Size-Dependent Effect of Silver Nanoparticles on the Tumor Necrosis Factor α-Induced DNA Damage Response
Abstract
1. Introduction
2. Results
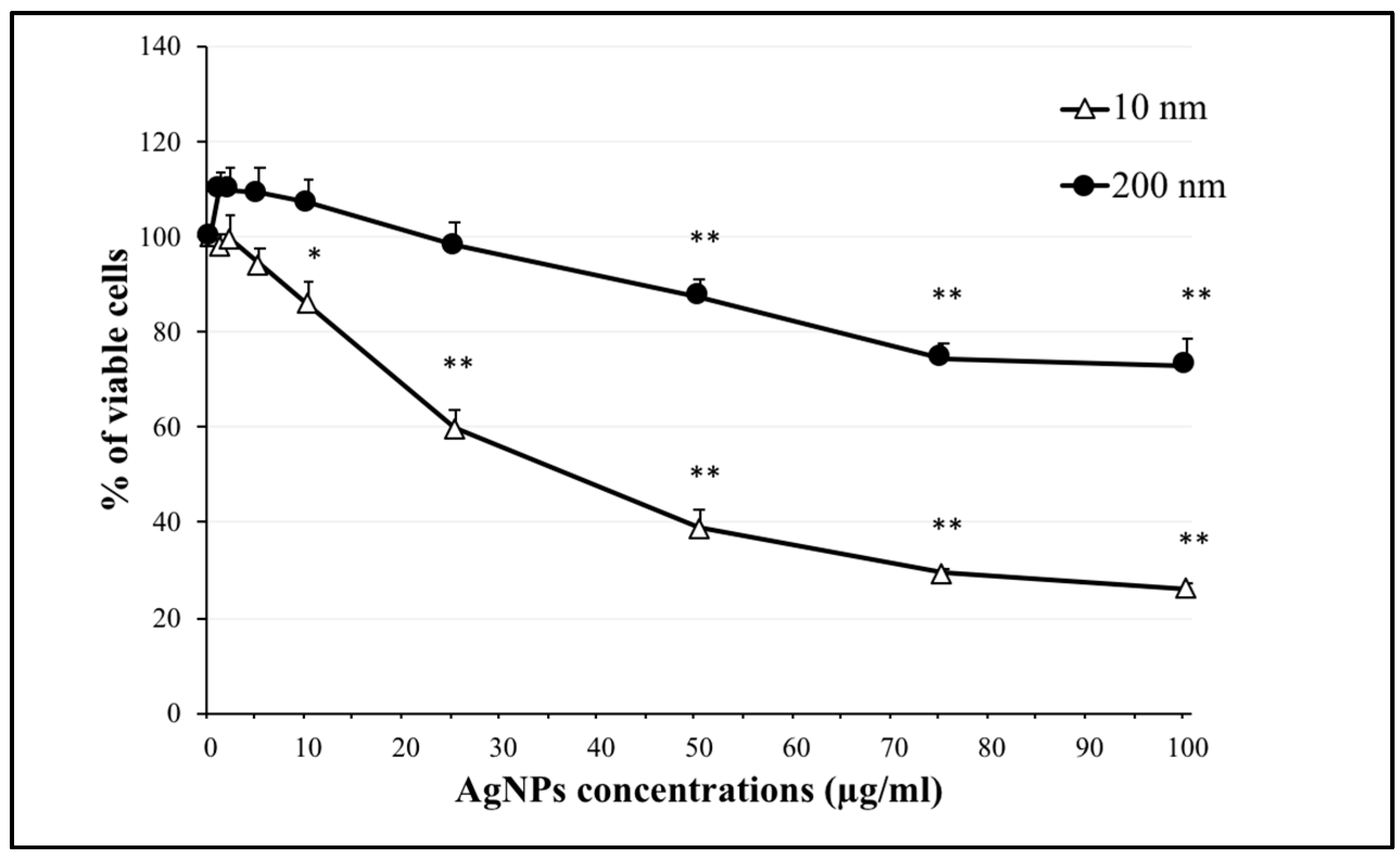
2.1. Effect of AgNPs on Cell Viability
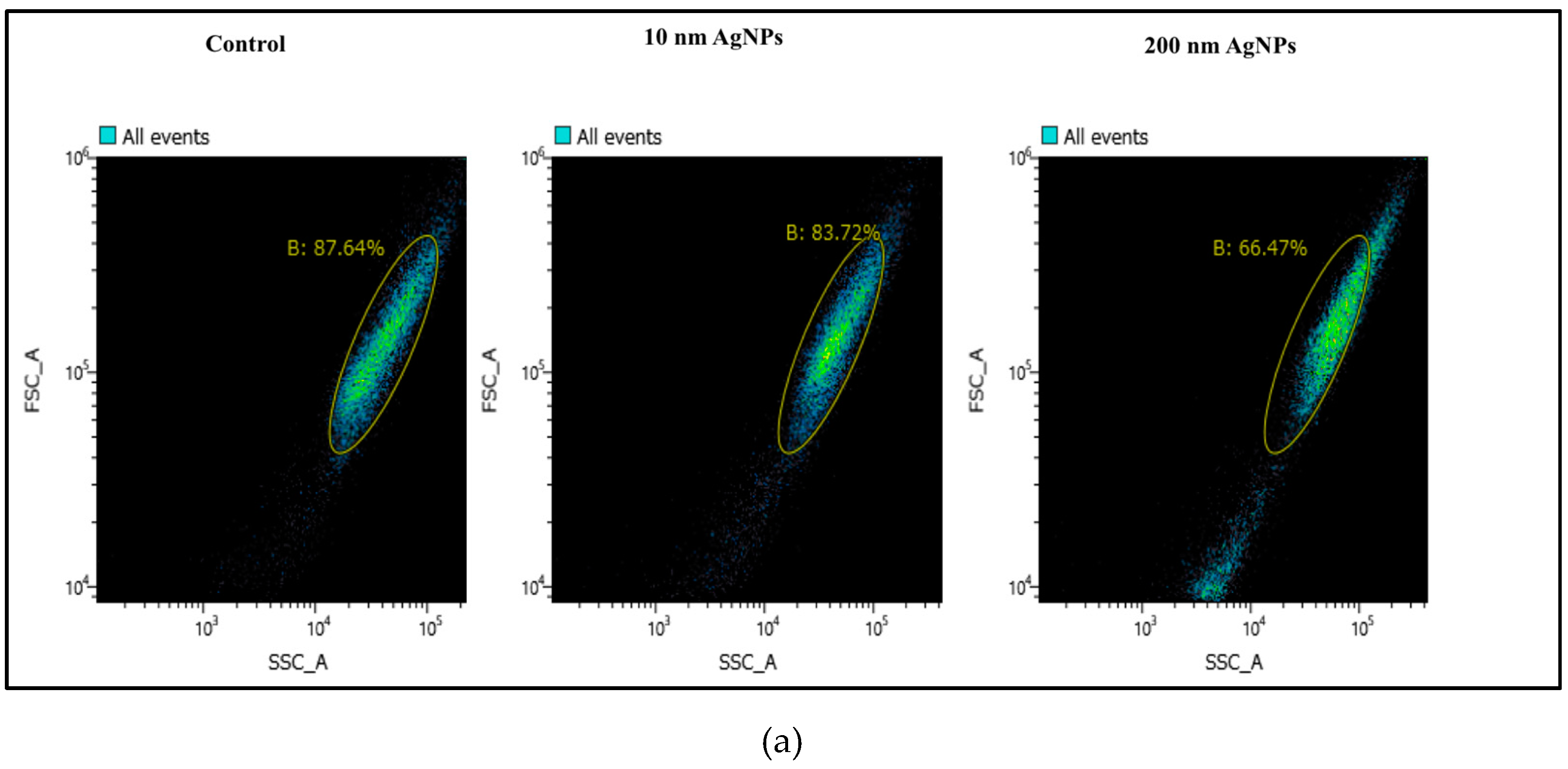
2.2. Cellular Uptake of AgNPs
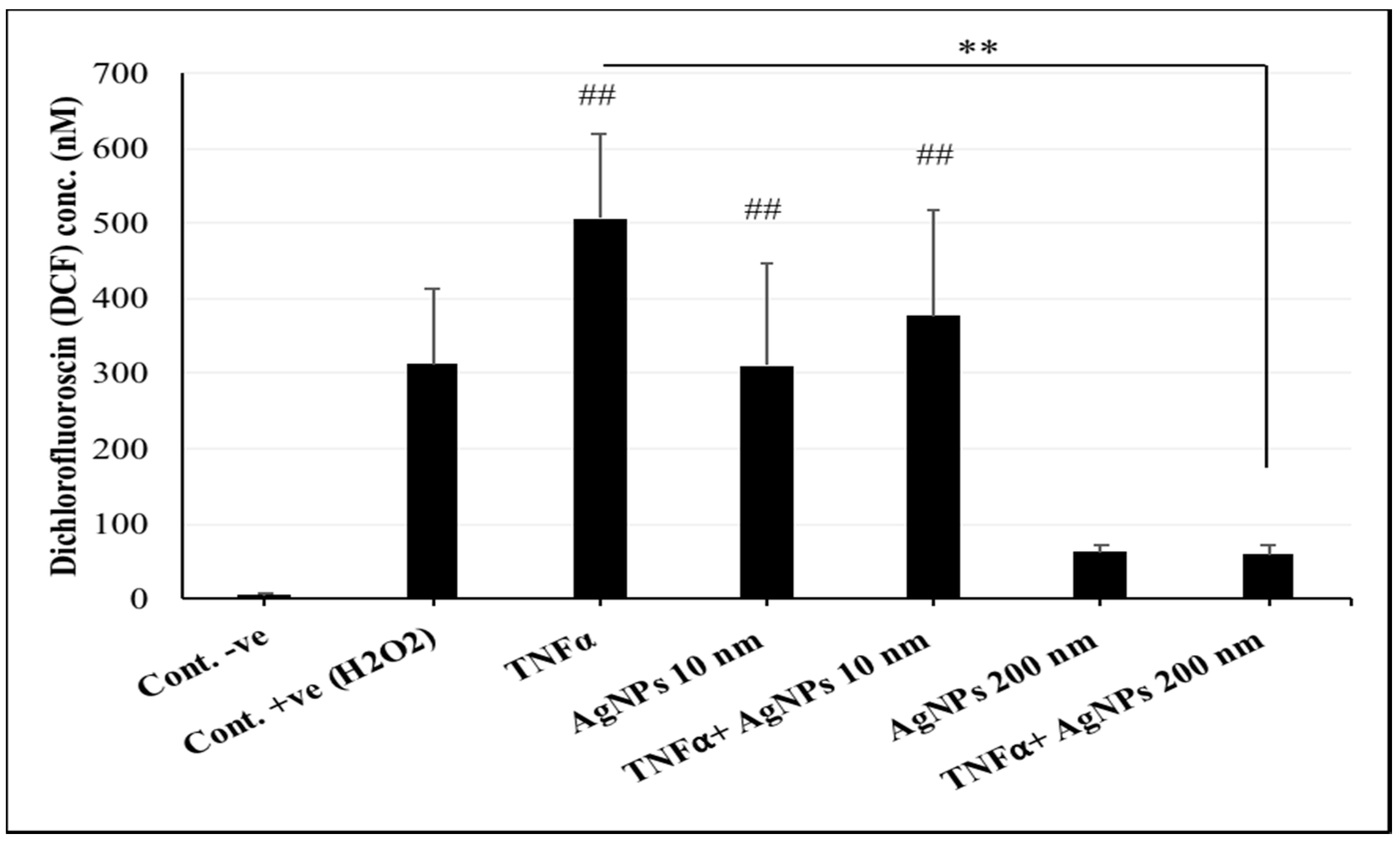
2.3. Interference of AgNPs with TNFα-Induced ROS Generation
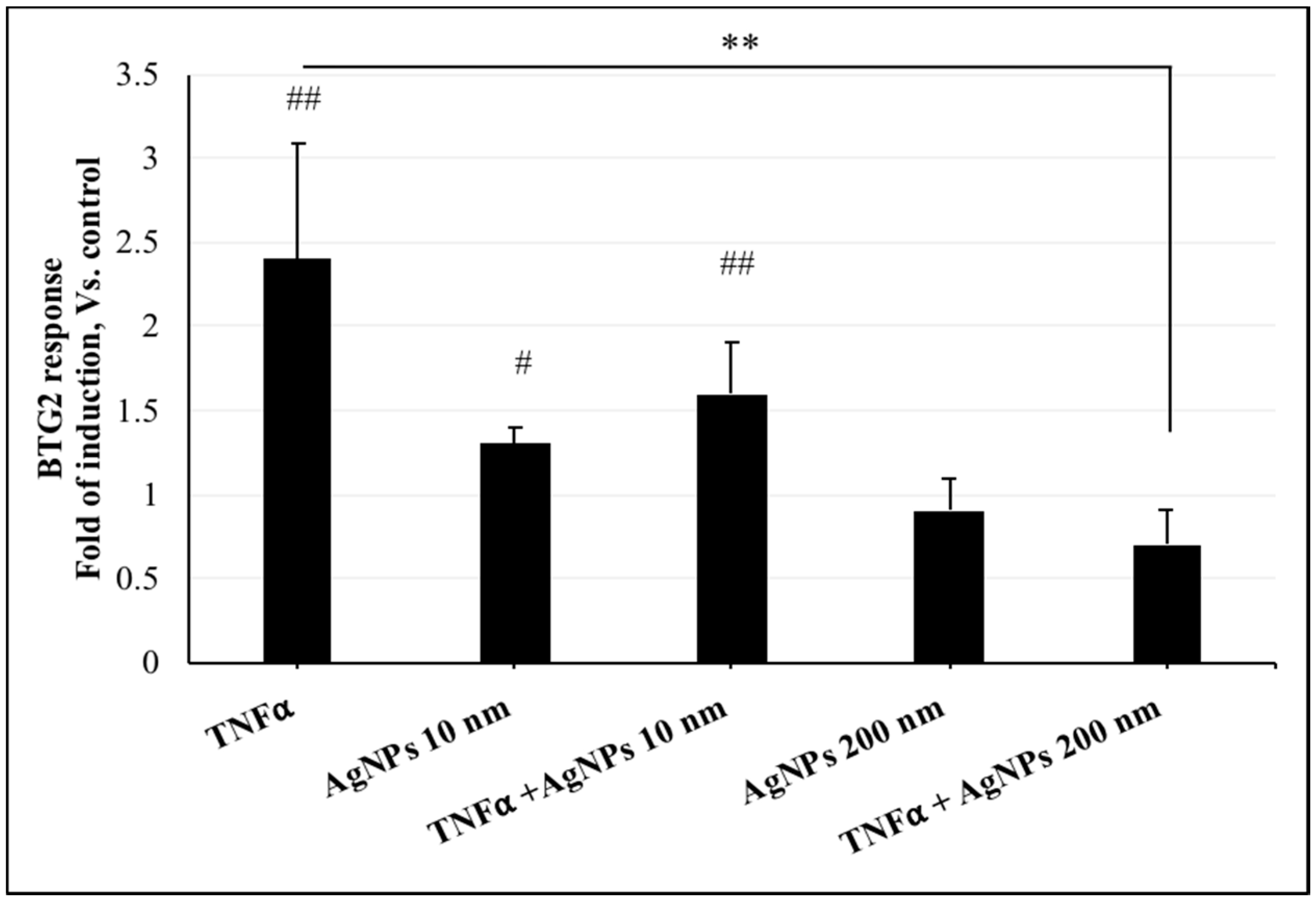
2.4. Effect of AgNPs on TNFα-Induced DNA Damage
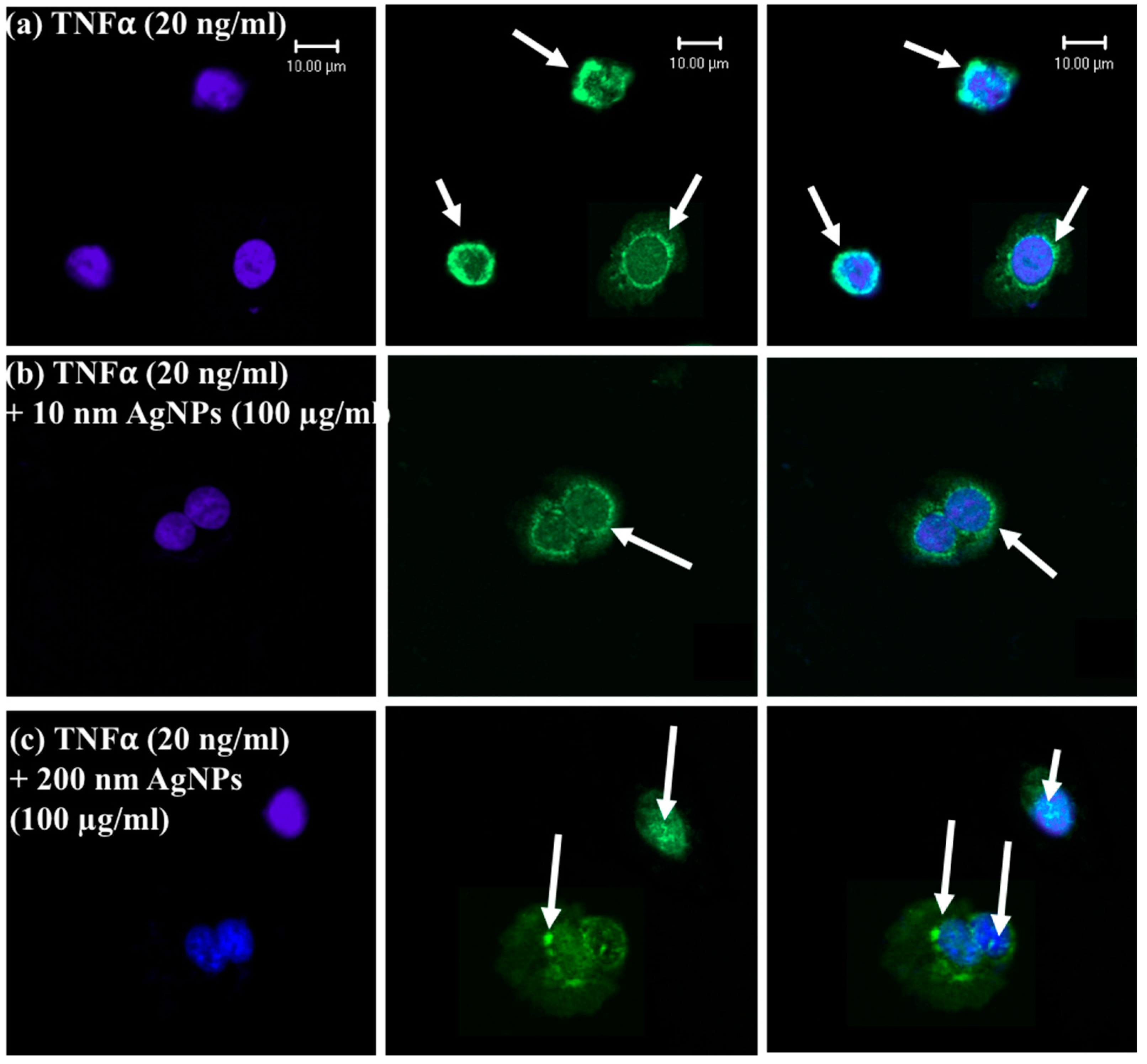
2.5. Localization of Tumor Necrosis Factor-α Receptor 1 (TNFR1)
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Silver Nanoparticles (AgNPs)
4.3. Tumor Necrosis Factor-α (TNFα)
4.4. Cell Viability Assay
4.5. Cellular Uptake Assay
4.6. DCF Assay for Oxidative Stress Determination
4.7. Dual-Luciferase Reporter Assay for BTG2 Response Assessment
4.7.1. Plasmids Employed
4.7.2. Transfection
4.7.3. Assessment of Luciferase Activity
4.8. Gene Expression Analysis
4.8.1. Polymerase Chain Reaction (PCR) Array
4.8.2. Real-Time (RT) PCR
4.9. Immunostaining and Confocal Laser Scanning Microscopy
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Nikalje, A.P. Nanotechnology and its Applications in Medicine. Med. Chem. 2015, 5, 81–89. [Google Scholar] [CrossRef]
- Edwards-Jones, V. The benefits of silver in hygiene, personal care and healthcare. Lett. Appl. Microbiol. 2009, 49, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Benn, T.M.; Westerhoff, P. Nanoparticle silver released into water from commercially available sock fabrics. Environ. Sci. Technol. 2008, 42, 4133–4139. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, M.; Karns, M.; Goodson, M.; Rowe, J.; Hussain, S.M.; Schlager, J.J.; Hong, Y. DNA damage response to different surface chemistry of silver nanoparticles in mammalian cells. Toxicol. Appl. Pharmacol. 2008, 233, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, C.; Heinze, T.; Hipler, U.C. Comparative in vitro study on cytotoxicity, antimicrobial activity, and binding capacity for pathophysiological factors in chronic wounds of alginate and silver-containing alginate. Wound Repair Regen. 2009, 17, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, J.; Güresir, E.; Seifert, V.; Raabe, A. Efficacy of silver- bearing external ventricular drainage catheters: A retro- spective analysis. J. Neurosurg. 2010, 112, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Xiong, L.; Zhou, G.; Wang, S.; Wang, J.; Liu, L.; Li, J.; Yuan, F.; Lu, S.; Wan, Z.; et al. Silver nanoparticles crossing through and distribution in the blood-brain barrier in vitro. J. Nanosci. Nanotechnol. 2010, 10, 6313–6317. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Lin, S.; Wang, P.C.; Sridhar, R. Techniques for physicochemical characterization of nanomaterials. Biotechnol. Adv. 2014, 32, 711–726. [Google Scholar] [CrossRef] [PubMed]
- Pleus, R. Nanotechnologies-Guidance on Physicochemical Characterization of Engineered Nanoscale Materials for Toxicologic Assessment; ISO: Geneva, Switzerland, 2012. [Google Scholar]
- Murdock, R.C.; Braydich-Stolle, L.; Schrand, A.M.; Schlager, J.J.; Hussain, S.M. Characterization of nanomaterial dispersion in solution prior to in vitro exposure using dynamic light scattering technique. Toxicol. Sci. 2008, 101, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.H.; Kim, J.H.; Lee, T.G.; Kim, J.H. Size, surface charge, and shape determine therapeutic effects of nanoparticles on brain and retinal diseases. Nanomedicine 2015, 11, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.P.; Li, Y.P. Physicochemical characteristics of nanoparticles affect circulation, biodistribution, cellular internalization, and trafficking. Small 2013, 9, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Tang, P.S.; Chan, W.C. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wu, Y.; Wang, C.; Li, H.C.; Wang, T.; Liao, C.Y.; Cui, L.; Zhou, Q.F.; Yan, B.; Jiang, G.B. Impact of silver nano- particles on human cells: Effect of particle size. Nanotoxicology 2010, 4, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Park, M.V.; Neigh, A.M.; Vermeulen, J.P.; de la Fonteyne, L.J.; Verharen, H.W.; Briedé, J.J.; van Loveren, H.; de Jong, W.H. The effect of particle size on the cytotoxicity, inflammation, developmental toxicity and genotoxicity of silver nanoparticles. Biomaterials 2011, 32, 9810–9817. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.G.; Han, J. Cellular responses to tumor necrosis factor. Curr. Issues Mol. Biol. 2001, 3, 79–90. [Google Scholar] [PubMed]
- Osbor, L.; Kunkel, S.; Nabel, G.J. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl. Acad. Sci. USA 1989, 86, 2336–2340. [Google Scholar] [CrossRef]
- McLeish, K.R.; Knall, C.; Ward, R.A.; Gerwins, P.; Coxon, P.Y.; Klein, J.B.; Johnson, G.L. Activation of mitogen-activated protein kinase cascades during priming of human neutrophils by TNF-alpha and GM-CSF. J. Leukoc. Biol. 1998, 64, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Choksi, S.; Shen, H.; Yang, Q.; Hur, G.M.; Kim, Y.S.; Tran, J.H.; Sergei, A.; Liu, Z. Tumor Necrosis Factor-induced Nonapoptotic Cell Death Requires Receptor-interacting Protein-mediated Cellular Reactive Oxygen Species Accumulation. J. Biol. Chem. 2004, 279, 10822–10828. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Farrah, T.; Goodwin, R.G. The TNF receptor superfamily of cellular and viral proteins: Activation, costimulation, and death. Cell 1994, 76, 959–962. [Google Scholar] [CrossRef]
- Nagata, S.; Golstein, P. The Fas death factor. Science 1995, 267, 1449–1456. [Google Scholar] [CrossRef] [PubMed]
- Wheelhouse, N.; Chan, Y.S.; Gillies, S.; Caldwell, H.; Ross, J.; Harrison, D.; Prost, S. TNF-α induced DNA damage in primary murine hepatocytes. Int. J. Mol. Med. 2003, 12, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Corda, S.; Laplace, C.; Vicaut, E.; Duranteau, J. Rapid reactive oxygen species production by mitochondria in endothelial cells exposed to tumor necrosis factor-alpha is mediated by ceramide. Am. J. Respir. Cell Mol. Biol. 2001, 24, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.B.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.F.; Shao, J.S.; Behrmann, A.; Krchma, K.; Cheng, S.L.; Towler, D.A. TNFR1-activated reactive oxidative species signals up-regulate osteogenic Msx2 programs in aortic myofibroblasts. Endocrinology 2012, 153, 3897–3910. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wong, K.K.; Ho, C.M.; Lok, C.N.; Yu, W.Y.; Che, C.M.; Chiu, J.F.; Tam, P.K. Topical delivery of silver nanoparticles promotes wound healing. Chem. Med. Chem. 2007, 2, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.K.; Cheung, S.O.; Huang, L.; Niu, J.; Tao, C.; Ho, C.M.; Che, C.M.; Tam, P.K. Further evidence of the anti-inflammatory effects of silver nanoparticles. Chem. Med. Chem. 2009, 4, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Fehaid, A.; Hamed, M.F.; Abouelmagd, M.M.; Taniguchi, A. Time- dependent toxic effect and distribution of silver nano- particles compared to silver nitrate after intratracheal instillation in rats. Am. J. Nanomater. 2016, 4, 12–19. [Google Scholar]
- Wu, T.; Tang, M. The inflammatory response to silver and titanium dioxide nanoparticles in the central nervous system. Nanomedicine 2018, 13, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Trickler, W.J.; Lantz, S.M.; Murdock, R.C.; Schrand, A.M.; Robinson, B.L.; Newport, G.D.; Schlager, J.J.; Oldenburg, S.J.; Paule, M.G.; Slikker, W.; et al. Silver Nanoparticle Induced Blood-Brain Barrier Inflammation and Increased Permeability in Primary Rat Brain Microvessel Endothelial Cells. Toxicol. Sci. 2010, 118, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Fehaid, A.; Taniguchi, A. Silver nanoparticles reduce the apoptosis induced by tumor necrosis factor-α. Sci. Technol. Adv. Mater. 2018, 19, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Li, Y.; Lin, Z.; Zhao, M.; Xu, T.; Wang, C.; Deng, N. Silver nanoparticles induce HePG-2 cells apoptosis through ROS-mediated signaling pathways. Nanoscale Res. Lett. 2016, 11, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhang, T.; Zhang, B.; Gong, F.; Huang, Y.; Tang, M. Cytotoxicity and apoptosis induced by silver nanoparticles in human liver HepG2 cells in different dispersion media. J. Appl. Toxicol. 2016, 36, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Choi, O.; Hu, Z. Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria. Environ. Sci. Technol. 2008, 42, 4583–4588. [Google Scholar] [CrossRef] [PubMed]
- Johnston, H.J.; Hutchison, G.; Christensen, F.M.; Peters, S.; Hankin, S.; Stone, V. A review of the in vivo and in vitro toxicity of silver and gold particulates: Particle attributes and biological mechanisms responsible for the observed toxicity. Crit. Rev. Toxicol. 2010, 4, 328–346. [Google Scholar] [CrossRef] [PubMed]
- Nymark, P.; Catalán, J.; Suhonen, S.; Järventaus, H.; Birkedal, R.; Clausen, P.A.; Jensen, K.A.; Vippola, M.; Savolainen, K.; Norppa, H. Genotoxicity of polyvinylpyrrolidone-coated silver nanoparticles in BEAS 2B cells. Toxicology 2013, 313, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Beutler, B.; Cerami, A. Tumor necrosis, cachexia, shock, and inflammation: A common mediator. Ann. Rev. Biochem. 1988, 57, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Tracey, K.J.; Cerami, A. Tumor necrosis factor, other cytokines and disease. Annu. Rev. Cell Biol. 1993, 9, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Santoro, C.M.; Duchsherer, N.L.; Grainger, D.W. Minimal in vitro antimicrobial efficacy and ocular cell toxicity from silver nanoparticles. Nanobiotechnology 2007, 3, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Ahlberg, S.; Antonopulos, A.; Diendorf, J.; Dringen, R.; Epple, M.; Flöck, R.; Goedecke, W.; Graf, C.; Haberl, N.; Helmlinger, J.; et al. PVP-coated, negatively charged silver nanoparticles: A multi-center study of their physicochemical characteristics, cell culture and in vivo experiments. Beilstein J. Nanotechnol. 2014, 5, 1944–1965. [Google Scholar] [CrossRef] [PubMed]
- Van Horssen, R.; Ten Hagen, T.L.; Eggermont, A.M. TNF-alpha in Cancer Treatment: Molecular Insights, Antitumor Effects, and Clinical Utility. Oncologist 2006, 11, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.; Nakada, M.T.; DeWitte, M. Tumor necrosis factoralpha in the pathogenesis and treatment of cancer. Curr. Opin. Pharmacol. 2004, 4, 314–320. [Google Scholar] [CrossRef] [PubMed]
- van Dullemen, H.M.; van Deventer, S.J.; Hommes, D.W.; Bijl, H.A.; Jansen, J.; Tytgat, G.N.; Woody, J. Treatment of Crohn’s disease with anti-tumor necrosis factor chimeric monoclonal antibody (cA2). Gastroenterology 1995, 109, 129–135. [Google Scholar] [CrossRef]
- Bathon, J.M.; Martin, R.W.; Fleischmann, R.M.; Tesser, J.R.; Schiff, M.H.; Keystone, E.C.; Genovese, M.C.; Wasko, M.C.; Moreland, L.W.; Weaver, A.L.; et al. A comparison of etanercept and methotrexate in patients with early rheumatoid arthritis. N. Engl. J. Med. 2000, 343, 1586–1593. [Google Scholar] [CrossRef] [PubMed]

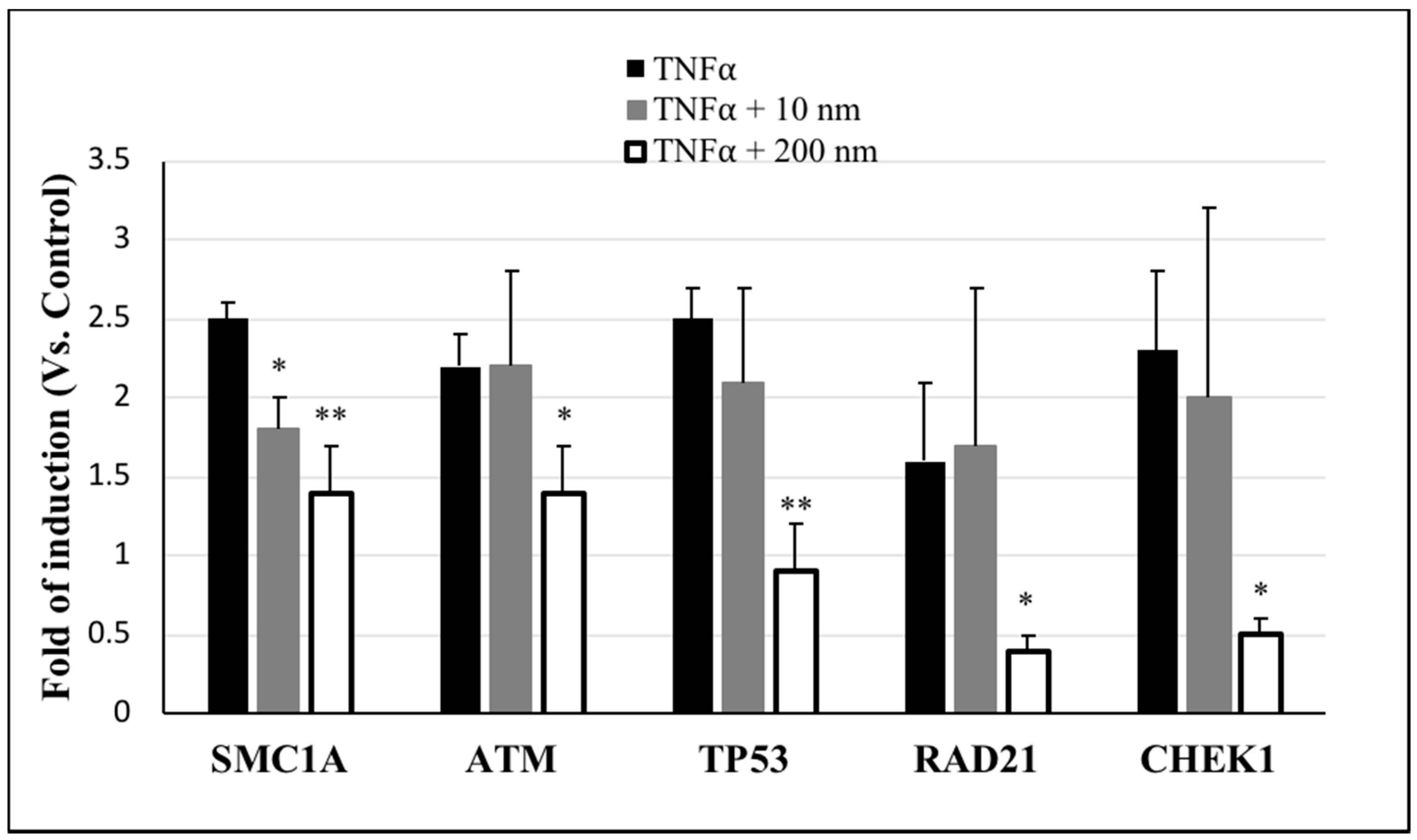

| Symbol of Genes | Description of the Genes | Fold Regulation Vs. Control | ||
|---|---|---|---|---|
| TNFα | TNFα + 10 nm AgNPs | TNFα + 200 nm AgNPs | ||
| ATM | Ataxia telangiectasia mutated | 2.1 | 1.9 | 0.8 |
| CDK7 | Cyclin-dependent kinase 7 | 3.8 | 6.9 | 1 |
| CHEK1 | CHK1 checkpoint homolog (S. pombe) | 2.9 | 4.4 | 0.4 |
| DDIT3 | DNA-damage-inducible transcript 3 | 2.1 | 2.7 | 1.8 |
| RAD21 | RAD21 homolog (S. pombe) | 1.9 | 2.9 | 0.3 |
| RAD51 | RAD51 homolog (S. cerevisiae) | 1.5 | 1.6 | 0.6 |
| SIRT1 | Sirtuin 1 | 1.3 | 3.5 | 0.8 |
| SMC1A | Structural maintenance of chromosomes 1A | 1.9 | 1.2 | 0.7 |
| SUMO1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | 2.5 | 4.1 | 1.6 |
| TP53 | Tumor protein p53 | 2.6 | 2.8 | 0.6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fehaid, A.; Taniguchi, A. Size-Dependent Effect of Silver Nanoparticles on the Tumor Necrosis Factor α-Induced DNA Damage Response. Int. J. Mol. Sci. 2019, 20, 1038. https://doi.org/10.3390/ijms20051038
Fehaid A, Taniguchi A. Size-Dependent Effect of Silver Nanoparticles on the Tumor Necrosis Factor α-Induced DNA Damage Response. International Journal of Molecular Sciences. 2019; 20(5):1038. https://doi.org/10.3390/ijms20051038
Chicago/Turabian StyleFehaid, Alaa, and Akiyoshi Taniguchi. 2019. "Size-Dependent Effect of Silver Nanoparticles on the Tumor Necrosis Factor α-Induced DNA Damage Response" International Journal of Molecular Sciences 20, no. 5: 1038. https://doi.org/10.3390/ijms20051038
APA StyleFehaid, A., & Taniguchi, A. (2019). Size-Dependent Effect of Silver Nanoparticles on the Tumor Necrosis Factor α-Induced DNA Damage Response. International Journal of Molecular Sciences, 20(5), 1038. https://doi.org/10.3390/ijms20051038






