Extracellular Vesicles from Thyroid Carcinoma: The New Frontier of Liquid Biopsy
Abstract
1. Introduction
2. Liquid Biopsy: History, Advantages
3. Potential Advantages of EVs for Liquid Biopsy
3.1. Protein Expression Profile
3.2. Non-Coding RNA Content
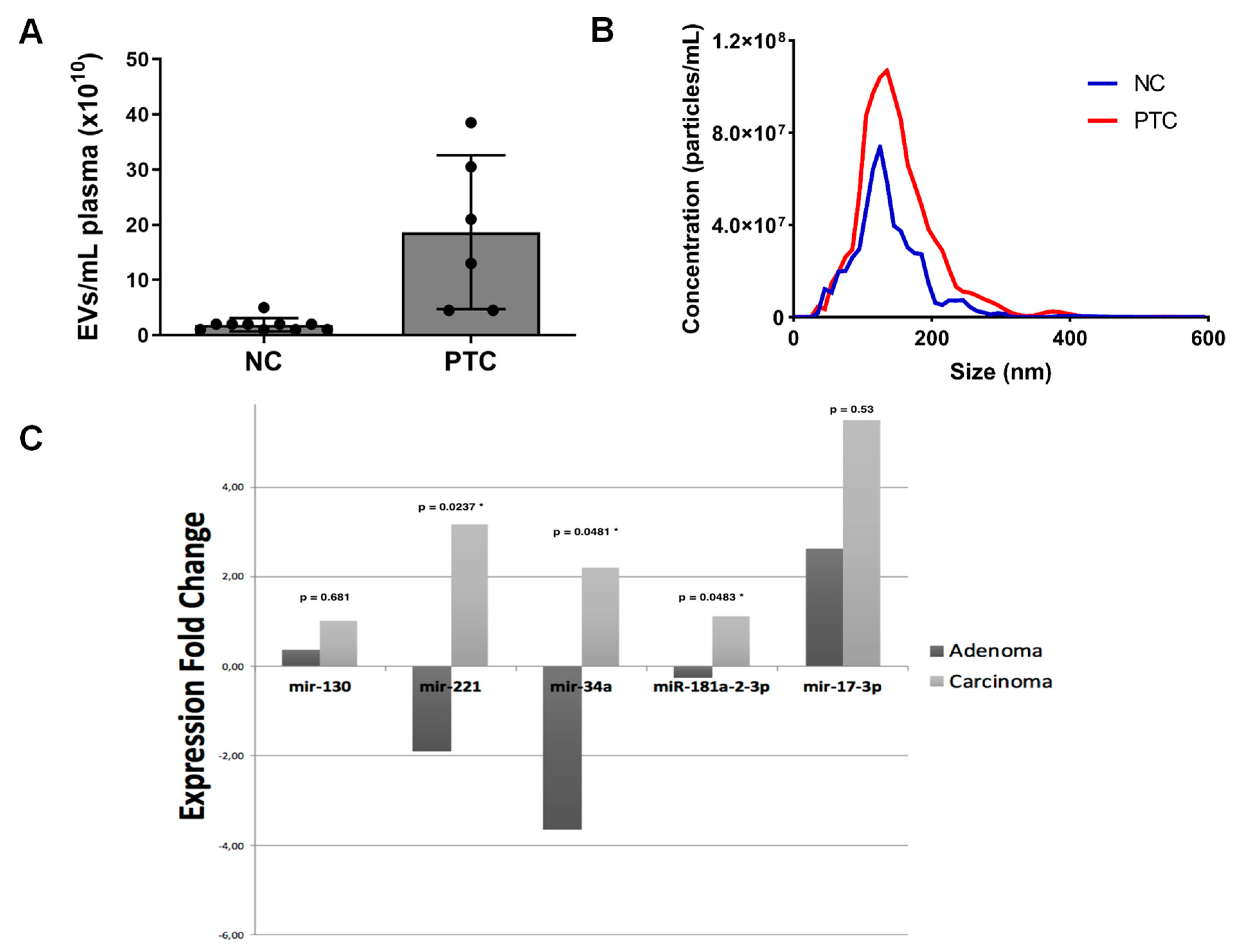
4. Potential Advantages of EVs for Liquid Biopsy in Patients with Thyroid Cancer
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| EVs | Extracellular vesicles |
| PTC | Papillary thyroid carcinoma |
| cfDNA | Cell-free DNA |
| MTC | Medullary thyroid carcinoma |
| NSCLC | Non-small cell lung cancer |
| CTCs | Circulating tumor cell |
| miRNA | Micro RNA |
| lncRNA | Long non-coding RNA |
| circRNA | Circular RNA |
| snoRNA | Small nucleolar RNA |
| snRNA | Small nuclear RNA |
| tRNA | Transfer RNA |
| piRNA | Piwi-interacting RNA |
| cfRNA | Cell-free RNA |
References
- Enewold, L.; Zhu, K.; Ron, E.; Marrogi, A.J.; Stojadinovic, A.; Peoples, G.E.; Devesa, S.S. Rising thyroid cancer incidence in the United States by demographic and tumor characteristics, 1980–2005. Cancer Epidemiol. Biomark. Prev. 2009, 18, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Mark, A.; Knox, M.D. Hawaii Island Family Medicine Residency, Hilo, Hawaii. Thyroid nodules. Am. Fam. Physician 2013, 88, 193–196. [Google Scholar]
- Brito, J.P.; Davies, L.; Zeballos-Palacios, C.; Morris, J.C.; Montori, V.M. Papillary lesions of indolent course: Reducing the overdiagnosis of indolent papillary thyroid cancer and unnecessary treatment. Future Oncol. 2014, 10, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.S.; Doherty, G.M.; Haugen, B.R.; Kloos, R.T.; Lee, S.L.; Mandel, S.J.; Mazzaferri, E.L.; McIver, B.; Pacini, F.; Sclumberger, M.; et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid 2009, 19, 1167–1214. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.T.; Lee, E.J.; Huang, M.G.; Park, Y.I.; Khullar, A.; Plodkowski, R.A. Diagnosis and treatment of patients with thyroid cancer. Am. Health Drug Benefits 2015, 8, 30–40. [Google Scholar] [PubMed]
- Halvaei, S.; Daryani, S.; Eslami-S, Z.; Samadi, T.; Jafarbeik-Iravani, N.; Bakhshayesh, T.O.; Majidzadeh-A, K.; Esmaeili, R. Exosomes in cancer liquid biopsy: A focus on breast cancer. Mol. Ther. Nucleic Acids 2018, 10, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Gyorgy, B.; Szabo, T.G.; Pasztoi, M.; Pal, Z.; Misjak, P.; Aradi, B.; Laszlo, V.; Pallinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. CMLS 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, 1413. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Tkach, M.; Thery, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yoshioka, Y.; Ochiya, T. Extracellular vesicle transfer of cancer pathogenic components. Cancer Sci. 2016, 107, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Laurenzana, I.; Lamorte, D.; Trino, S.; De Luca, L.; Ambrosino, C.; Zoppoli, P.; Ruggieri, V.; Del Vecchio, L.; Musto, P.; Caivano, A.; et al. Extracellular Vesicles: A New Prospective in Crosstalk between Microenvironment and Stem Cells in Hematological Malignancies. Stem Cells Int. 2018, 2018, 9863194. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.P.; Kim, E.Y.; Badr, C.E.; Weissleder, R.; Mempel, T.R.; Tannous, B.A.; Breakefield, X.O. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015, 6, 7029. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Liu, Y.; Wang, J.; Guo, Z.; Zhang, Q.; Yu, F.; Zhang, Y.; Huang, K.; Li, Y.; Song, E.; Zheng, X.L. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2012, 97, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Osti, D.; Del Bene, M.; Rappa, G.; Santos, M.; Matafora, V.; Richichi, C.; Faletti, S.; Beznoussenko, G.V.; Mironov, A.; Bachi, A.; et al. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin. Cancer Res. 2019, 25, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. C. R. Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar] [PubMed]
- Kwapisz, D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann. Transl. Med. 2017, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Cote, G.J.; Evers, C.; Hu, M.I.; Grubbs, E.G.; Williams, M.D.; Hai, T.; Duose, D.Y.; Houston, M.R.; Bui, J.H.; Mehrotra, M.; et al. Prognostic significance of circulating RET M918T mutated tumor DNA in patients with advanced medullary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2017, 102, 3591–3599. [Google Scholar] [CrossRef] [PubMed]
- Sandulache, V.C.; Williams, M.D.; Lai, S.Y.; Lu, C.; William, W.N.; Busaidy, N.L.; Cote, G.J.; Singh, R.R.; Luthra, R.; Cabanillas, M.E. Real-time genomic characterization utilizing circulating cell-free DNA in patients with anaplastic thyroid carcinoma. Thyroid 2017, 27, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Trino, S.; Lamorte, D.; Caivano, A.; Laurenzana, I.; Tagliaferri, D.; Falco, G.; Del Vecchio, L.; Musto, P.; De Luca, L. MicroRNAs as New Biomarkers for Diagnosis and Prognosis, and as Potential Therapeutic Targets in Acute Myeloid Leukemia. Int. J. Mol. Sci. 2018, 19, 460. [Google Scholar] [CrossRef] [PubMed]
- Fuji, T.; Umeda, Y.; Nyuya, A.; Taniguchi, F.; Kawai, T.; Yasui, K.; Toshima, T.; Yoshida, K.; Fujiwara, T.; Goel, A.; et al. Detection of circulating microRNAs with Ago2 complexes to monitor the tumor dynamics of colorectal cancer patients during chemotherapy. Int. J. Cancer 2018, 142, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabières, C.; Riethdorf, S. Cancer micrometastases. Nat. Rev. Clin. Oncol. 2009, 6, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.G.; Chianese, D.; Doyle, G.V.; Miller, M.C.; Russell, T.; Sanders, R.A., Jr.; Terstappen, L.W. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int. J. Oncol. 2005, 27, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and miRNA is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating miRNA as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of miRNA in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Kogure, T.; Lin, W.L.; Yan, I.K.; Braconi, C.; Patel, T. Intercellular nanovesicle-mediated miRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 2011, 54, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Ma, L.; Gong, M.; Su, G.; Zhu, S.; Zhang, W.; Wang, S.; Li, Z.; Chen, C.; Li, L.; et al. Protein Profiling and Sizing of Extracellular Vesicles from Colorectal Cancer Patients via Flow Cytometry. ACS Nano 2018, 12, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.J.; Jin, D.D.; Jiang, F.; Liu, J.X.; Qu, L.S.; Ni, W.K.; Liu, Z.X.; Lu, C.H.; Ni, R.Z.; Zhu, J.; et al. Characterization and proteomic profiling of pancreatic cancer-derived serum exosomes. J. Cell Biochem. 2019, 120, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Lim, J.W.; Tauro, B.J.; Ji, H.; Moritz, R.L.; Simpson, R.J. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteom. 2010, 9, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Ji, H.; Chen, M.; Robinson, B.W.; Dick, I.M.; Creaney, J.; Simpson, R.J. Secreted primary human malignant mesothelioma exosome signature reflects oncogenic cargo. Sci. Rep. 2016, 6, 32643. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Rubio, D.; Martin-Burriel, I.; Gil, A.; Cubero, P.; Forner, M.; Khalyfa, A.; Marin, J.M. Stability of Circulating Exosomal miRNAs in Healthy Subjects. Sci. Rep. 2018, 8, 10306. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Alegre, E.; Sanmamed, M.F.; Rodriguez, C.; Carranza, O.; Martín-Algarra, S.; González, A. Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Arch. Pathol. Lab. Med. 2014, 138, 828–832. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, B.; Yue, S.; Galli, U.; Rana, S.; Gross, W.; Müller, M.; Giese, N.A.; Kalthoff, H.; Becker, T.; Büchler, M.W.; et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer 2015, 136, 2616–2627. [Google Scholar] [CrossRef] [PubMed]
- Dejima, H.; Iinuma, H.; Kanaoka, R.; Matsutani, N.; Kawamura, M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 2017, 13, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, F.; Zhao, Q.; Zhan, F.; Wang, R.; Wang, L.; Zhang, Y.; Huang, X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci. Rep. 2017, 7, 4150. [Google Scholar] [CrossRef] [PubMed]
- Manterola, L.; Guruceaga, E.; Gállego Pérez-Larraya, J.; González-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Samprón, N.; Barrena, C.; et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014, 16, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.; Di Bella, S.; Nigita, G.; Macca, V.; Laganà, A.; Rosalba, G.; Alfredo, P.; Alfredo, F. miRandola: Extracellular circulating microRNAs database. PLoS ONE 2012, 7, e47786. [Google Scholar] [CrossRef] [PubMed]
- Alhasan, A.H.; Scott, A.W.; Wu, J.J.; Feng, G.; Meeks, J.J.; Thaxton, C.S.; Mirkin, C.A. Circulating miRNA signature for the diagnosis of very high-risk prostate cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 10655–10660. [Google Scholar] [CrossRef] [PubMed]
- Warnecke-Eberz, U.; Chon, S.H.; Hölscher, A.H.; Drebber, U.; Bollschweiler, E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: Comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015, 36, 4643–4653. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Rappa, G.; Mercapide, J.; Anzanello, F.; Pope, R.M.; Lorico, A. Biochemical and biological characterization of exosomes containing prominin-1/CD133. Mol. Cancer 2013, 12, 62. [Google Scholar] [CrossRef] [PubMed]
- Nabet, B.Y.; Qiu, Y.; Shabason, J.E.; Wu, T.J.; Yoon, T.; Kim, B.C.; Benci, J.L.; DeMichele, A.M.; Tchou, J.; Marcotrigiano, J.; et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 2017, 170, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Rappa, G.; Conigliaro, A.; Santos, M.F.; Alessandro, R.; Lorico, A. Cancer relevance of signal recognition particle and other non-coding RNAs in extracellular vesicles. Transl. Cancer Res. 2017, 6, S1257–S1260. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; t Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gu, Y.; Han, Y.; Zhang, Q.; Jiang, Z.; Zhang, X.; Huang, B.; Xu, X.; Zheng, J.; Cao, X. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell 2016, 30, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Rodriguez, R.; Garcia, J.M.; Muñoz, C.; Silva, J.; Dominguez, G.; Provencio, M.; España, P.; Bonilla, F. Detection of epithelial tumor RNA in the plasma of colon cancer patients is associated with advanced stages and circulating tumour cells. Gut 2002, 50, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Dominguez, G.; Silva, J.; Garcia, J.M.; Sanchez, A.; Rodriguez, O.; Provencio, M.; España, P.; Bonilla, F. Detection of epithelial messenger RNA in the plasma of breast cancer patients is associated with poor prognosis tumor characteristics. Clin. Cancer Res. 2001, 7, 2821–2825. [Google Scholar] [PubMed]
- Kopreski, M.S.; Benko, F.A.; Gocke, C.D. Circulating RNA as a tumor marker: Detection of 5T4 mRNA in breast and lung cancer patient serum. Ann. N. Y. Acad. Sci. 2001, 945, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Gal, S.; Fidler, C.; Lo, Y.M.; Chin, K.; Moore, J.; Harris, A.L.; Wainscoat, J.S. Detection of mammaglobin mRNA in the plasma of breast cancer patients. Ann. N. Y. Acad. Sci. 2001, 945, 192–194. [Google Scholar] [CrossRef] [PubMed]
- Fleischhacker, M.; Beinert, T.; Ermitsch, M.; Seferi, D.; Possinger, K.; Engelmann, C.; Jandrig, B. Detection of amplifiable messenger RNA in the serum of patients with lung cancer. Ann. N. Y. Acad. Sci. 2001, 945, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Hasselmann, D.O.; Rappl, G.; Rössler, M.; Ugurel, S.; Tilgen, W.; Reinhold, U. Detection of tumor-associated circulating mRNA in serum, plasma and blood cells from patients with disseminated malignant melanoma. Oncol. Rep. 2001, 8, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Keup, C.; Mach, P.; Aktas, B.; Tewes, M.; Kolberg, H.C.; Hauch, S.; Sprenger-Haussels, M.; Kimmig, R.; Kasimir-Bauer, S. RNA profiles of circulating tumor cells and extracellular vesicles for therapy stratification of metastatic breast cancer patients. Clin. Chem. 2018, 64, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.S.; Doherty, G.M.; Haugen, B.R.; Kloos, R.T.; Lee, S.L.; Mandel, S.J.; Mazzaferri, E.L.; McIver, B.; Sherman, S.I.; Tuttle, R.M. Management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid 2006, 16, 109–142. [Google Scholar] [CrossRef] [PubMed]
- Pacini, F.; Schlumberger, M.; Dralle, H.; Elisei, R.; Smit, J.W.; Wiersinga, W.; European Thyroid Cancer Taskforce. European consensus for the management of patients with differentiated thyroid carcinoma of the follicular epithelium. Eur. J. Endocrinol. 2006, 154, 787–803. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Gundara, J.S.; Glover, A.; Serpell, J.; Sidhu, S.B. MicroRNA expression profiles in the management of papillary thyroid cancer. Oncologist 2014, 19, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Zhao, J.T.; Gundara, J.; Serpell, J.; Bach, L.A.; Sidhu, S. Papillary thyroid cancer derived exosomes contain miRNA-146b and miRNA-222. J. Surg. Res. 2015, 196, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Rosignolo, F.; Sponziello, M.; Giacomellietal, L. Identification of thyroid-associated serum microRNA profiles and their potential use in thyroid cancer follow-up. J. Endocr. Soc. 2017, 1, 3–13. [Google Scholar] [PubMed]
- Lee, J.C.; Zhao, J.T.; Clifton-Bligh, R.J.; Gill, A.; Gundara, J.S.; Ip, J.C.; Glover, A.; Sywak, M.S.; Delbridge, L.W.; Robinson, B.G.; et al. MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer 2013, 119, 4358. [Google Scholar] [CrossRef] [PubMed]
- Cantara, S.; Pilli, T.; Sebastiani, G.; Cevenini, G.; Busonero, G.; Cardinale, S.; Dotta, F.; Pacini, F. Circulating miRNA95 and miRNA190 are sensitive markers for the differential diagnosis of thyroid nodules in a Caucasian population. J. Clin. Endocrinol. Metab. 2014, 99, 4190–4198. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Song, Q.; Li, H.; Lou, Y.; Wang, L. Circulating miR- 25-3p and miR-451a may be potential biomarkers for the diagnosis of papillary thyroid carcinoma. PLoS ONE 2015, 10, e0135549. [Google Scholar]
- Samsonov, R.; Burdakov, V.; Shtam, T.; Radzhabovа, Z.; Vasilyev, D.; Tsyrlina, E.; Titov, S.; Ivanov, M.; Berstein, L.; Filatov, M.; et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumour Biol. 2016, 37, 12011–12021. [Google Scholar] [CrossRef] [PubMed]
- Yoruker, E.E.; Terzioglu, D.; Teksoz, S.; Uslu, F.E.; Gezer, U.; Dalay, N. MicroRNA expression profiles in papillary thyroid carcinoma, benign thyroid nodules and healthy controls. J. Cancer 2016, 7, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Rosignolo, F.; Memeo, L.; Monzani, F.; Colarossi, C.; Pecce, V.; Verrienti, A.; Durante, C.; Grani, G.; Lamartina, L.; Forte, S.; et al. MicroRNA-based molecular classification of papillary thyroid carcinoma. Int. J. Oncol. 2017, 50, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Celano, M.; Rosignolo, F.; Maggisano, V.; Pecce, V.; Iannone, M.; Russo, D.; Bulotta, S. MicroRNAs as biomarkers in thyroid carcinoma. Int. J. Genom. 2017, 2017, 11. [Google Scholar] [CrossRef] [PubMed]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D.; Diorio, D.; Nikiforov, Y.E. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J. Clin. Endocr. Metab. 2008, 93, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, M.; Perren, A.; Moch, H.; Komminoth, P.; Nikiforov, Y.E.; Nikiforova, M.N. Comprehensive microRNA expression profiling identifies novel markers in follicular variant of papillary thyroid carcinoma. Thyroid 2013, 23, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, H.; Schnitzer-Perlman, T.; Shtabsky, A.; VandenBussche, C.J.; Ali, S.Z.; Kolar, Z.; Pagni, F.; Rosetta Genomics Group; Bar, D.; Meiri, E. Analytical validity of a microRNA-based assay for diagnosing indeterminate thyroid FNA smears from routinely prepared cytology slides. Cancer Cytopathol. 2016, 124, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Zhan, S.; Xia, W.; Huang, L.; Ge, W.; Wang, T. Proteomics study of serum exosomes from papillary thyroid cancer patients. Endocr. Relat. Cancer 2018, 25, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Gangadaran, P.; Li, X.J.; Kalimuthu, S.K.; Min, O.J.; Hong, C.M.; Rajendran, R.L.; Lee, H.W.; Zhu, L.; Baek, S.H.; Jeong, S.Y.; et al. New optical imaging reporter-labeled anaplastic thyroid cancer-derived extracellular vesicles as a platform for in vivo tumor targeting in a mouse model. Sci. Rep. 2018, 8, 13509. [Google Scholar] [CrossRef] [PubMed]
- Degosserie, J.; Heymans, C.; Spourquet, C.; Halbout, M.; D’Auria, L.; Van Der Smissen, P.; Vertommen, D.; Courtoy, P.J.; Tyteca, D.; Pierreux, C.E. Extracellular vesicles from endothelial progenitor cells promote thyroid follicle formation. J. Extracell. Vesicles 2018, 7, 1487250. [Google Scholar] [CrossRef] [PubMed]
- Hardin, H.; Helein, H.; Meyer, K.; Robertson, S.; Zhang, R.; Zhong, W.; Lloyd, R.V. Thyroid cancer stem-like cell exosomes: Regulation of EMT via transfer of lncRNAs. Lab. Investig. 2018, 98, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Sriramareddy, S.N.; Hamoir, E.; Chavez, M.; Louis, R.; Beckers, A.; Willems, L. Tumor cells may circulate in medullary thyroid cancer patients independenty of serum calcitonin. Endocr. Relat. Cancer 2018, 25, 12. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.L.; Wei, W.J.; Sun, Z.K.; Shen, C.T.; Song, H.J.; Zhang, X.Y.; Zhang, G.Q.; Chen, X.Y.; Luo, Q.Y. Circulating tumor cells correlate with clinicopathological features and outcomes in differentiated thyroid cancer. Cell. Physiol. Biochem. 2018, 48, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.Y.; Handy, B.; Michaelis, C.L.; Waguespack, S.G.; Hu, M.I.; Busaidy, N.; Jimenez, C.; Cabanillas, M.E.; Fritsche, H.A., Jr.; Cote, G.J.; et al. Detection and prognostic significance of circulating tumor cells in patients with metastatic thyroid cancer. J. Clin. Endocrinol. Metab. 2016, 101, 4461–4467. [Google Scholar] [CrossRef] [PubMed]
- Sorg, S.; Pachmann, K.; Brede-Hekimian, K.; Freesmeyer, M.; Winkens, T. Determining tissue origin of circulating epithelial cells (CEC) in patients with differentiated thyroid cancer by real-time PCR using thyroid mRNA probes. Cancer Lett. 2015, 356, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Ghossein, R.A.; Carusone, L.; Bhattacharya, S. Polymerase chain reaction detection of micrometastases and circulating tumor cells. Diagn. Mol. Pathol. 1999, 8, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Perdas, E.; Stawski, R.; Nowak, D.; Zubrzycka, M. Potential of liquid biopsy in papillary thyroid carcinoma in context of miRNA, BRAF and p53 mutation. Curr. Drug Targets 2018, 19, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Salvianti, F.; Giuliani, C.; Petrone, L.; Mancini, I.; Vezzosi, V.; Pupilli, C.; Pinzani, P. Integrity and quantity of total cell-free DNA in the diagnosis of thyroid cancer: Correlation with cytological classification. Int. J. Mol. Sci. 2017, 18, 1350. [Google Scholar] [CrossRef] [PubMed]
- Fussey, J.M.; Bryant, J.L.; Batis, N.; Spruce, R.J.; Hartley, A.; Good, J.S.; McCabe, C.J.; Boelaert, K.; Sharma, N.; Mehanna, H. The clinical utility of cell-free DNA measurement in differentiated thyroid cancer: A systematic review. Front. Oncol. 2018, 8, 132. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yang, Z. Whole exome sequencing of circulating cell-free tumor DNA in a follicular thyroid carcinoma patient with lung and bone metastases. J. Circ. Biomark. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Condello, V.; Macerola, E.; Ugolini, C.; De Napoli, L.; Romei, C.; Materazzi, G.; Elisei, R.; Basolo, F. Analysis of circulating tumor DNA does not improve the clinical management of patients with locally advanced and metastatic papillary thyroid carcinoma. Head Neck 2018, 40, 1752–1758. [Google Scholar] [CrossRef] [PubMed]
- Lupo, M.; Guttler, R.; Geck, Z.; Tonozzi, T.R.; Kammesheidt, A.; Braunstein, G.D. Is measurement of circulating tumor DNA of diagnostic use in patients with thyroid nodules? Endocr. Pract. 2018, 24, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Pupilli, C.; Pinzani, P.; Salvianti, F.; Fibbi, B.; Rossi, M.; Petrone, L.; Perigli, G.; De Feo, M.L.; Vezzosi, V.; Pazzagli, M.; et al. Circulating BRAFV600E in the diagnosis and follow-up of differentiated papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, 3359–3365. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.C.; Chuang, A.Y.; Poeta, L.; Koch, W.M.; Califano, J.A.; Tufano, R.P. Detectable BRAF mutation in serum DNA samples from patients with papillary thyroid carcinomas. Head Neck 2010, 32, 229–234. [Google Scholar] [PubMed]
- Cradic, K.W.; Milosevic, D.; Rosenberg, A.M.; Erickson, L.A.; McIver, B.; Grebe, S.K. Mutant BRAF(T1799A) can be detected in the blood of papillary thyroid carcinoma patients and correlates with disease status. J. Clin. Endocrinol. Metab. 2009, 94, 5001–5009. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ewertz, M.; Tufano, R.P.; Brait, M.; Carvalho, A.L.; Liu, D.; Tufaro, A.P.; Basaria, S.; Cooper, D.S.; Sidransky, D.; et al. Detection of serum deoxyribonucleic acid methylation markers: A novel diagnostic tool for thyroid cancer. J. Clin. Endocrinol. Metab. 2006, 91, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Kim, I.J.; Lee, B.J.; Lee, J.C.; Kim, I.S.; Kim, S.J.; Kim, W.J.; Jeon, Y.K.; Kim, S.S.; Kim, Y.K. Detection of plasma BRAF(V600E) mutation is associated with lung metastasis in papillary thyroid carcinomas. Yonsei Med. J. 2015, 56, 630–640. [Google Scholar]
- Zane, M.; Agostini, M.; Enzo, M.V.; Casal Ide, E.; Del Bianco, P.; Torresan, F.; Merante Boschin, I.; Pennelli, G.; Saccani, A.; Rubello, D.; et al. Circulating cell-free DNA, SLC5A8 and SLC26A4 hypermethylation, BRAF(V600E): A non-invasive tool panel for early detection of thyroid cancer. Biomed. Pharmacother. 2013, 67, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.B. Detection of Circulating Thyroid Tumor DNA in Patients with Thyroid Nodules. Master’s Thesis, The University of Western Ontario, London, ON, Canada, 2015. [Google Scholar]
- Graham, M.E.; Hart, R.D.; Douglas, S.; Makki, F.M.; Pinto, D.; Butler, A.L.; Bullock, M.; Rigby, M.H.; Trites, J.R.; Taylor, S.M.; et al. Serum microRNA profiling to distinguish papillary thyroid cancer from benign thyroid masses. J. Otolaryngol. Head Neck Surg. 2015, 44, 33. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Qin, H.; Cui, Y. MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway. Biochem. Biophys. Res. Commun. 2013, 441, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Takakura, S.; Mitsutake, N.; Nakashima, M.; Namba, H.; Saenko, V.A.; Rogounovitch, T.I.; Nakazawa, Y.; Hayashi, T.; Ohtsuru, A.; Yamashita, S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008, 99, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
| Liquid Biopsy in Thyroid Cancer | ||||
|---|---|---|---|---|
| Sample Type | Object | Up-/Down-Regulation | Histotype | References |
| EV | miR-146b, miR-222 | Up | PTC | [66] |
| miR-222, miR-142 | Up | PTC | [68] | |
| miR-25-3p, miR-451a, miR-140-3p, let-7 | Up | PTC | [70] | |
| miR-31-5p, miR-126-3p, miR-145-5p, miR-181a miR-21 | Up | PTCFTC | [71] | |
| miR-21, miR-181a-5p | Up | PTC | [73] | |
| SRC, TLN1, ITGB2, CAPNS1 | - | PTC | [78] | |
| Drug delivery system | - | TC | [79] | |
| The increase of EPC-EVs and laminins involves folliculogenesis | Up | FTC | [80] | |
| lncRNAs, linc-ROR | - | PTC | [74] | |
| lncRNA MALAT1, SLUG, SOX2,and induced EMT | - | PTC | [81] | |
| CTC | Calcitonin-positive CTCs after 12 years | Up | MTC | [82] |
| High number of CTCs | Up | DTC DM+ | [83] | |
| CTCs ≥ 5 is worse OS | - | TC | [84] | |
| High number of CTCs → progressive cancer disease | - | TC | [85] | |
| PCR detection | - | TC | [86] | |
| cfDNA/ctDNA | BRAF mutation and deregulation miRNA | Up/Down | PTC | [87] |
| RETM91PT mutation | - | MTC | [20] | |
| cfDNA integrity index | - | TC | [88] | |
| BRAF mutation | - | TC | [89] | |
| 95% common alteration between cfDNA and tissue DNA | - | FTC | [90] | |
| BRAF, PIK3CA, NRAS, PTEN, TP53 mutation in cfDNA and tissue DNA | - | ATC | [21] | |
| BRAF mutation | - | PTC | [91] | |
| ctDNA panel: 9 cancer gene driver | - | TC | [92] | |
| BRAFV600 | Up | PTC | [93] | |
| BRAFV600 | Up | PTC | [94] | |
| BRAFV600 | Up | DTC | [95] | |
| cfDNA methylation of β-actin, CDH1,DAPK, CALCA, and RARβ2 | - | DTC | [96] | |
| BRAFV600 | Up | PTC | [97] | |
| High number | Up | DTC | [98] | |
| BRAFV600 | Up | PTC | [99] | |
| cfRNA | miR-146a-5p, miR221-3p | Up | PTC | [67] |
| let-7e, miR-151-5p, miR-222 | Up | PTC | [16] | |
| miR-579, miR-95, miR-29b, miR-190 | Down Up | PTC | [69] | |
| miR-21, miR-151-5p, miR-222, miR-221 | Up | PTC | [72] | |
| let-7e, miR-151-5p, miR-222 | Up | PTC | [16] | |
| miR-146a-5p, miR-150-5p, miR-199b-3p, miR-342-3p | Down | PTC | [100] | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rappa, G.; Puglisi, C.; Santos, M.F.; Forte, S.; Memeo, L.; Lorico, A. Extracellular Vesicles from Thyroid Carcinoma: The New Frontier of Liquid Biopsy. Int. J. Mol. Sci. 2019, 20, 1114. https://doi.org/10.3390/ijms20051114
Rappa G, Puglisi C, Santos MF, Forte S, Memeo L, Lorico A. Extracellular Vesicles from Thyroid Carcinoma: The New Frontier of Liquid Biopsy. International Journal of Molecular Sciences. 2019; 20(5):1114. https://doi.org/10.3390/ijms20051114
Chicago/Turabian StyleRappa, Germana, Caterina Puglisi, Mark F. Santos, Stefano Forte, Lorenzo Memeo, and Aurelio Lorico. 2019. "Extracellular Vesicles from Thyroid Carcinoma: The New Frontier of Liquid Biopsy" International Journal of Molecular Sciences 20, no. 5: 1114. https://doi.org/10.3390/ijms20051114
APA StyleRappa, G., Puglisi, C., Santos, M. F., Forte, S., Memeo, L., & Lorico, A. (2019). Extracellular Vesicles from Thyroid Carcinoma: The New Frontier of Liquid Biopsy. International Journal of Molecular Sciences, 20(5), 1114. https://doi.org/10.3390/ijms20051114








