Identification and Expression Profiling of Protein Phosphatases (PP2C) Gene Family in Gossypium hirsutum L.
Abstract
1. Introduction
2. Results
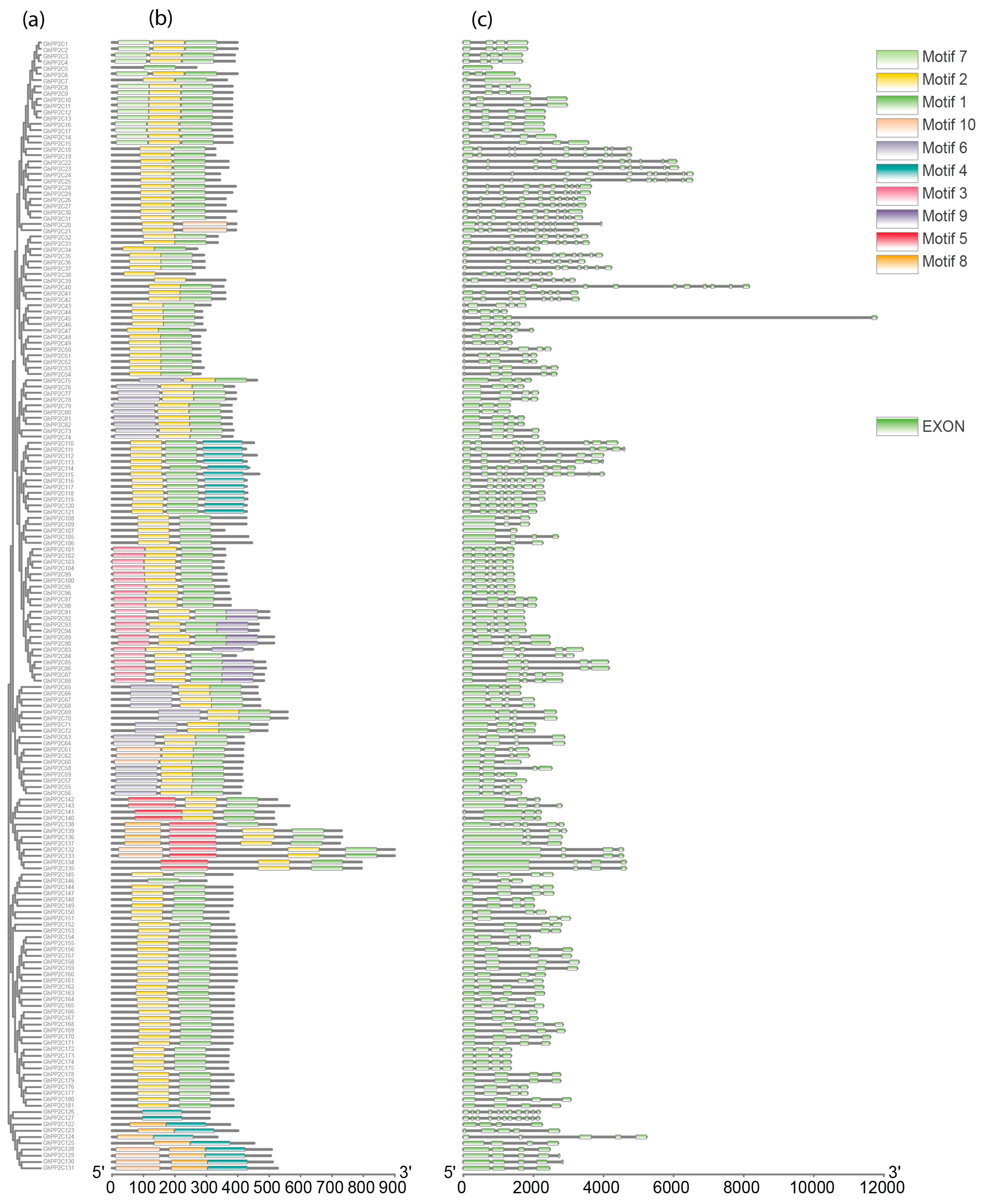
2.1. Characterization of PP2C Gene Family in Cotton
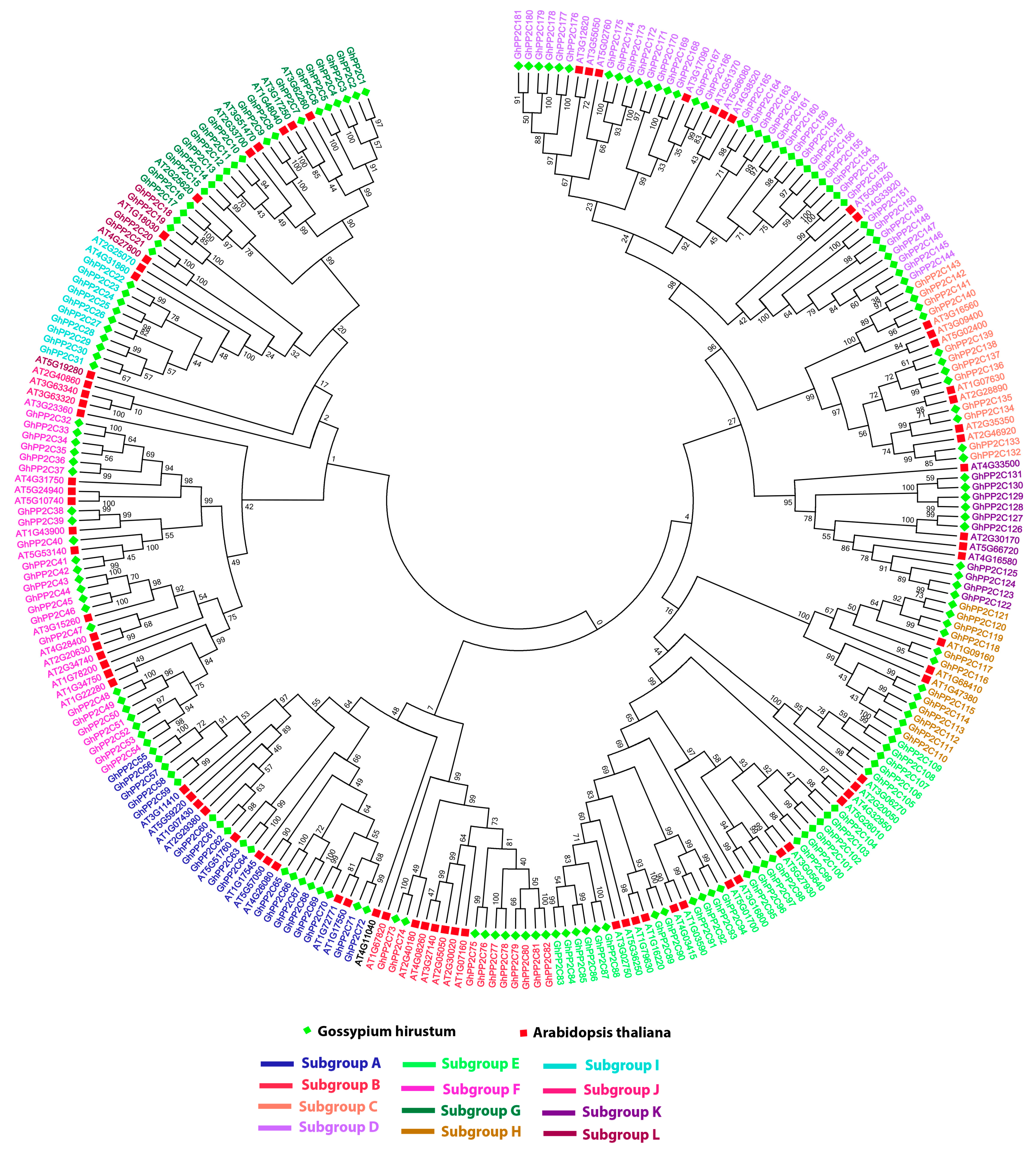
2.2. Phylogenetic and Genomic Distribution and Organization Analysis of PP2C Gene Family
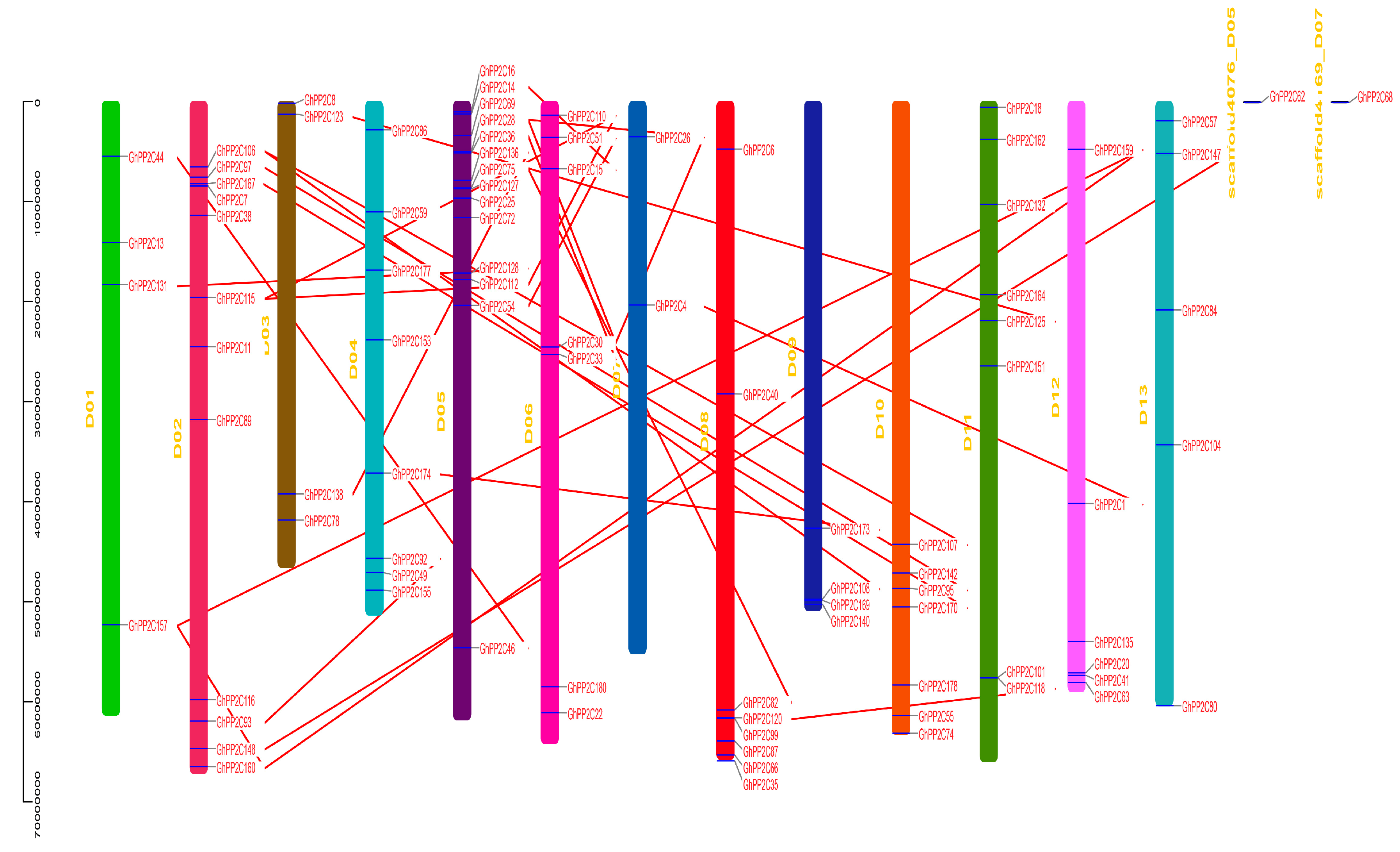
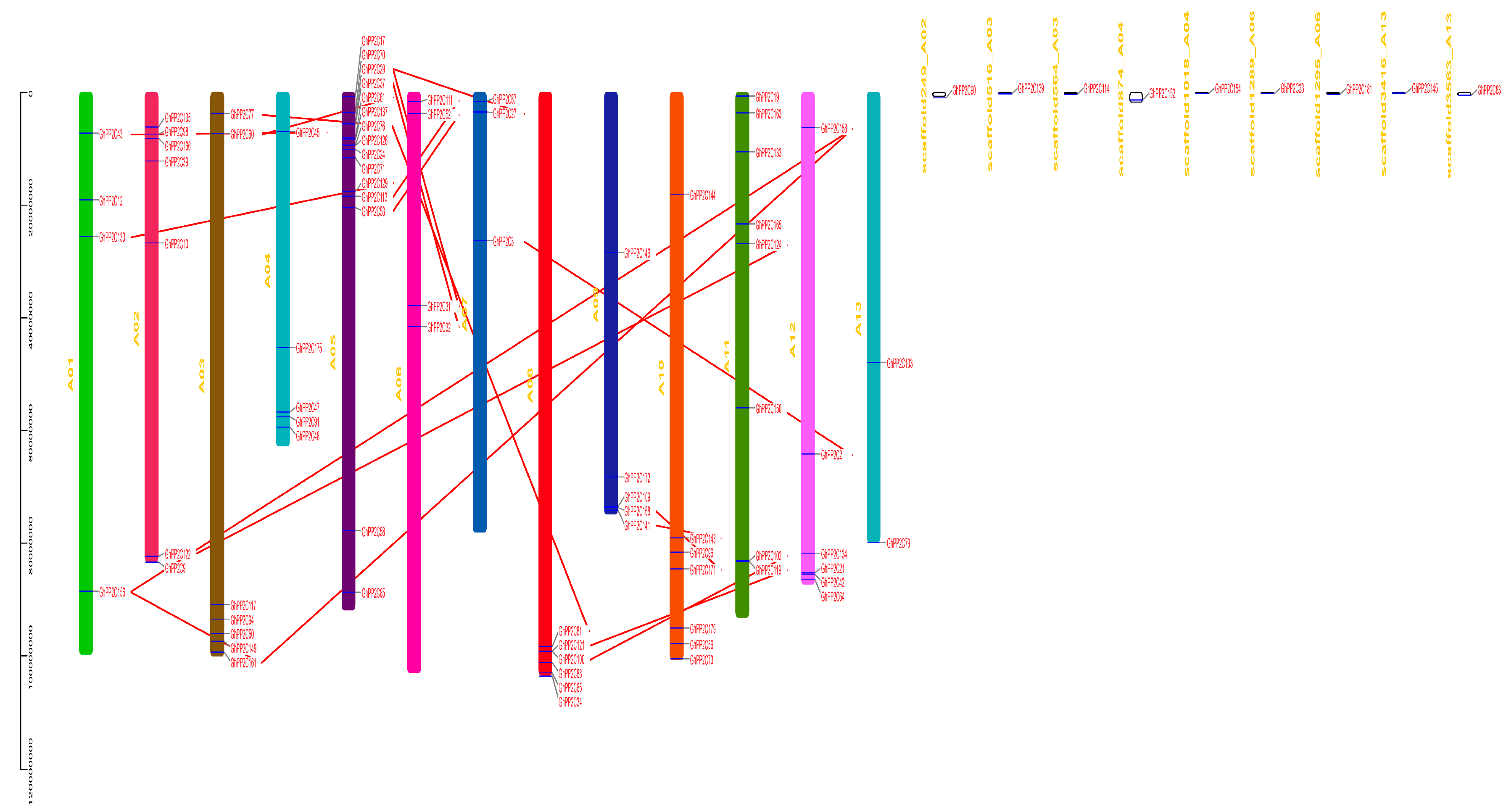
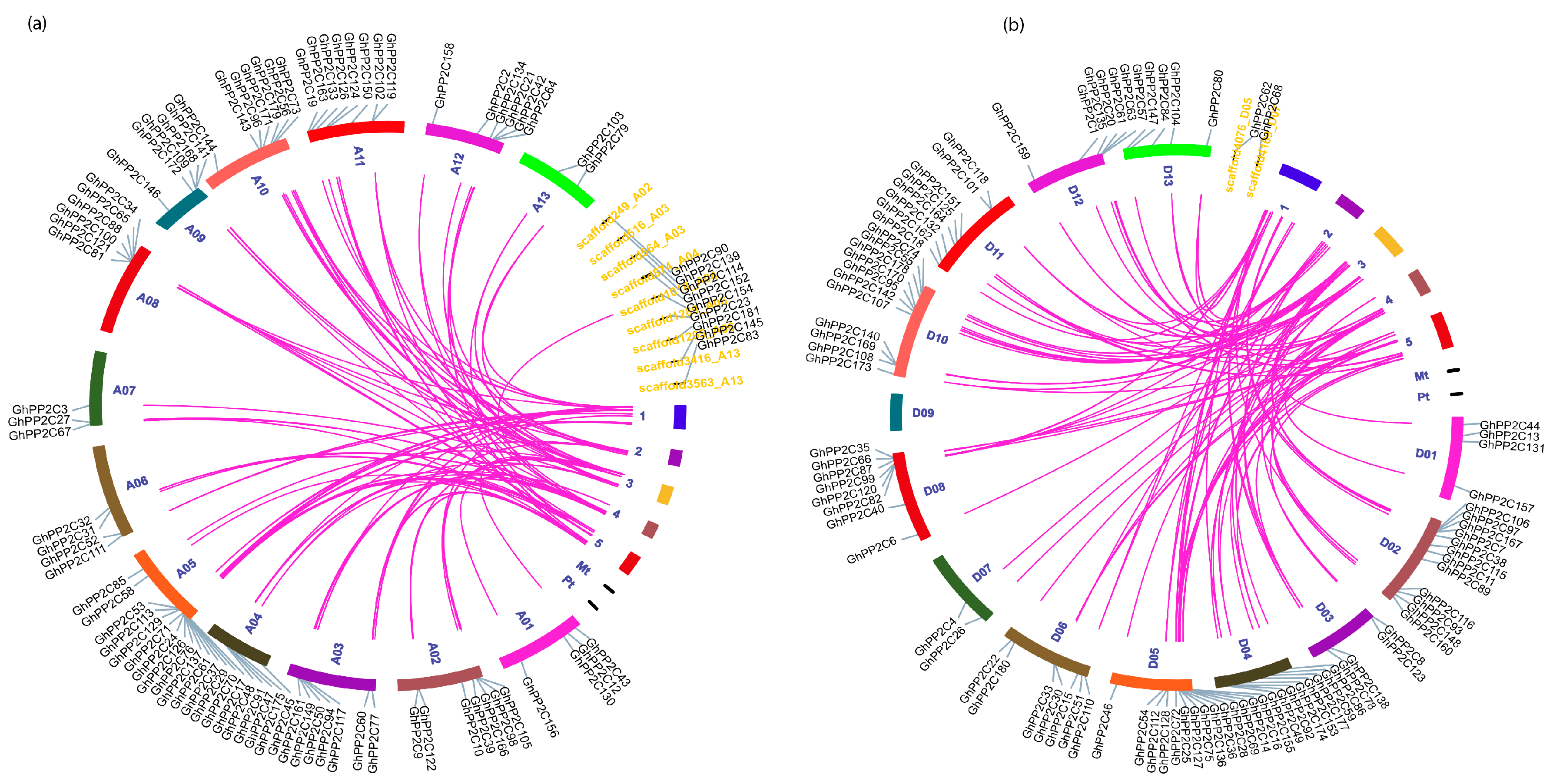
2.3. Chromosomal Localization and Syntenic Relationships of PP2C Gene Family
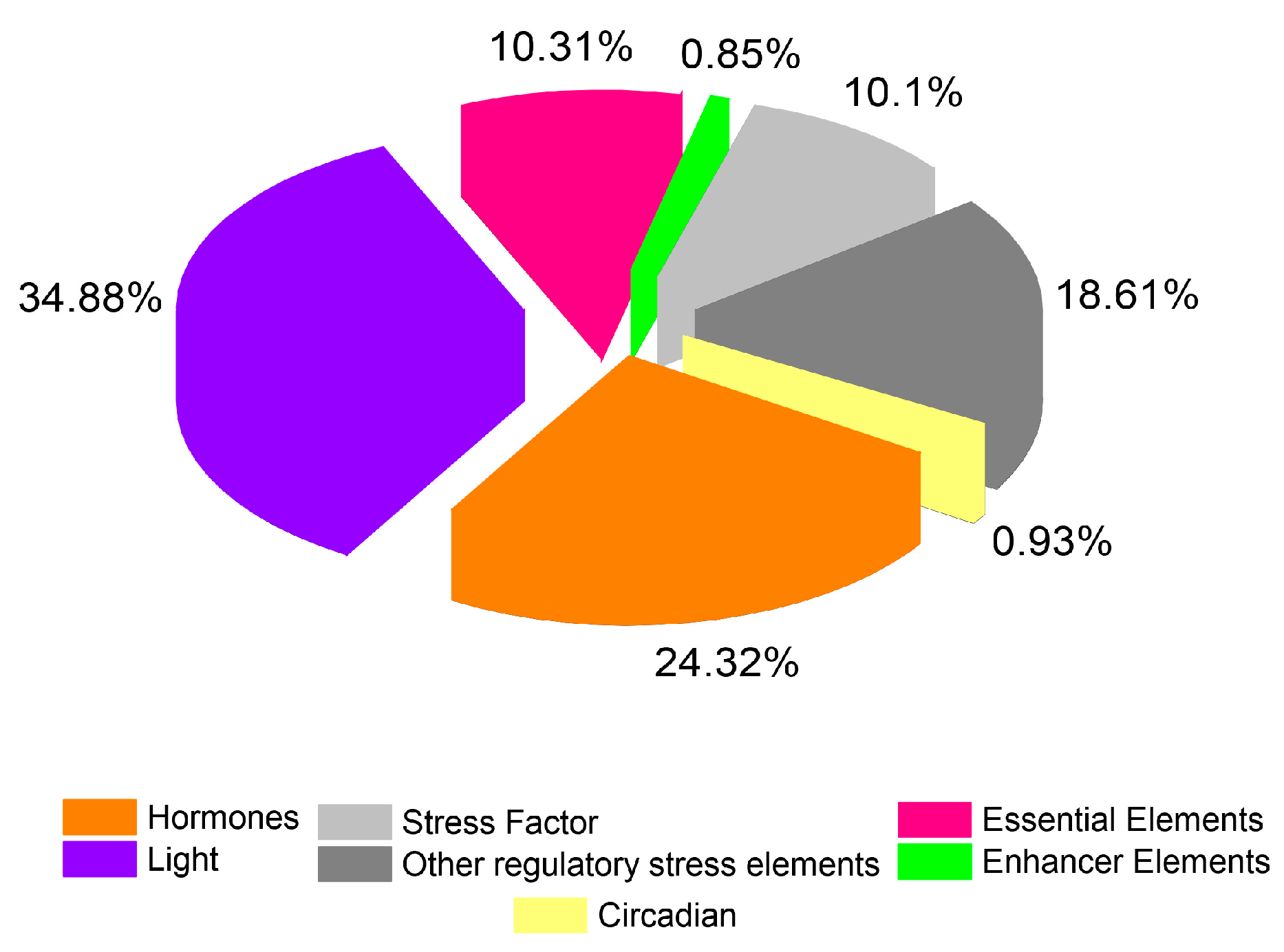
2.4. Analysis of Putative Regulatory Cis-Element and Gene Duplication Analysis of PP2C Gene Family in Cotton
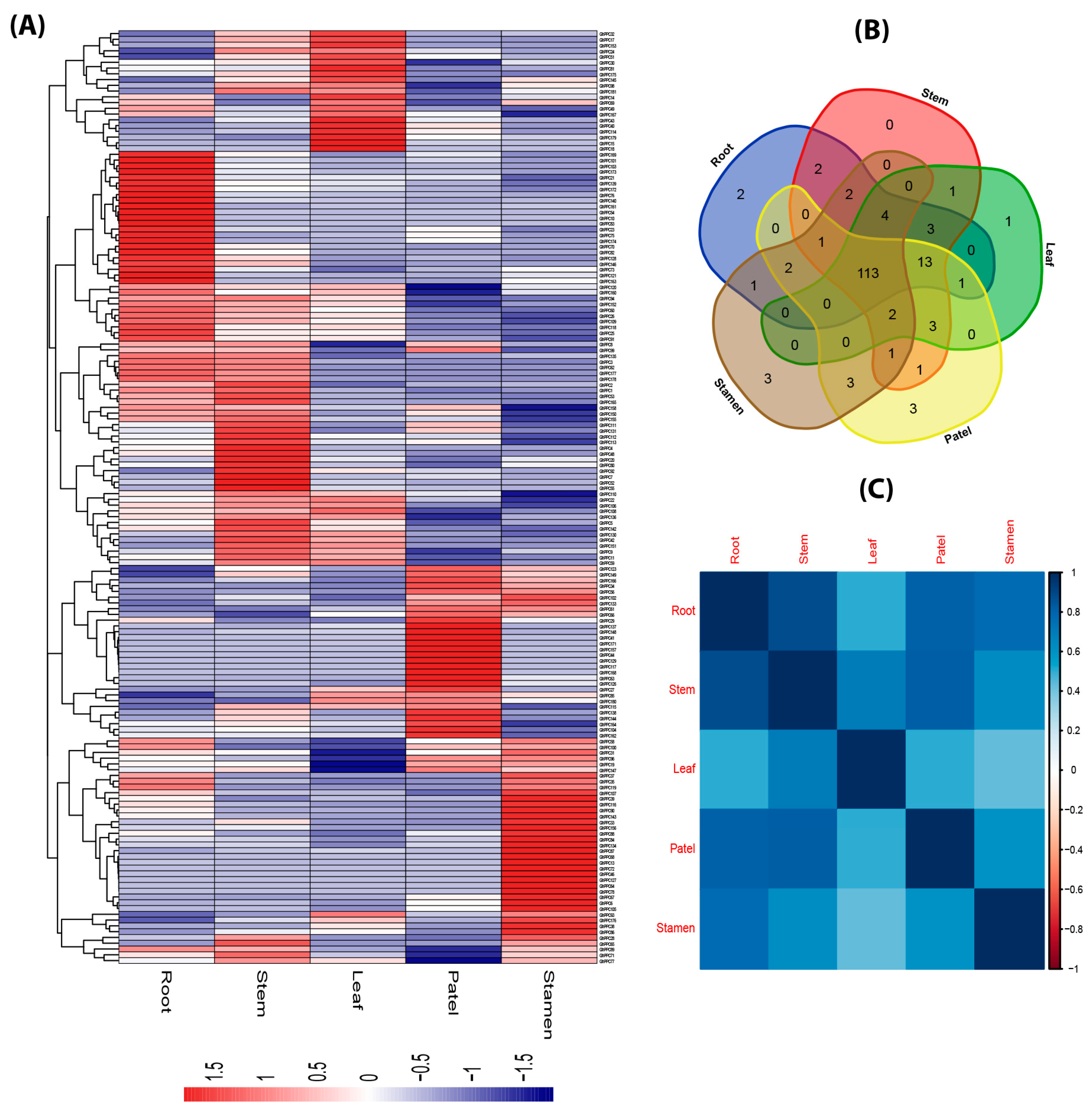
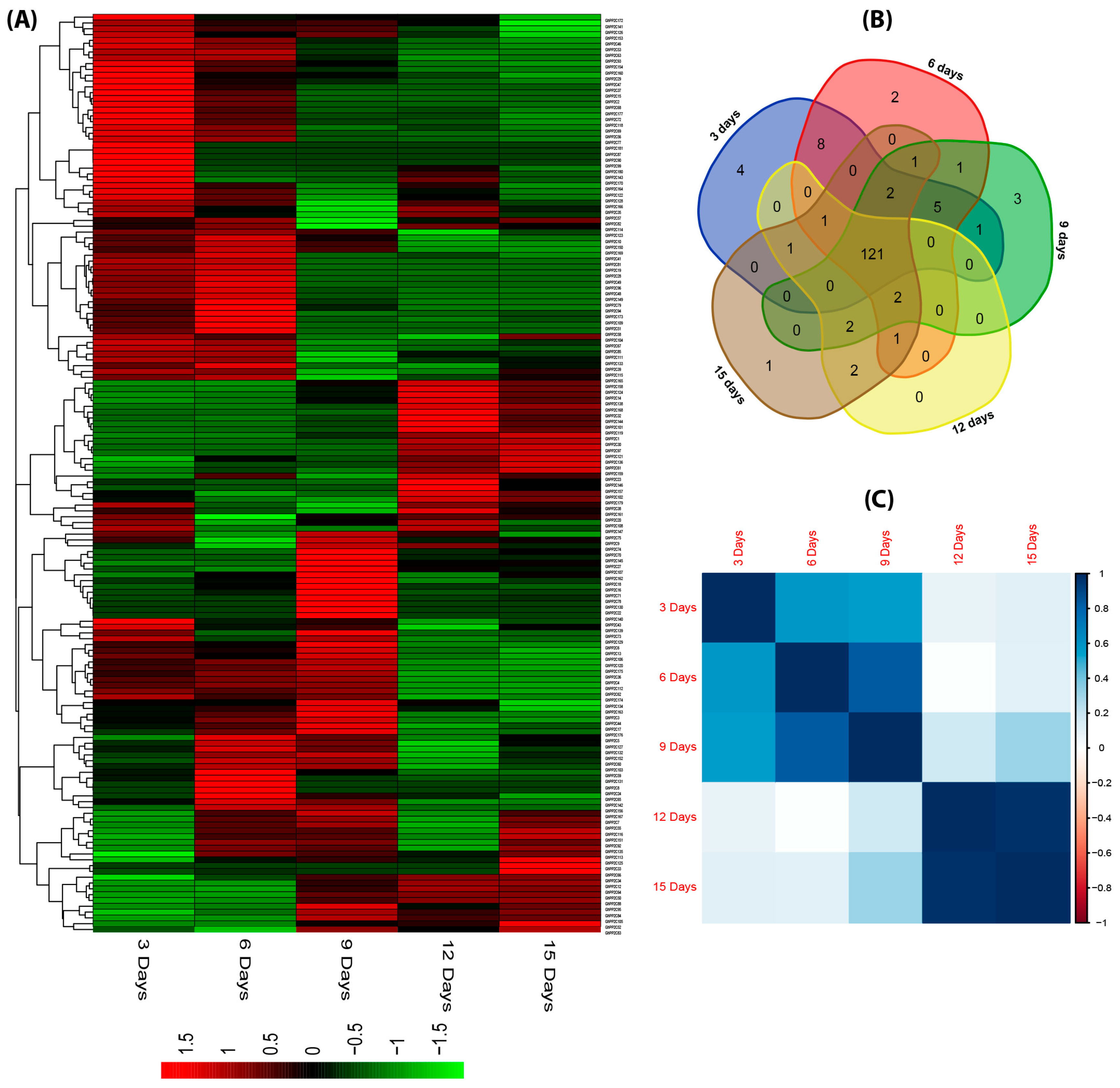
2.5. Expression Profiling of PP2C Gene Family in Different Tissues of G. hirsutum
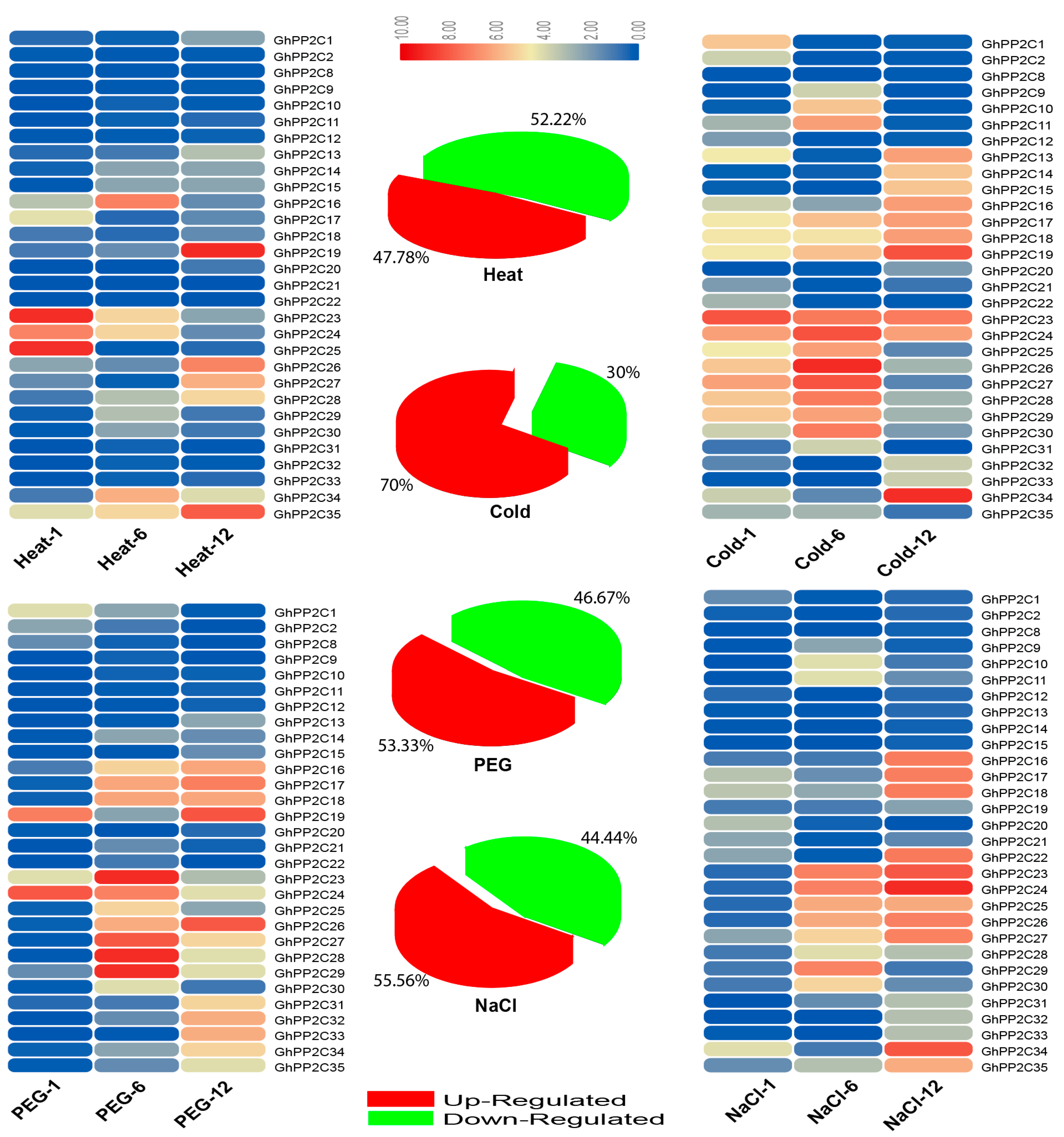
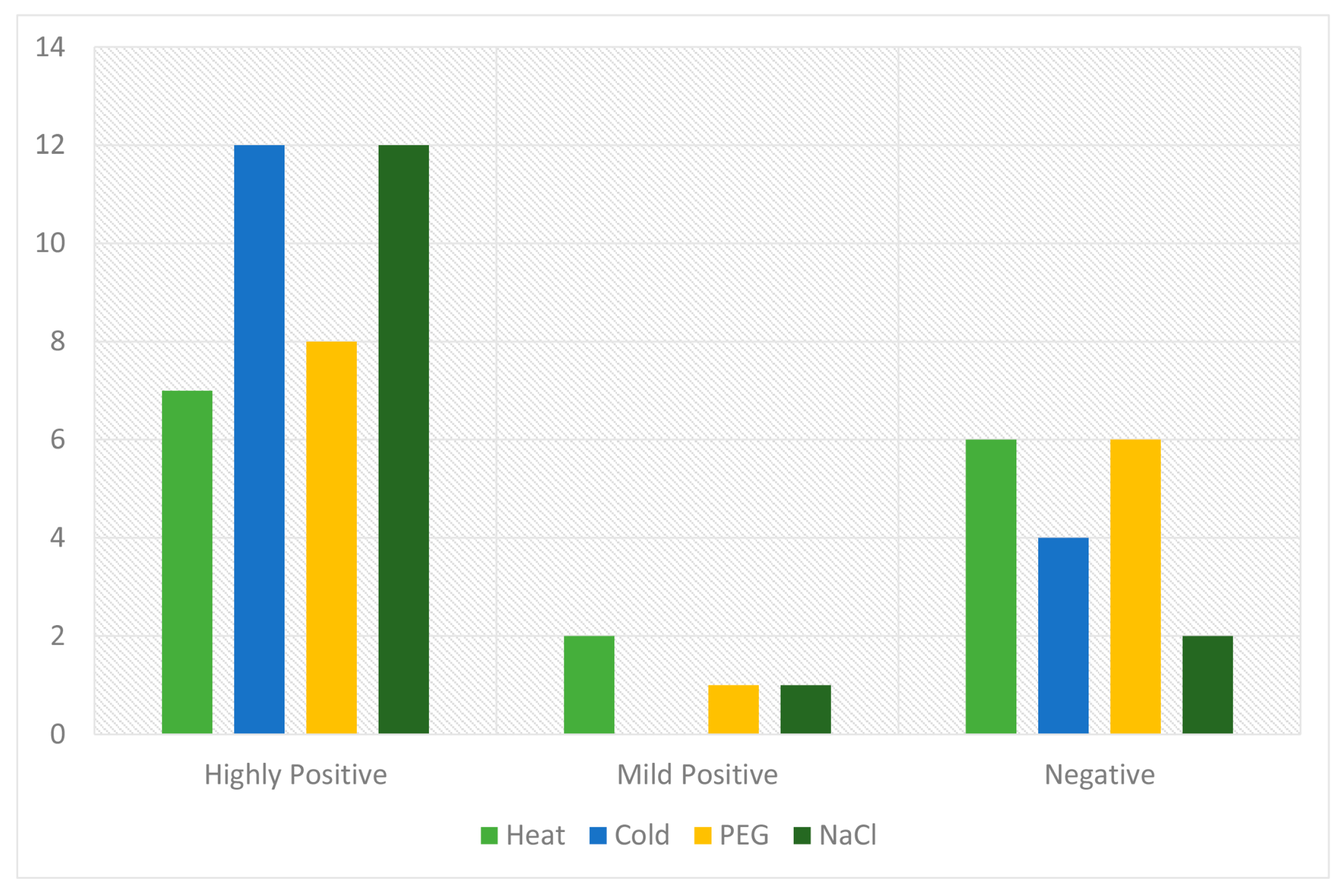
2.6. qRT-PCR Analysis of the Candidate PP2C Gene Family in Response to Various Stresses
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Data Resources for Sequence Retrieval
5.2. Multiple Sequence Alignment and Phylogenetic Analysis
5.3. Calculation of the Ka/Ks for Duplicated Genes
5.4. Conserved Motifs, Exon-Intron Structure Analysis, and Physicochemical Parameters of PP2C Proteins
5.5. Cis-Elements Predictions of GhPP2C
5.6. Chromosomal Location and Synteny Correlation Analysis
5.7. Pearson Correlation Analyses (PCC)
5.8. Plant Material and Treatments
5.9. RNA Isolation and Transcriptional Profiling of GhPP2C under Various Stresses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lessard, P.; Kreis, M.; Thomas, M. Protein phosphatases and protein kinases in higher plants. Comptes Rendus de l’Academie des Sciences. Serie III, Sciences de la Vie 1997, 320, 675–688. [Google Scholar] [PubMed]
- Li, F.; Fan, G.; Wang, K.; Sun, F.; Yuan, Y.; Song, G.; Li, Q.; Ma, Z.; Lu, C.; Zou, C.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Min, J.; Peng, L.; Chu, Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genom. 2016, 17, 175. [Google Scholar] [CrossRef] [PubMed]
- Kerk, D.; Templeton, G.; Moorhead, G.B.G. Evolutionary Radiation Pattern of Novel Protein Phosphatases Revealed by Analysis of Protein Data from the Completely Sequenced Genomes of Humans, Green Algae, and Higher Plants. Plant Physiol. 2008, 146, 351–367. [Google Scholar] [CrossRef]
- Cohen, P. The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 2003, 58, 453–508. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.W.; Delong, A. Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 2004, 16, 709–722. [Google Scholar] [CrossRef]
- Boudsocq, M.; Barbier-Brygoo, H.; Lauriere, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Boil. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef]
- Ma, S.Y.; Wu, W.H. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 2007, 65, 511–518. [Google Scholar] [CrossRef]
- Song, S.K.; Clark, S.E. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev. Boil. 2005, 285, 272–284. [Google Scholar] [CrossRef]
- Kerk, D.; Bulgrien, J.; Smith, D.W.; Barsam, B.; Veretnik, S.; Gribskov, M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002, 129, 908–925. [Google Scholar] [CrossRef]
- Singh, A.; Giri, J.; Kapoor, S.; Tyagi, A.K.; Pandey, G.K. Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genom. 2010, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Pan, S. Maize protein phosphatase gene family: Identification and molecular characterization. BMC Genom. 2014, 15, 773. [Google Scholar] [CrossRef] [PubMed]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Garcia, M.P.; Rodriguez, D.; Nicolas, C.; Rodriguez, P.L.; Nicolas, G.; Lorenzo, O. Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol. 2003, 133, 135–144. [Google Scholar] [CrossRef]
- Saez, A.; Apostolova, N.; Gonzalez-Guzman, M.; Gonzalez-Garcia, M.P.; Nicolas, C.; Lorenzo, O.; Rodriguez, P.L. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004, 37, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Haider, M.S.; Zhang, C.; Kurjogi, M.M.; Pervaiz, T.; Zheng, T.; Zhang, C.; Lide, C.; Shangguan, L.; Fang, J. Insights into grapevine defense response against drought as revealed by biochemical, physiological and RNA-Seq analysis. Sci. Rep. 2017, 7, 13134. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl. Acad. Sci. USA 1998, 95, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Soderman, E.; Mattsson, J.; Engstrom, P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 1996, 10, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Strizhov, N.; Abraham, E.; Okresz, L.; Blickling, S.; Zilberstein, A.; Schell, J.; Koncz, C.; Szabados, L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997, 12, 557–569. [Google Scholar] [CrossRef]
- Murata, Y.; Pei, Z.M.; Mori, I.C.; Schroeder, J. Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 2001, 13, 2513–2523. [Google Scholar] [CrossRef]
- Haider, M.S.; Kurjogi, M.M.; Khalilurrehman, M.; Fiaz, M.; Pervaiz, T.; Jiu, S.; Haifeng, J.; Chen, W.; Fang, J. Grapevine immune signaling network in response to drought stress as revealed by transcriptomic analysis. Plant Physiol. Biochem. 2017, 121, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Tahtiharju, S.; Palva, T. Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J. 2010, 26, 461–470. [Google Scholar] [CrossRef]
- Yuan, D.; Tang, Z.; Wang, M.; Gao, W.; Tu, L.; Jin, X.; Chen, L.; He, Y.; Zhang, L.; Zhu, L.; et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 2015, 5, 17662. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liu, K.; Niu, X.; Wang, Q.; Wan, Y.; Yang, F.; Li, G.; Wang, Y.; Wang, R. Genome-wide Identification of PP2C Genes and Their Expression Profiling in Response to Drought and Cold Stresses in Medicago truncatula. Sci. Rep. 2018, 8, 12841. [Google Scholar] [CrossRef]
- Xue, T.; Wang, D.; Zhang, S.; Ehlting, J.; Ni, F.; Jakab, S.; Zheng, C.; Zhong, Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom. 2008, 9, 550. [Google Scholar] [CrossRef]
- Wei, L.; Xiao, C.; Zhaolu, M.; Xiahe, H.; Qi, X.; Heng, W.; Hailing, J.; Dabing, Z.; Wanqi, L. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012, 158, 1279–1292. [Google Scholar]
- Maestrini, P.; Cavallini, A.; Rizzo, M.; Giordani, T.; Bernardi, R.; Durante, M.; Natali, L. Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba). J. Plant Physiol. 2009, 166, 1544–1556. [Google Scholar] [CrossRef]
- Wen, F.; Zhu, H.; Li, P.; Jiang, M.; Mao, W.; Ong, C.; Chu, Z. Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res. 2014, 21, 327–339. [Google Scholar] [CrossRef]
- Prince, V.E.; Pickett, F.B. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 2002, 3, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Vandepoele, K.; Simillion, C.; Van de Peer, Y. Evidence that rice and other cereals are ancient aneuploids. Plant Cell 2003, 15, 2192–2202. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Yan, Y.; Shi, H.; Liu, J.; Miao, H.; Tie, W.; Ding, Z.; Ding, X.; Wu, C.; Liu, Y.; et al. The core regulatory network of the abscisic acid pathway in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. BMC Plant Boil. 2017, 17, 145. [Google Scholar] [CrossRef] [PubMed]
- Hurles, M. Gene Duplication: The Genomic Trade in Spare Parts. PLoS Boil. 2004, 2, e206. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009, 17, 699–717. [Google Scholar] [CrossRef]
- Jiao, Y.; Shi, C.; Edil, B.H.; de Wilde, R.F.; Klimstra, D.S.; Maitra, A.; Schulick, R.D.; Tang, L.H.; Wolfgang, C.L.; Choti, M.A.; et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011, 331, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Wolf, U.; Atkin, N.B. Evolution from fish to mammals by gene duplication. Hereditas 1968, 59, 169–187. [Google Scholar] [CrossRef]
- Juretic, N.; Hoen, D.R.; Huynh, M.L.; Harrison, P.M.; Bureau, T.E. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005, 15, 1292–1297. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.-S.; Wang, J. Correlation Between Ka/Ks and Ks is Related to Substitution Model and Evolutionary Lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, Y.; Zhai, H.; Cai, H.; Ji, W.; Luo, X.; Li, J.; Bai, X. AtPP2CG1, a protein phosphatase 2C, positively regulates salt tolerance of Arabidopsis in abscisic acid-dependent manner. Biochem. Biophys. Res. Commun. 2012, 422, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Chonnanit, C. Characterization and functional analysis of a novel PP2C phosphatase AP2C2 from Arabidopsis. Ph.D. Thesis, University of Vienna, Viennam, Austria, 2008. [Google Scholar]
- Williams, R.W.; Wilson, J.M.; Meyerowitz, E.M. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 10467–10472. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Boil. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucl. Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl. Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. bioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Khan, N.; Hu, C.-M.; Amjad Khan, W.; Hou, X. Genome-Wide Identification, Classification, and Expression Divergence of Glutathione-Transferase Family in Brassica rapa under Multiple Hormone Treatments. BioMed Res. Int. 2018, 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Hu, C.-M.; Amjad Khan, W.; Naseri, E.; Ke, H.; Huijie, D.; Hou, X. Evolution and Expression Divergence of E2 Gene Family under Multiple Abiotic and Phytohormones Stresses in Brassica rapa. BioMed. Res. Int. 2018, 2018, 5206758. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Hu, C.-M.; Khan, W.A.; Wang, W.; Ke, H.; Huijie, D.; Zhishuo, Z.; Hou, X. Genome-wide Identification, Classification, and Expression Pattern of Homeobox Gene Family in Brassica rapa under Various Stresses. Sci. Rep. 2018, 8, 16265. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, X.; Tian, L.; Wang, G.; Zhang, X.; Wang, X.; Guo, W. Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci. Rep. 2016, 6, 29022. [Google Scholar] [CrossRef]
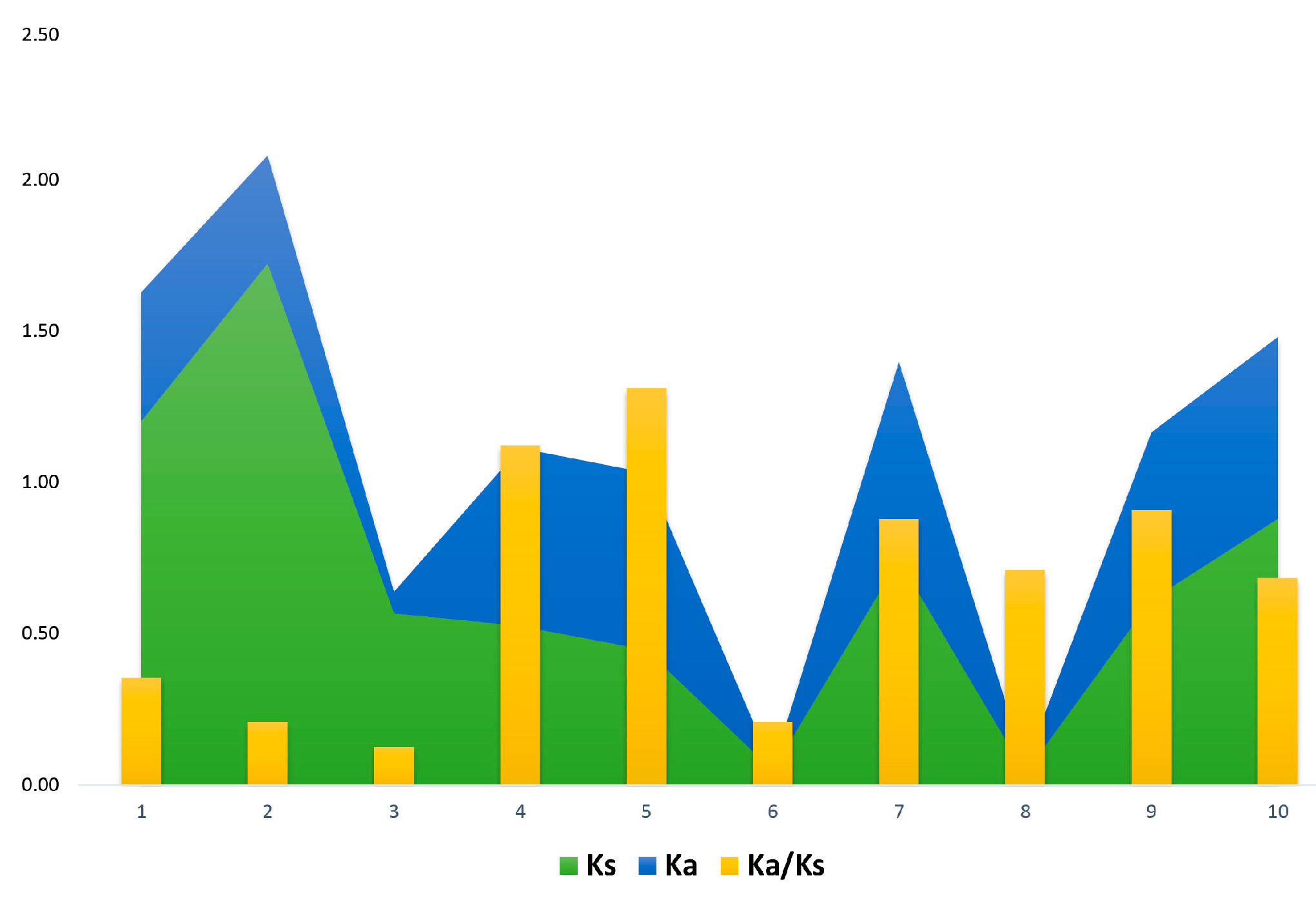
| Gene 1 | Gene 2 | Ks | Ka | Ka/Ks | Selection Pressure | Gene Duplications |
|---|---|---|---|---|---|---|
| GhPP2C9 | GhPP2C3 | 1.20 | 0.43 | 0.354114713 | Purifying Selection | Segmental |
| GhPP2C10 | GhPP2C6 | 1.72 | 0.36 | 0.207076566 | Purifying Selection | Segmental |
| GhPP2C4 | GhPP2C1 | 0.57 | 0.07 | 0.125220459 | Purifying Selection | Segmental |
| GhPP2C7 | GhPP2C8 | 0.53 | 0.59 | 1.121904762 | Positive Selection | Segmental |
| GhPP2C11 | GhPP2C2 | 0.45 | 0.59 | 1.311659193 | Positive Selection | Segmental |
| GhPP2C89 | GhPP2C90 | 0.05 | 0.01 | 0.208333333 | Purifying Selection | Dispersed |
| GhPP2C114 | GhPP2C146 | 0.74 | 0.65 | 0.879032258 | Purifying Selection | Dispersed |
| GhPP2C83 | GhPP2C84 | 0.04 | 0.03 | 0.710526316 | Purifying Selection | Dispersed |
| GhPP2C139 | GhPP2C180 | 0.61 | 0.56 | 0.908346972 | Purifying Selection | Dispersed |
| GhPP2C62 | GhPP2C23 | 0.88 | 0.60 | 0.683731513 | Purifying Selection | Dispersed |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shazadee, H.; Khan, N.; Wang, J.; Wang, C.; Zeng, J.; Huang, Z.; Wang, X. Identification and Expression Profiling of Protein Phosphatases (PP2C) Gene Family in Gossypium hirsutum L. Int. J. Mol. Sci. 2019, 20, 1395. https://doi.org/10.3390/ijms20061395
Shazadee H, Khan N, Wang J, Wang C, Zeng J, Huang Z, Wang X. Identification and Expression Profiling of Protein Phosphatases (PP2C) Gene Family in Gossypium hirsutum L. International Journal of Molecular Sciences. 2019; 20(6):1395. https://doi.org/10.3390/ijms20061395
Chicago/Turabian StyleShazadee, Hamna, Nadeem Khan, Jingjing Wang, Chencan Wang, Jianguo Zeng, Zhongyi Huang, and Xinyu Wang. 2019. "Identification and Expression Profiling of Protein Phosphatases (PP2C) Gene Family in Gossypium hirsutum L." International Journal of Molecular Sciences 20, no. 6: 1395. https://doi.org/10.3390/ijms20061395
APA StyleShazadee, H., Khan, N., Wang, J., Wang, C., Zeng, J., Huang, Z., & Wang, X. (2019). Identification and Expression Profiling of Protein Phosphatases (PP2C) Gene Family in Gossypium hirsutum L. International Journal of Molecular Sciences, 20(6), 1395. https://doi.org/10.3390/ijms20061395





