Developmental Stage- and Genotype-Dependent Regulation of Specialized Metabolite Accumulation in Fruit Tissues of Different Citrus Varieties
Abstract
1. Introduction
2. Results and Discussion
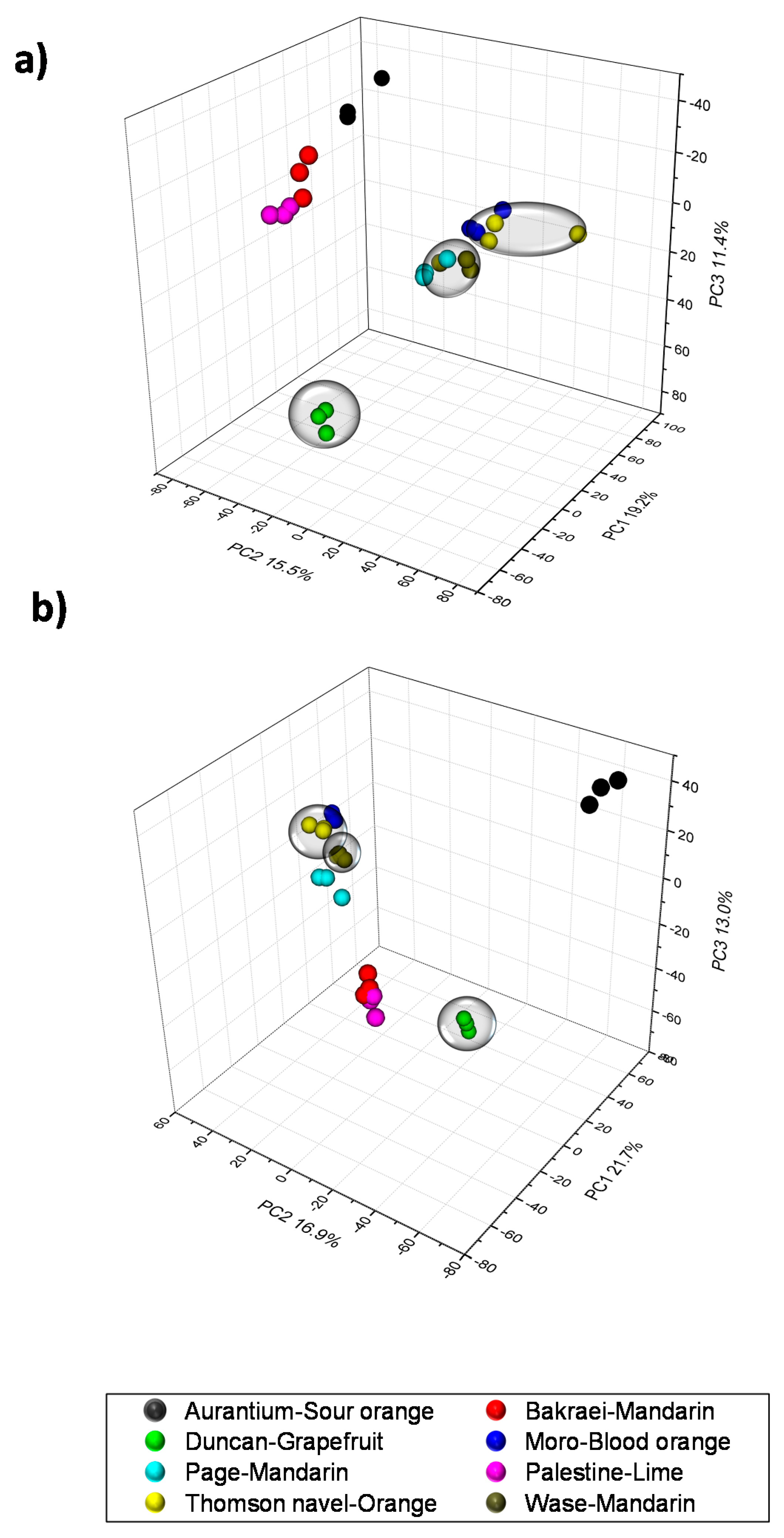
2.1. Validation of Results, Variable Selection and Annotation of Compounds
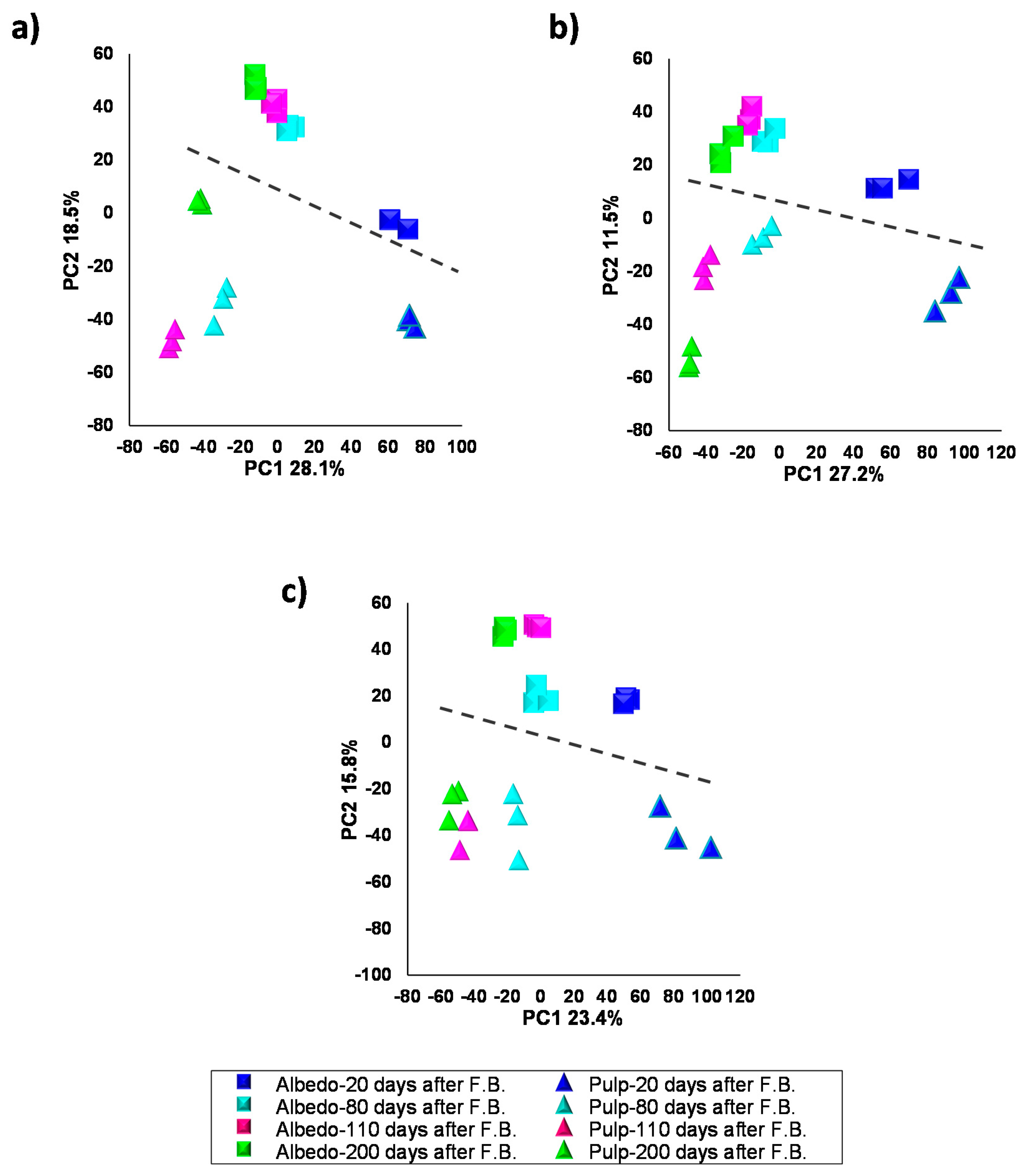
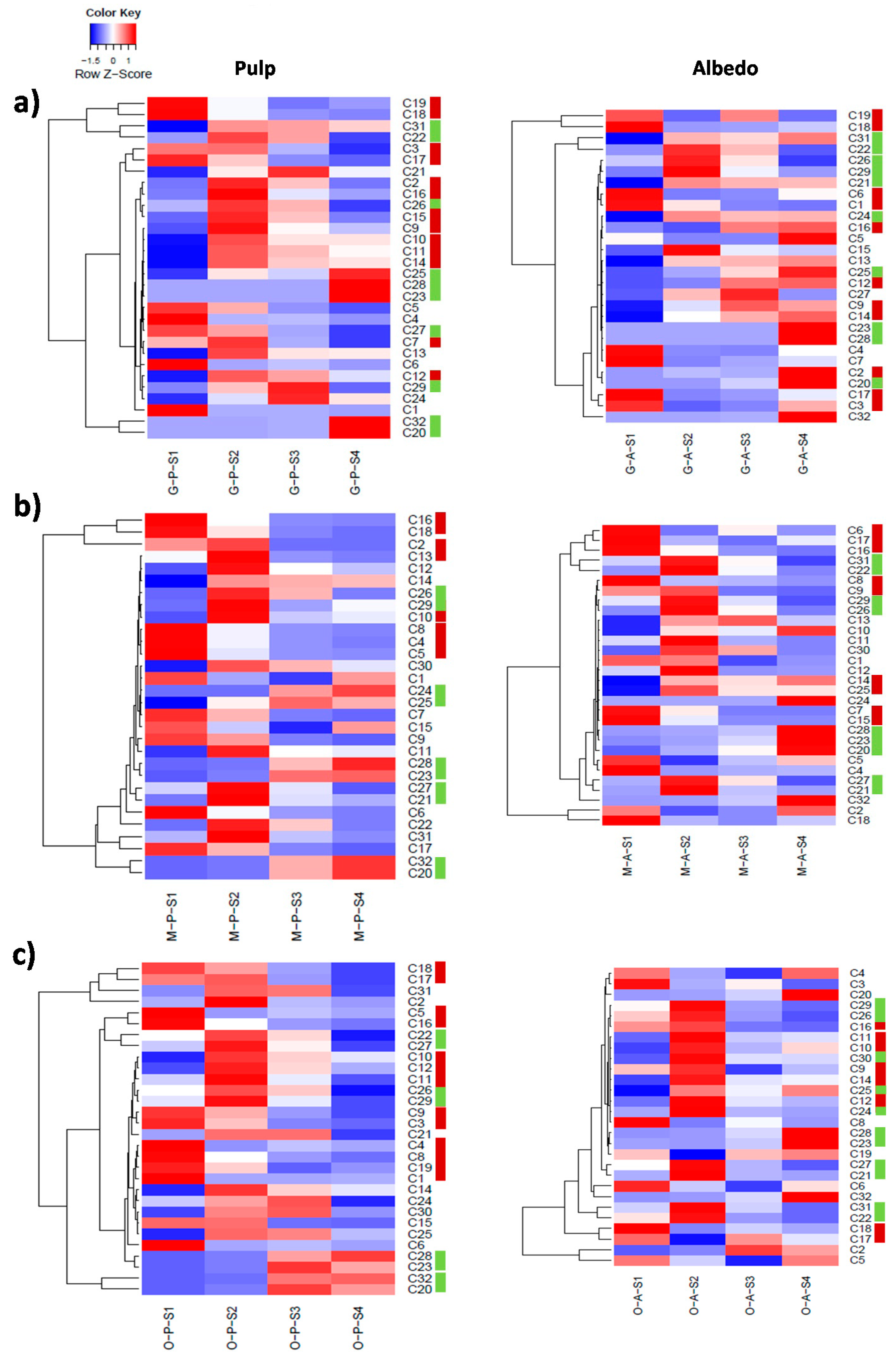
2.2. Characterization of Metabolite Profiles in Albedo and Pulp Tissues
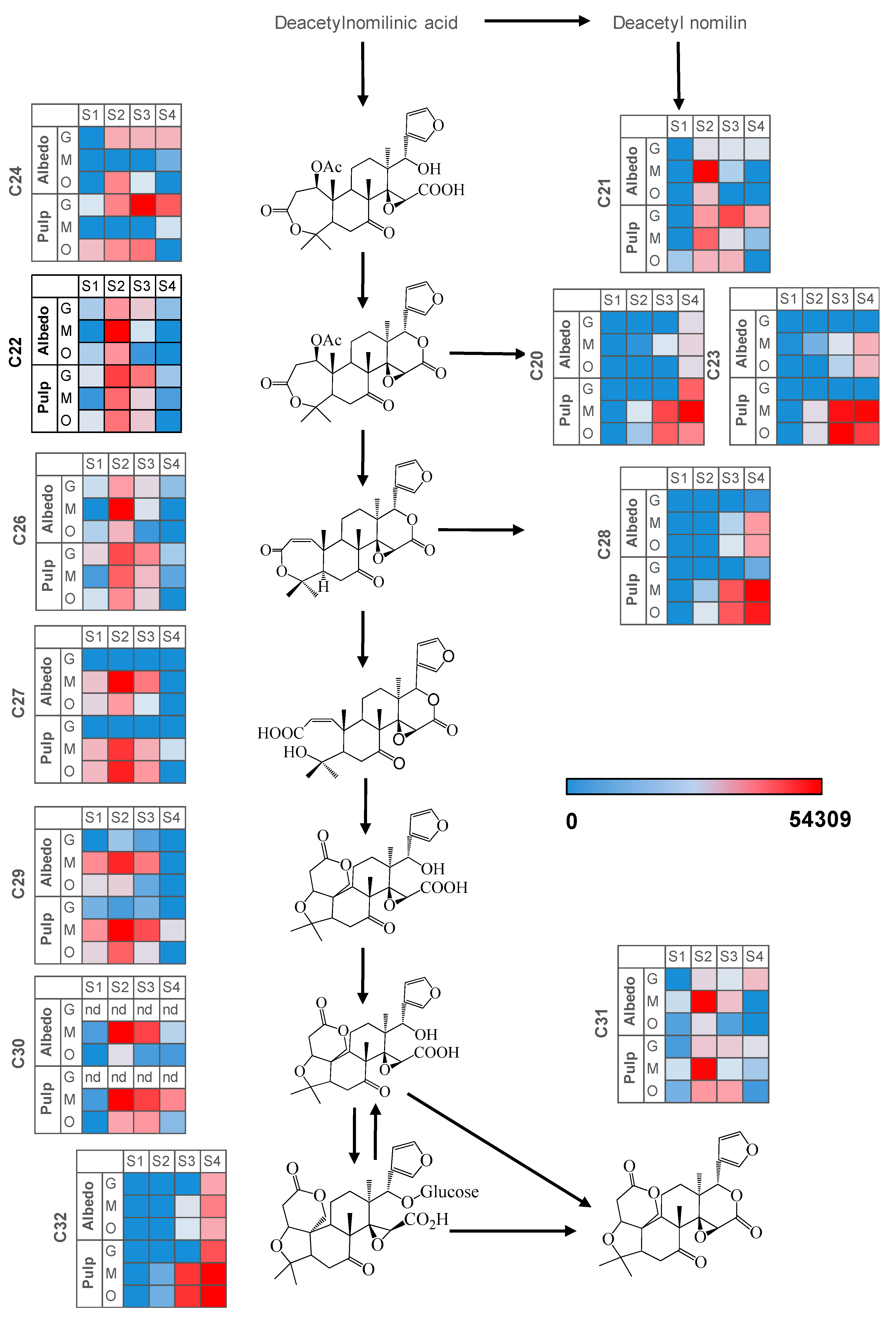
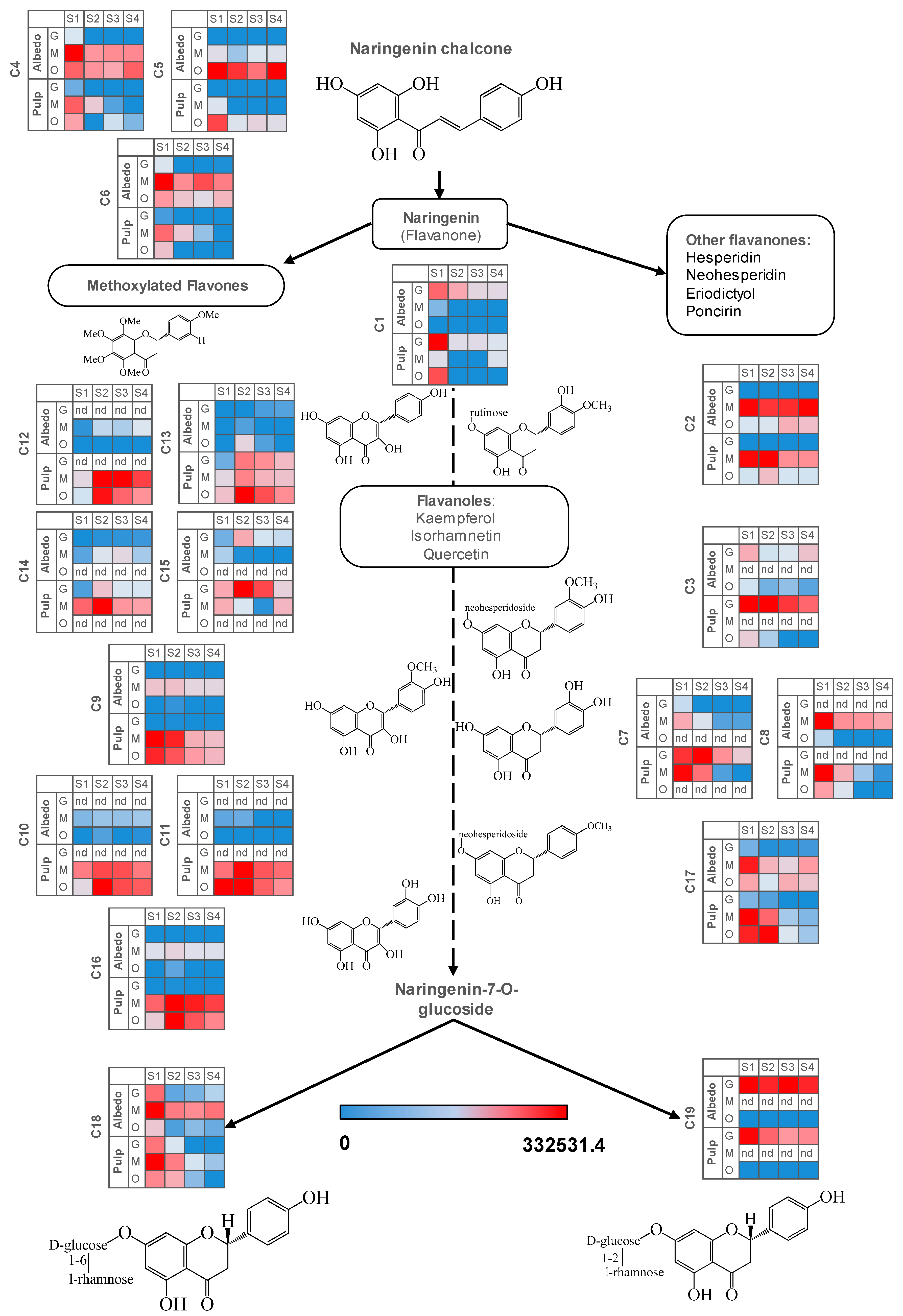
2.3. Fruit Ripening: Co-Regulation of Specialized Metabolites in Citrus Fruit Tissues
3. Materials and Methods
3.1. Plant Material and Sample Preparation
3.2. Extraction of Samples for Chromatographic Analyses
3.3. Chromatographic and QqTOF-MS Conditions
3.4. Mass Spectrometry (MS) Data Processing, Statistical Analyses and Compound Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Terol, J.; Soler, G.; Talon, M.; Cercos, M. The aconitate hydratase family from Citrus. BMC Plant Biol. 2010, 10, 222. [Google Scholar] [CrossRef]
- Dugrand-Judek, A.; Olry, A.; Hehn, A.; Costantino, G.; Ollitrault, P.; Froelicher, Y.; Bourgaud, F. The distribution of coumarins and furanocoumarins in Citrus species closely matches Citrus phylogeny and reflects the organization of biosynthetic pathways. PLoS ONE 2015, 10, e0142757. [Google Scholar] [CrossRef]
- Patil, B.S.; Jayaprakasha, G.K.; Chidambara Murthy, K.N.; Vikram, A. Bioactive compounds: Historical perspectives, opportunities and challenges. J. Agric. Food Chem. 2009, 57, 8142–8160. [Google Scholar] [CrossRef]
- Manners, G.D. Citrus limonoids: Analysis, bioactivity, and biomedical prospects. J. Agric. Food Chem. 2007, 55, 8285–8294. [Google Scholar] [CrossRef]
- Frydman, A.; Liberman, R.; Huhman, D.V.; Carmeli-Weissberg, M.; Sapir-Mir, M.; Ophir, R.; W Sumner, L.; Eyal, Y. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: Evolution of branch-forming rhamnosyltransferases under domestication. Plant J. 2012, 73, 166–178. [Google Scholar] [CrossRef]
- Wang, F.; Wang, M.; Liu, X.; Xu, Y.; Zhu, S.; Shen, W.; Zhao, X. Identification of putative genes involved in limonoids biosynthesis in citrus by comparative transcriptomic analysis. Front. Plant Sci. 2017, 8, 782. [Google Scholar] [CrossRef]
- Akyildiz, A.; Erdal, A. Food Processing: Strategies for Quality Assessment; Malik, A., Erginkaya, Z., Ahmad, S., Erten, H., Eds.; Food Engineering Series; Springer: New York, NY, USA, 2014; ISBN 978-1-4939-1377-0. [Google Scholar]
- Dala Paula, B.M.; Raithore, S.; Manthey, J.A.; Baldwin, E.A.; Bai, J.; Zhao, W.; Glória, M.B.A.; Plotto, A. Active taste compounds in juice from oranges symptomatic for Huanglongbing (HLB) citrus greening disease. LWT Food Sci. Technol. 2018, 91, 518–525. [Google Scholar] [CrossRef]
- Zaare-Nahandi, F.; Hosseinkhani, S.; Zamani, Z.; Asadi-Abkenar, A.; Omidbaigi, R. Delay expression of limonoid UDP-glucosyltransferase makes delayed bitterness in citrus. Biochem. Biophys. Res. Commun. 2008, 371, 59–62. [Google Scholar] [CrossRef]
- Ververidis, F.; Trantas, E.; Douglas, C.; Vollmer, G.; Kretzschmar, G.; Panopoulos, N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotechnol. J. 2007, 2, 1214–1234. [Google Scholar] [CrossRef]
- Wang, S.; Tu, H.; Wan, J.; Chen, W.; Liu, X.; Luo, J.; Xu, J.; Zhang, H. Spatio-temporal distribution and natural variation of metabolites in citrus fruits. Food Chem. 2016, 199, 8–17. [Google Scholar] [CrossRef]
- Tundis, R.; Loizzo, M.R.; Menichini, F. An overview on chemical aspects and potential health benefits of limonoids and their derivatives. Crit. Rev. Food Sci. Nutr. 2014, 54, 225–250. [Google Scholar] [CrossRef]
- Drewnowski, A.; Gomez-Carneros, C. Bitter taste, phytonutrients, and the consumer: A review 1–3. Am. J. Clin. Nutr. 2000, 72, 1424–1435. [Google Scholar] [CrossRef]
- Arbona, V.; Iglesias, D.J.; Talón, M.; Gómez-Cadenas, A. Plant phenotype demarcation using nontargeted LC-MS and GC-MS metabolite profiling. J. Agric. Food Chem. 2009, 57, 7338–7347. [Google Scholar] [CrossRef]
- Arbona, V.; Iglesias, D.J.; Gómez-Cadenas, A. Non-targeted metabolite profiling of citrus juices as a tool for variety discrimination and metabolite flow analysis. BMC Plant Biol. 2015, 15, 38. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Sales, C.; Beltrán, J.; Gómez-Cadenas, A.; Arbona, V. Activation of secondary metabolism in citrus plants is associated to sensitivity to combined drought and high temperatures. Front. Plant Sci. 2016, 7, 1954. [Google Scholar] [CrossRef]
- Durand-Hulak, M.; Dugrand, A.; Duval, T.; Bidel, L.P.R.; Jay-Allemand, C.; Froelicher, Y.; Bourgaud, F.; Fanciullino, A.L. Mapping the genetic and tissular diversity of 64 phenolic compounds in Citrus species using a UPLC-MS approach. Ann. Bot. 2015, 115, 861–877. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Xia, R. Expression of chalcone synthase and chalcone isomerase genes and accumulation of corresponding flavonoids during fruit maturation of Guoqing No. 4 satsuma mandarin (Citrus unshiu Marcow). Sci. Hortic. (Amsterdam) 2010, 125, 110–116. [Google Scholar] [CrossRef]
- Barreca, D.; Bellocco, E.; Caristi, C.; Leuzzi, U.; Gattuso, G. Flavonoid composition and antioxidant activity of juices from Chinotto (Citrus x myrtifolia Raf.) fruits at different ripening stages. J. Agric. Food Chem. 2010, 58, 3031–3036. [Google Scholar] [CrossRef]
- Chaudhary, P.R.; Bang, H.; Jayaprakasha, G.K.; Patil, B.S. Variation in key flavonoid biosynthetic enzymes and phytochemicals in “Rio Red” grapefruit (Citrus paradisi Macf.) during fruit development. J. Agric. Food Chem. 2016, 64, 9022–9032. [Google Scholar] [CrossRef]
- Asikin, Y.; Taira, I.; Inafuku, S.; Sumi, H.; Sawamura, M.; Takara, K.; Wada, K. Volatile aroma components and antioxidant activities of the flavedo peel extract of unripe shiikuwasha (Citrus depressa Hayata). J. Food Sci. 2012, 77, 469–475. [Google Scholar] [CrossRef]
- Liu, C.; Yan, F.; Gao, H.; He, M.; Wang, Z.; Cheng, Y.; Deng, X.; Xu, J. Features of citrus terpenoid production as revealed by carotenoid, limonoid and aroma profiles of two pummelos (Citrus maxima) with different flesh color. J. Sci. Food Agric. 2015, 95, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.R.; Lafuente, M.T.; González-Candelas, L. Spatial study of antioxidant enzymes, peroxidase and phenylalanine ammonia-lyase in the citrus fruit-Penicillium digitatum interaction. Postharvest Biol. Technol. 2006, 39, 115–124. [Google Scholar] [CrossRef]
- Ben-Yehoshua, S.; Rodov, V.; Nafussi, B.; Feng, X.; Yen, J.; Koltai, T.; Nelkenbaum, U. Involvement of limonene hydroperoxides formed after oil gland injury in the induction of defense response against Penicillium digitatum in lemon fruit. J. Agric. Food Chem. 2008, 56, 1889–1895. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.Q.; Azrina, A. Antioxidant content and activity in different parts of pomelo [Citrus grandis (L.) Osbeck] by-products. Acta Hortic. 2017, 27–34. [Google Scholar] [CrossRef]
- Cronjé, P.J.R.; Zacarías, L.; Alférez, F. Susceptibility to postharvest peel pitting in Citrus fruits as related to albedo thickness, water loss and phospholipase activity. Postharvest Biol. Technol. 2017, 123, 77–82. [Google Scholar] [CrossRef]
- Chen, J.; Li, S.; Xu, J.; Ding, F.; Wang, Z.; Cheng, Y.; Deng, X. Concentration and distribution of main bitter compounds in fruit tissues of “Oroblanco” (Citrus grandis L.× Citrus paradisi Macf.). Sci. Hortic. (Amsterdam) 2015, 193, 84–89. [Google Scholar] [CrossRef]
- Breksa, A.P.; Manners, G.D. Determination of limonin D-ring lactone hydrolase activity by solid phase extraction with indirect fluorescence detection. J. Agric. Food Chem. 2004, 52, 3772–3775. [Google Scholar] [CrossRef]
- Hamdan, D.; El-Readi, M.Z.; Tahrani, A.; Herrmann, F.; Kaufmann, D.; Farrag, N.; El-Shazly, A.; Wink, M. Chemical composition and biological activity of Citrus jambhiri Lush. Food Chem. 2011, 127, 394–403. [Google Scholar] [CrossRef]
- Pan, Z.; Li, Y.; Deng, X.; Xiao, S. Non-targeted metabolomic analysis of orange (Citrus sinensis [L.] Osbeck) wild type and bud mutant fruits by direct analysis in real-time and HPLC-electrospray mass spectrometry. Metabolomics 2014, 10, 508–523. [Google Scholar] [CrossRef]
- Zhang, A.; Zhou, X.; Zhao, H.; Guan, Y.; Zou, S.; Yan, G.; Ma, C.W.; Liu, Q.; Wang, X. Correction: Rapidly improved determination of metabolites from biological data sets using the high-efficient TransOmics tool. Mol. BioSyst. 2015, 11, 317. [Google Scholar] [CrossRef]
- De Castro, W.V.; Mertens-Talcott, S.; Rubner, A.; Butterweck, V.; Derendorf, H. Variation of flavonoids and furanocoumarins in grapefruit juices: A potential source of variability in grapefruit juice-drug interaction studies. J. Agric. Food Chem. 2006, 54, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Böttcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2012, 84, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Nueda, M.J.; Ferrer, A.; Talón, M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics 2006, 22, 1096–1102. [Google Scholar] [CrossRef]
| Compound Class | Compound Name | Taste Trait |
|---|---|---|
| Flavanones | Naringin (O-neohesperidoside) | Bitter |
| Narirutin (O-rutinoside) | Tasteless | |
| Diosmin (O-rutinoside) | Tasteless | |
| Neodiosmin (O-neohesperidoside) | Bitter | |
| Flavones | Tangeretin | Bitter |
| Nobiletin | Bitter | |
| Sinensetin | Bitter | |
| Flavonols | Rutin | Tasteless |
| Limonoid aglycones | Limonin D-ring lactone | Tasteless |
| Nomilin | Bitter | |
| Limonin A-ring lactone | Bitter | |
| Limonoid glycosides | Limonin D-ring glycoside | Tasteless |
| Compound | Chemical Formula | Quantifier Ion ESI+ | Annotation Positive | Quantifier Ion ESI− | Annotation Negative | Retention Time (min) | |
|---|---|---|---|---|---|---|---|
| Flavonoids | |||||||
| C1 | Naringenin | C15H12O5 | 273.07 | [M+H]+ | 271.06 | [M−H]− | 9.05 |
| C2 | Hesperidin | C28H34O15 | 303.09 | [M-hesperidoside]+ | 301.07 | [M-hesperidoside]− | 6.92 |
| C3 | Neohesperidin | C28H34O15 | 303.09 | [M-neohesperidoside]+ | 301.07 | [M-hesperidoside]− | 7.7 |
| C4 | Isosinensetin | C20H20O7 | 373.13 | [M+H]+ | nd | nd | 9.75 |
| C5 | Sinensetin | C20H20O7 | 373.13 | [M+H]+ | nd | nd | 10.4 |
| C6 | Tangeretin | C20H20O7 | 373.13 | [M+H]+ | nd | nd | 11.6 |
| C7 | Eriodictyol rutinoside #1 | C27H32O15 | 597.17 | [M+H]+ | 595.17 | [M−H]− | 5.31 |
| C8 | Eriodictyol rutinoside #2 | C27H32O15 | 597.17 | [M+H]+ | 595.175 | [M−H]− | 6.14 |
| C9 | Isorhamnetin-3-O-rutinoside | C28H32O16 | 625.19 | [M+H]+ | 623.18 | [M−H]− | 6.71 |
| C10 | Isorhamnetin rutinoside hexoside | C34H42O21 | 787.22 | [M+H]+ | 785.21 | [M−H]− | 5.36 |
| C11 | Isorhamnetin rutinoside deoxyhexoside | C34H42O20 | 771.23 | [M+H]+ | 769.22 | [M−H]− | 6.07 |
| C12 | Kaempferol diDeoxyhexoside hexoside | C33H40O19 | 595.16 | [M-Hexose]+ | 739.21 | [M−H]− | 6.03 |
| C13 | Kaempferol Deoxyhexoside hexoside | C27H30O15 | 595.17 | [M+H]+ | nd | nd | 6.62 |
| C14 | Kaempferol Caffeoyl Hexoside Deoxyhexoside | C36H36O18 | 757.23 | [M+H]+ | 755.21 | [M−H]− | 5.28 |
| C15 | Kaempferol hidoxymethyl glutaryl (HMG)-glucoside tentative | C27H28O15 | 593.15 | [M+H]+ | 591.13 | [M−H]− | 7.12 |
| C16 | Quercetin hexoside rutinoside | C33H40O21 | 773.21 | [M+H]+ | 771.26 | [M−H]− | 5.01 |
| C17 | Poncirin | C28H34O14 | 595.20 | [M+H]+ | 593.19 | [M−H]− | 7.94 |
| C18 | Narirutin | C27H32O14 | 581.18 | [M+H]+ | 579.17 | [M−H]− | 5.53 |
| C19 | Naringin | C27H32O14 | 581.18 | [M+H]+ | 579.17 | [M−H]− | 6.65 |
| Limonoids | |||||||
| C20 | Deacetyl Nomilinic acid glycoside tentative | C32H46O15 | nd | nd | 669.27 | [M−H]− | 6.55 |
| C21 | Deacetyl Nomilin glycoside | C32H44O14 | nd | nd | 651.27 | [M−H]− | 6.96 |
| C22 | Nomilin | C28H34O9 | 515.23 | [M+H]+ | 513.21 | [M−H]− | 11.14 |
| C23 | Nomilinic acid glycoside | C34H48O16 | 735.29 | [M+Na]+ | 711.29 | [M−H]− | 7.37 |
| C24 | Nomilin A-ring lactone | C28H36O10 | 533.24 | [M+H]+ | 531.23 | [M−H]− | 10.53 |
| C25 | Nomilin glycoside tentative | C34H44O14 | nd | nd | 675.27 | [M−H]− | 9.51 |
| C26 | Obacunone | C26H30O7 | 455.23 | [M+H]+ | nd | nd | 11.16 |
| C27 | Obacunoic acid | C26H32O8 | 473.21 | [M+H]+ | 471.20 | [M−H]− | 10.20 |
| C28 | Obacunone glycoside | C32H42O13 | nd | nd | 633.25 | [M−H]− | 7.71 |
| C29 | Ichangin | C26H32O9 | 489.21 | [M+H]+ | 487.19 | [M−H]− | 9.63 |
| C30 | Limonoate A-ring lactone | C26H32O9 | 489.21 | [M+H]+ | 487.19 | [M−H]− | 9.81 |
| C31 | Limonin | C26H30O8 | 471.20 | [M+H]+ | 469.18 | [M−H]− | 10.57 |
| C32 | Limonin 17-β-d-glucopyranoside | C32H42O14 | 471.20 | [M-Hexose]+ | 649.25 | [M−H]− | 6.43 |
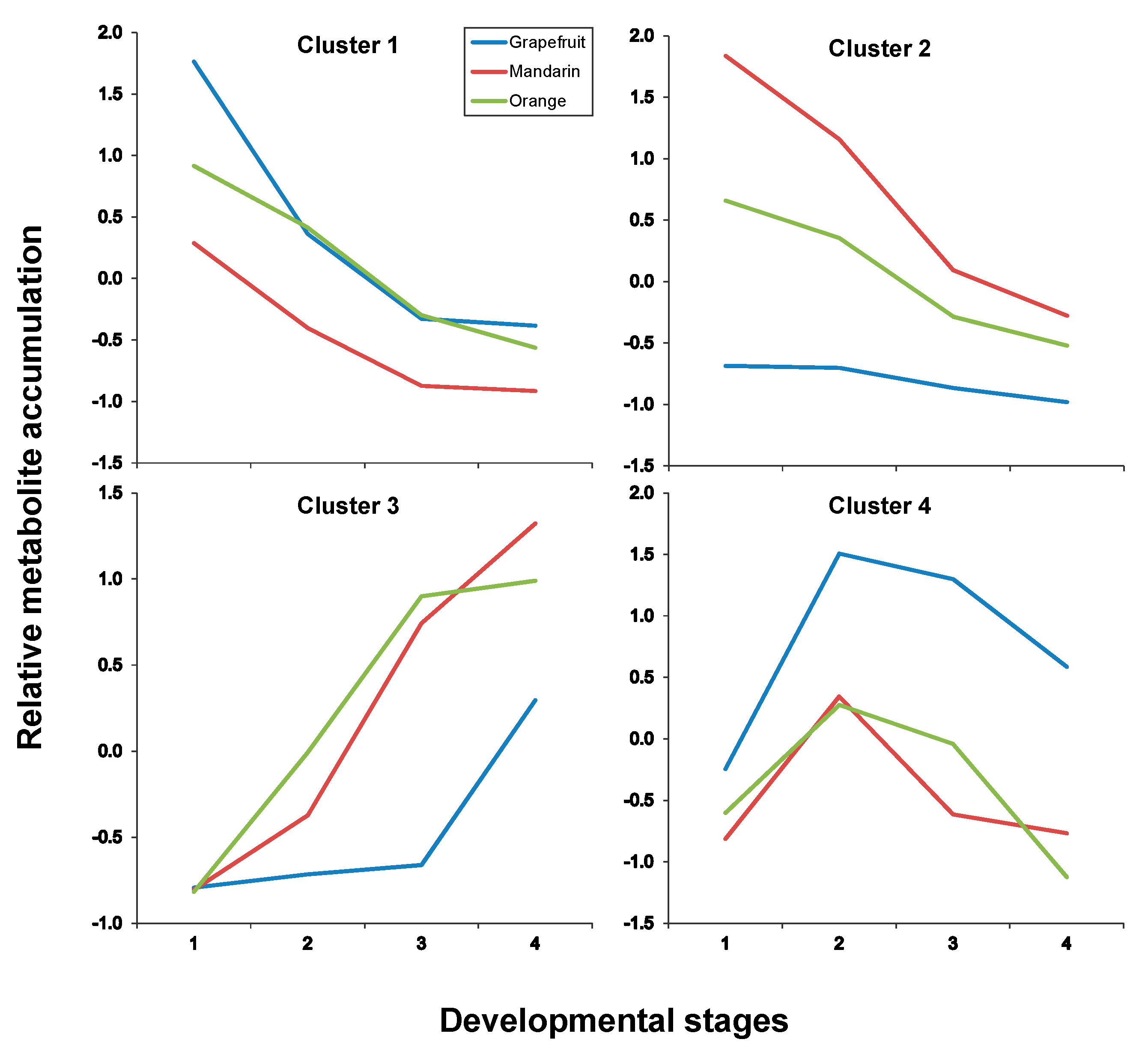
| Pulp | |
|---|---|
| Cluster # | Metabolites * |
| Cluster 1 | C1, C7, C8, C13, C17, C19 |
| Cluster 2 | C2, C3, C4, C5, C6, C9, C27, C29, C30 |
| Cluster 3 | C14, C20, C23, C28, C32 |
| Cluster 4 | C15, C21, C22, C24, C25, C26, C31 |
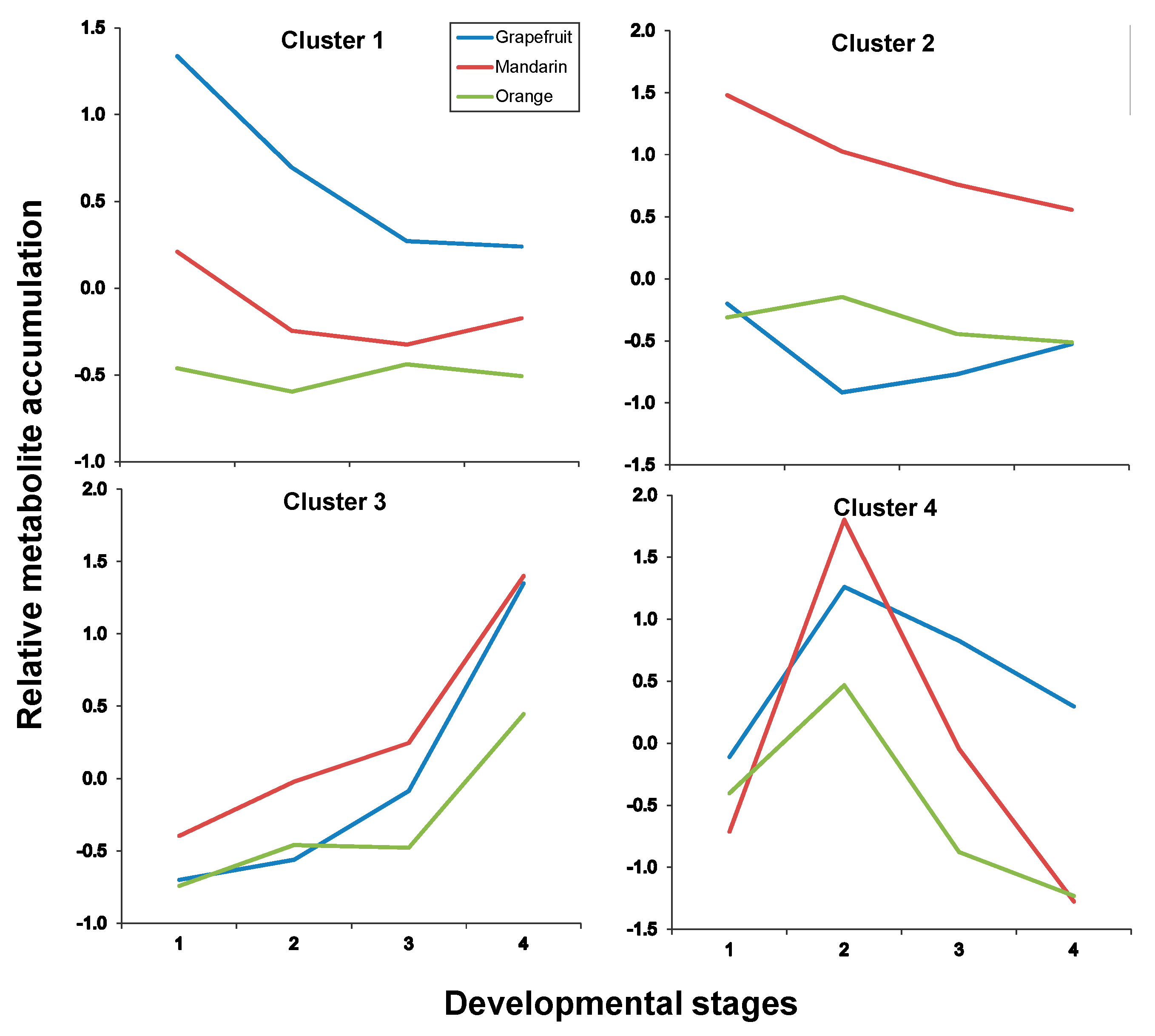
| Albedo | |
| Cluster # | Metabolites |
| Cluster 1 | C1, C15, C17, C19 |
| Cluster 2 | C2, C3, C6, C7, C8, C12, C30 |
| Cluster 3 | C14, C18, C23, C25, C28, C32 |
| Cluster 4 | C21, C22, C26, C31 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nadi, R.; Golein, B.; Gómez-Cadenas, A.; Arbona, V. Developmental Stage- and Genotype-Dependent Regulation of Specialized Metabolite Accumulation in Fruit Tissues of Different Citrus Varieties. Int. J. Mol. Sci. 2019, 20, 1245. https://doi.org/10.3390/ijms20051245
Nadi R, Golein B, Gómez-Cadenas A, Arbona V. Developmental Stage- and Genotype-Dependent Regulation of Specialized Metabolite Accumulation in Fruit Tissues of Different Citrus Varieties. International Journal of Molecular Sciences. 2019; 20(5):1245. https://doi.org/10.3390/ijms20051245
Chicago/Turabian StyleNadi, Roya, Behrouz Golein, Aurelio Gómez-Cadenas, and Vicent Arbona. 2019. "Developmental Stage- and Genotype-Dependent Regulation of Specialized Metabolite Accumulation in Fruit Tissues of Different Citrus Varieties" International Journal of Molecular Sciences 20, no. 5: 1245. https://doi.org/10.3390/ijms20051245
APA StyleNadi, R., Golein, B., Gómez-Cadenas, A., & Arbona, V. (2019). Developmental Stage- and Genotype-Dependent Regulation of Specialized Metabolite Accumulation in Fruit Tissues of Different Citrus Varieties. International Journal of Molecular Sciences, 20(5), 1245. https://doi.org/10.3390/ijms20051245







