Genome-Wide Comparative Analyses of Polyadenylation Signals in Eukaryotes Suggest a Possible Origin of the AAUAAA Signal
Abstract
1. Introduction
2. Results
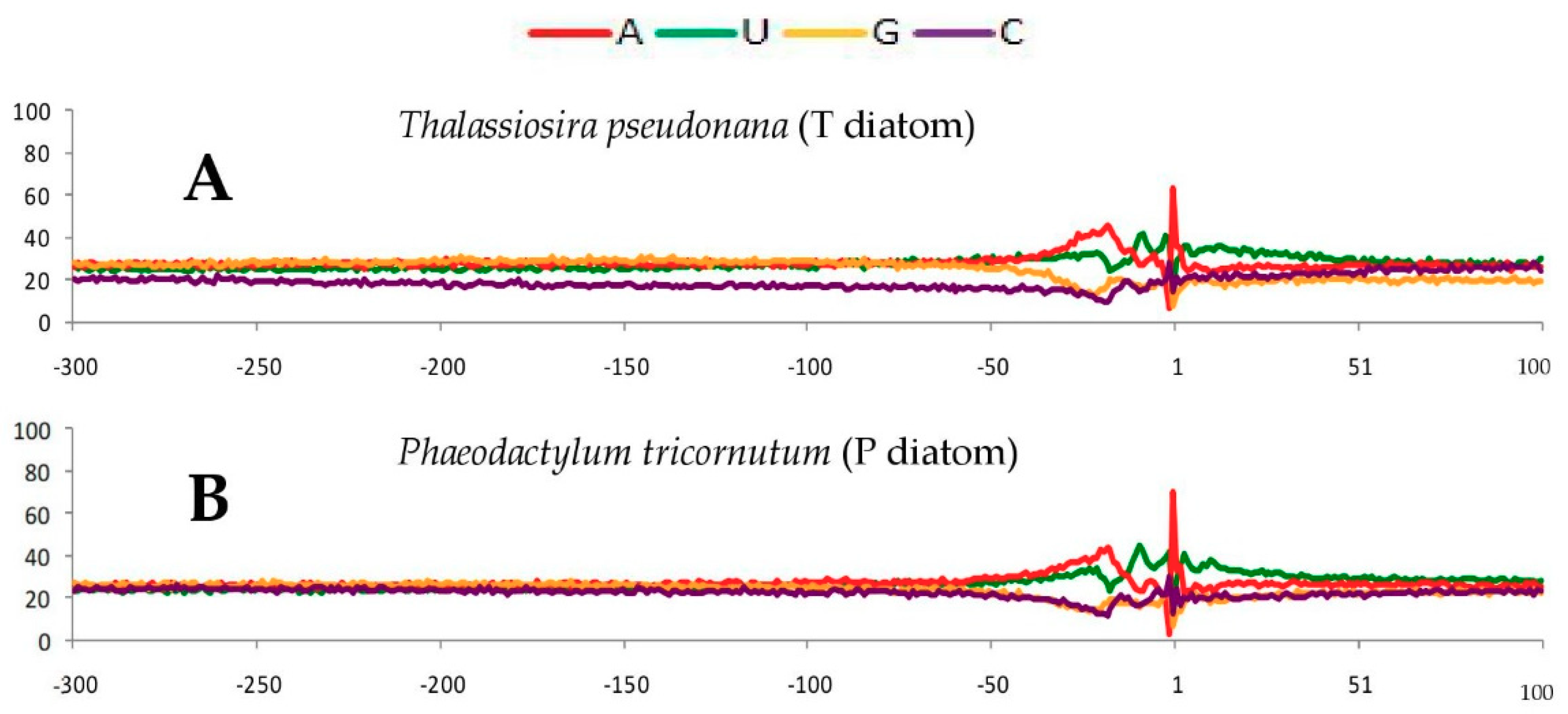
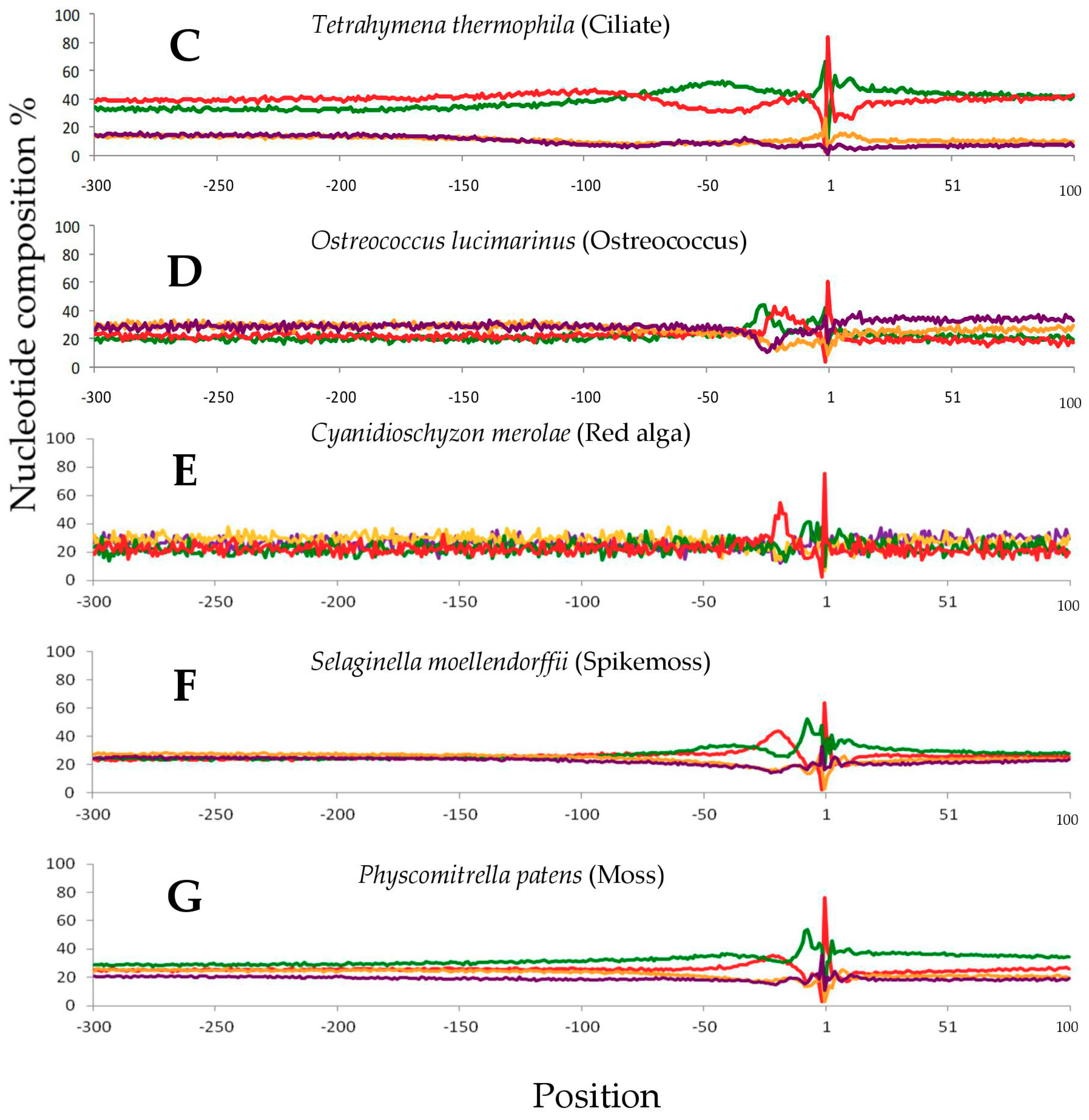
2.1. Single Nucleotide Profiles Around Poly(A) Sites in Seven Species
2.2. Putative Polyadenylation Signals Revealed in the Seven Species
2.3. Conservative Nucleotide Composition Around Poly(A) Sites in the Seven Species
2.4. The Frequency Variations of Polyadenylation Signals in NUE
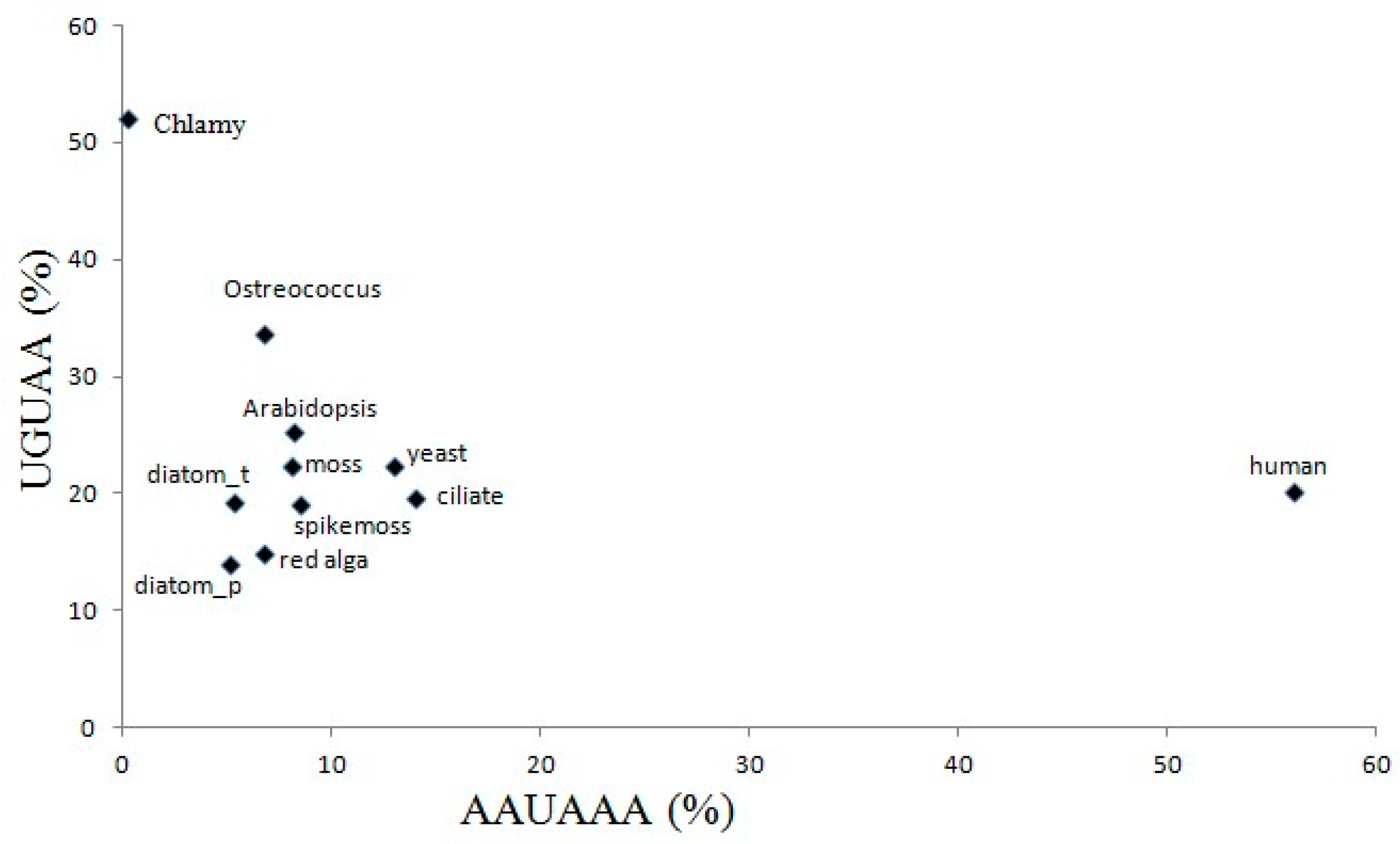
2.5. The Variation of Canonical Polyadenylation Signals (UGUAA and AAUAAA) in the 11 Species
3. Discussion
3.1. The Usage and Distribution of Poly(A) Signals in the NUE Region
3.2. The Poly(A) Signals in the FUE and CE Regions
3.3. The Relationships Between Single Nucleotide Profiles and Poly(A) Signals
3.4. The Comparative Analysis of Poly(A) Signals in 11 Species Suggested an Evolutionary Pathway of Poly(A) Signal Variation
4. Material and Methods
4.1. Data Collection and Polyadenylation Site Definition
4.2. Poly(A) Signal Elements Definition and Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| B | nucleotides U, C, or G |
| CE | cleavage element |
| EST | Expressed sequence tag |
| FUE | far upstream element |
| Nt | nucleotide |
| NUE | near upstream element |
| Y | nucleotides U or C |
References
- Rothnie, H.M. Plant mRNA 3′-end formation. Plant Mol. Biol. 1996, 32, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hyman, L.; Moore, C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. Mmbr 1999, 63, 405–445. [Google Scholar] [PubMed]
- Birse, C.E.; Minvielle-Sebastia, L.; Lee, B.A.; Keller, W.; Proudfoot, N.J. Coupling termination of transcription to messenger RNA maturation in yeast. Science 1998, 280, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Danckwardt, S.; Hentze, M.W.; Kulozik, A.E. 3′ end mRNA processing: Molecular mechanisms and implications for health and disease. Embo J. 2008, 27, 482–498. [Google Scholar] [CrossRef] [PubMed]
- Millevoi, S.; Vagner, S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010, 38, 2757–2774. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Hunt, A.G. The Polyadenylation of RNA in Plants. Plant Physiol. 1997, 115, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Graber, J.H. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. RNA 2011, 3, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, K.; Brown, K.M.; Gilmartin, G.M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005, 19, 1315–1327. [Google Scholar] [CrossRef]
- Xing, D.; Li, Q.Q. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip. Rev. RNA 2011, 2, 445–458. [Google Scholar] [CrossRef]
- Hunt, A.G.; Xu, R.; Addepalli, B.; Rao, S.; Forbes, K.P.; Meeks, L.R.; Xing, D.; Mo, M.; Zhao, H.; Bandyopadhyay, A.; et al. Arabidopsis mRNA polyadenylation machinery: Comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genom. 2008, 9, 220. [Google Scholar] [CrossRef]
- Kamasawa, M.; Horiuchi, J. Identification and characterization of polyadenylation signal (PAS) variants in human genomic sequences based on modified EST clustering. In Silico Biol. (Gedrukt) 2008, 8, 347–361. [Google Scholar]
- Tian, B.; Hu, J.; Zhang, H.; Lutz, C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005, 33, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Mangone, M.; Manoharan, A.P.; Thierry-Mieg, D.; Thierry-Mieg, J.; Han, T.; Mackowiak, S.D.; Mis, E.; Zegar, C.; Gutwein, M.R.; Khivansara, V.; et al. The landscape of C. elegans 3′UTRs. Science 2010, 329, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Loke, J.C.; Stahlberg, E.A.; Strenski, D.G.; Haas, B.J.; Wood, P.C.; Li, Q.Q. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 2005, 138, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Ji, G.; Haas, B.J.; Wu, X.; Zheng, J.; Reese, G.J.; Li, Q.Q. Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucleic Acids Res. 2008, 36, 3150–3161. [Google Scholar] [CrossRef] [PubMed]
- Graber, J.H.; Cantor, C.R.; Mohr, S.C.; Smith, T.F. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. USA 1999, 96, 14055–14060. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Kapranov, P.; Foissac, S.; Kim, S.W.; Fishilevich, E.; Monaghan, A.P.; John, B.; Milos, P.M. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 2010, 143, 1018–1029. [Google Scholar] [CrossRef]
- Mages, W.; Cresnar, B.; Harper, J.F.; Bruderlein, M.; Schmitt, R. Volvox carteri alpha 2- and beta 2-tubulin-encoding genes: Regulatory signals and transcription. Gene 1995, 160, 47–54. [Google Scholar] [CrossRef]
- Terauchi, M.; Kato, A.; Nagasato, C.; Motomura, T. Research note: Analysis of expressed sequence tags from the chrysophycean alga Ochromonas danica (Heterokontophyta). Phycol. Res. 2010, 58, 217–221. [Google Scholar] [CrossRef]
- Shen, Y.; Liu, Y.; Liu, L.; Liang, C.; Li, Q.Q. Unique features of nuclear mRNA poly(A) signals and alternative polyadenylation in Chlamydomonas reinhardtii. Genetics 2008, 179, 167–176. [Google Scholar] [CrossRef]
- Zhao, Z.; Wu, X.; Kumar, P.K.R.; Dong, M.; Ji, G.; Li, Q.Q.; Liang, C. Bioinformatics analysis of alternative polyadenylation in green alga Chlamydomonas reinhardtii using transcriptome sequences from three different sequencing platforms. G3 (Bethesda) 2014, 4, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Wodniok, S.; Simon, A.; Glockner, G.; Becker, B. Gain and loss of polyadenylation signals during evolution of green algae. BMC Evol. Biol. 2007, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Coseno, M.; Gilmartin, G.M.; Doublié, S. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 2011, 19, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Gilmartin, G.M.; Doublié, S. The structure of human Cleavage Factor Im hints at functions beyond UGUA-specific RNA binding: A role in alternative polyadenylation and a potential link to 5′ capping and splicing. RNA Biol. 2011, 8, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, N.; Hernández, R.; López-Griego, L.; López-Villaseñor, I. Separable putative polyadenylation and cleavage motifs in Trichomonas vaginalis mRNAs. Gene 2002, 289, 81–86. [Google Scholar] [CrossRef]
- Fuentes, V.; Barrera, G.; Sánchez, J.; Hernández, R.; López-Villaseñor, I. Functional Analysis of Sequence Motifs Involved in the Polyadenylation of Trichomonas vaginalis mRNAs. Eukaryot. Cell 2012, 11, 725–734. [Google Scholar] [CrossRef]
- Van Helden, J.; Andre, B.; Collado-Vides, J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998, 281, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, M.; Downie, B.; Liang, C.; Ji, G.; Li, Q.Q.; Hunt, A.G. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 2011, 108, 12533–12538. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Li, H.; Tong, S.; Li, X.; Shi, H.; Ying, Z.; Gao, Y.; Ge, H.; Niu, L.; Teng, M. Structural basis of pre-mRNA recognition by the human cleavage factor Im complex. Cell Res. 2011, 21, 1039–1051. [Google Scholar] [CrossRef]
- Rothnie, H.M.; Reid, J.; Hohn, T. The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′-end formation in plants. Embo J. 1994, 13, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Sheets, M.D.; Ogg, S.C.; Wickens, M.P. Point mutations in AAUAAA and the poly (A) addition site: Effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990, 18, 5799–5805. [Google Scholar] [CrossRef] [PubMed]
- Beaudoing, E.; Freier, S.; Wyatt, J.R.; Claverie, J.M.; Gautheret, D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000, 10, 1001–1010. [Google Scholar] [CrossRef]
- Eisen, J.A.; Coyne, R.S.; Wu, M.; Wu, D.; Thiagarajan, M.; Wortman, J.R.; Badger, J.H.; Ren, Q.; Amedeo, P.; Jones, K.M.; et al. Macronuclear Genome Sequence of the Ciliate Tetrahymena thermophila, a Model Eukaryote. PLoS Biol. 2006, 4, e286. [Google Scholar] [CrossRef]
- Xiong, J.; Lu, X.; Zhou, Z.; Chang, Y.; Yuan, D.; Tian, M.; Zhou, Z.; Wang, L.; Fu, C.; Orias, E.; et al. Transcriptome Analysis of the Model Protozoan, Tetrahymena thermophila, Using Deep RNA Sequencing. PLoS ONE 2012, 7, e30630. [Google Scholar] [CrossRef] [PubMed]
- Hunt, A.G.; Xing, D.; Li, Q.Q. Plant polyadenylation factors: Conservation and variety in the polyadenylation complex in plants. BMC Genom. 2012, 13, 641. [Google Scholar] [CrossRef]
- Liang, C.; Liu, Y.; Liu, L.; Davis, A.C.; Shen, Y.; Li, Q.Q. Expressed sequence tags with cDNA termini: Previously overlooked resources for gene annotation and transcriptome exploration in Chlamydomonas reinhardtii. Genetics 2008, 179, 83–93. [Google Scholar] [CrossRef]
- Maheswari, U.; Mock, T.; Armbrust, E.V.; Bowler, C. Update of the Diatom EST Database: A new tool for digital transcriptomics. Nucleic Acids Res. 2009, 37, D1001–D1005. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinform. (Oxf. Engl.) 2005, 21, 1859–1875. [Google Scholar] [CrossRef]
- Graber, J.H.; Cantor, C.R.; Mohr, S.C.; Smith, T.F. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999, 27, 888–894. [Google Scholar] [CrossRef] [PubMed]


| Species Name (Common Name) | Conserved Signal | Frequency (%) | Z-Score |
|---|---|---|---|
| T. pseudonana (T diatom) | UAA | 86.51 | 19.12 1 |
| P. tricornutum (P diatom) | UAA | 78.21 | 6.94 1 |
| T. thermophila (Ciliate) | - | - | - |
| O. lucimarinus (Ostreococcus) | UGUAA | 30.4 | 30.21 1 |
| C. merolae (Red alga) | UAAA | 86.45 | 15.33 1 |
| GUAA | 43.23 | 8.78 1 | |
| S. moellendorffii (Spikemoss) | AAUAAA | 7.83 | 7.18 3 |
| P. patens (Moss) | AAUAAA | 7.25 | 15.46 3 |
| Species Name (Common Name) | Canonical Signal (Frequency) | Variant (Frequency) | Overall Frequency | |
|---|---|---|---|---|
| UGUAA Group | C. reinhardtii (Chlamydomonas) | UGUAA (50.38%) | UGCAA (6.42%) UGUUA (3.38%) UUUAA (3.28%) UGUAG (2.92%) UGUAU (2.43%) UGUGA (2.34%) UGAAA (2.33%) UGUAC (2.14%) | 72.86% |
| O. lucimarinus (Ostreococcus) | UGUAA (30.40%) | UUUAA (10.47%) UGAAA (5.75%) UGUAU (5.24%) UGUUA (4.60%) UGUGA (4.09%) UGCAA (4.09%) | 59.64% | |
| AAUAAA Group | P. patens (Moss) | AAUAAA (7.25%) | AGUAAA (2.73%) UAUAAA (2.25%) | 11.93% |
| S. moellendorffii (Spikemoss) | AAUAAA (7.83%) | UAUAAA (3.31%) AUUAAA (2.08%) AGUAAA (1.67%) | 14.41% | |
| A. thaliana (Arabidopsis) | AAUAAA (8.59%) | UAUAAA (3.44%) AUUAAA (2.17%) | 13.71% | |
| H. sapiens (Human) | AAUAAA (64.92%) | AUUAAA (16.68%) UAUAAA (4.30%) AGUAAA (3.76%) | 85.19% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Wu, X.; Ji, G.; Liang, C.; Li, Q.Q. Genome-Wide Comparative Analyses of Polyadenylation Signals in Eukaryotes Suggest a Possible Origin of the AAUAAA Signal. Int. J. Mol. Sci. 2019, 20, 958. https://doi.org/10.3390/ijms20040958
Zhao Z, Wu X, Ji G, Liang C, Li QQ. Genome-Wide Comparative Analyses of Polyadenylation Signals in Eukaryotes Suggest a Possible Origin of the AAUAAA Signal. International Journal of Molecular Sciences. 2019; 20(4):958. https://doi.org/10.3390/ijms20040958
Chicago/Turabian StyleZhao, Zhixin, Xiaohui Wu, Guoli Ji, Chun Liang, and Qingshun Quinn Li. 2019. "Genome-Wide Comparative Analyses of Polyadenylation Signals in Eukaryotes Suggest a Possible Origin of the AAUAAA Signal" International Journal of Molecular Sciences 20, no. 4: 958. https://doi.org/10.3390/ijms20040958
APA StyleZhao, Z., Wu, X., Ji, G., Liang, C., & Li, Q. Q. (2019). Genome-Wide Comparative Analyses of Polyadenylation Signals in Eukaryotes Suggest a Possible Origin of the AAUAAA Signal. International Journal of Molecular Sciences, 20(4), 958. https://doi.org/10.3390/ijms20040958






