Turabian StyleHuang, Qi, Jocelyn M. Rodgers, Russell J. Hemley, and Toshiko Ichiye. 2019. "Effects of Pressure and Temperature on the Atomic Fluctuations of Dihydrofolate Reductase from a Psychropiezophile and a Mesophile" International Journal of Molecular Sciences 20, no. 6: 1452. https://doi.org/10.3390/ijms20061452
APA StyleHuang, Q., Rodgers, J. M., Hemley, R. J., & Ichiye, T. (2019). Effects of Pressure and Temperature on the Atomic Fluctuations of Dihydrofolate Reductase from a Psychropiezophile and a Mesophile. International Journal of Molecular Sciences, 20(6), 1452. https://doi.org/10.3390/ijms20061452