Identification of Quantitative Trait Loci Associated with Nutrient Use Efficiency Traits, Using SNP Markers in an Early Backcross Population of Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation of NuUE Traits and Their Correlation among Traits
2.2. ANOVA and Interaction with the Environment
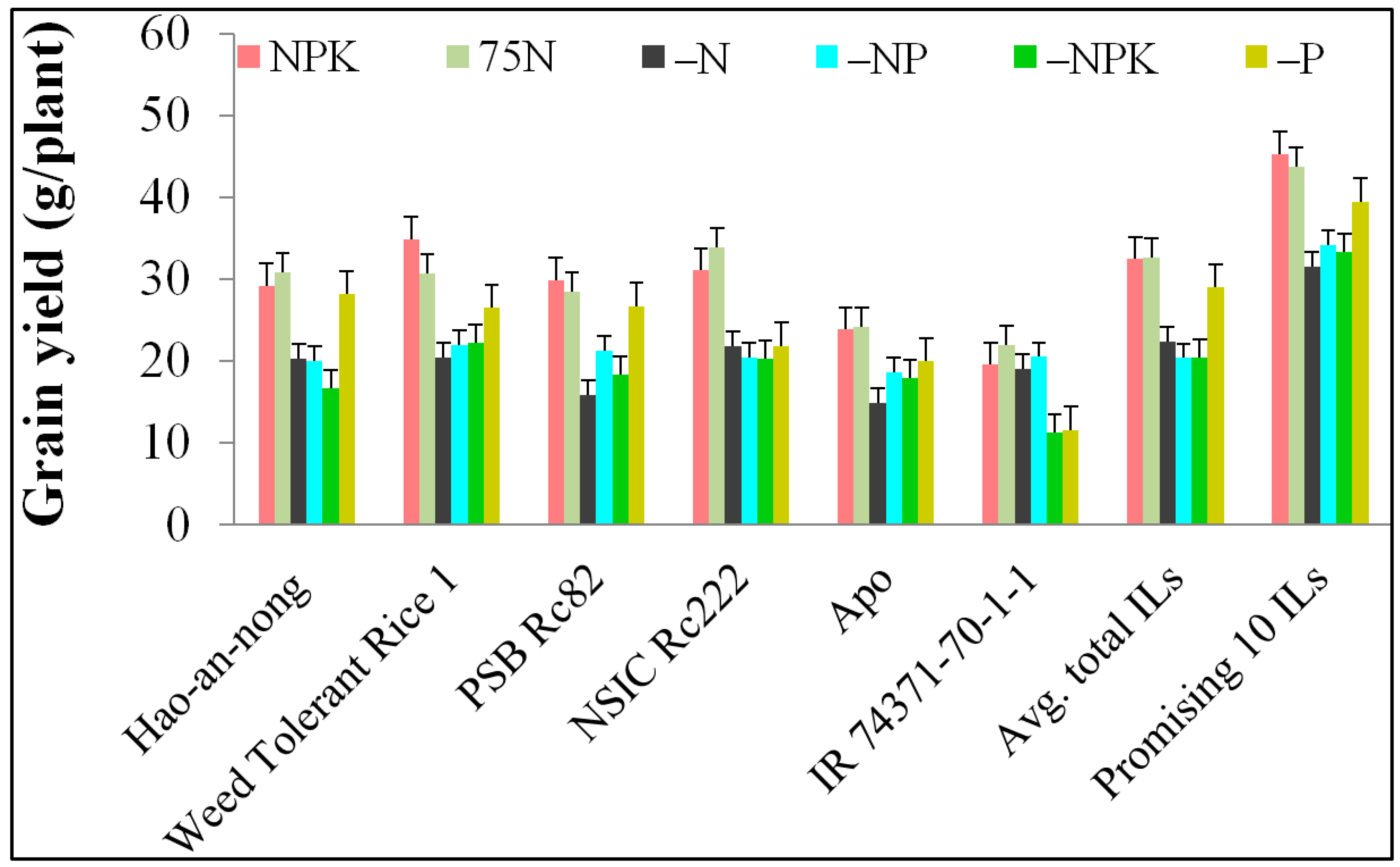
2.3. Analysis of Agronomic Efficiency (AE) and Partial Factor Productivity (PFP)
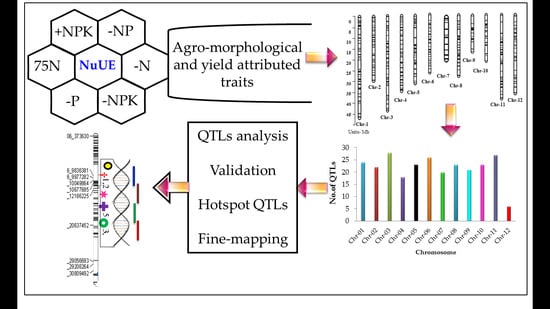
2.4. Construction of Linkage Map and Segregation of SNP Markers
2.5. QTLs for NuUE Traits
2.5.1. Agronomic Efficiency (AE)
2.5.2. Partial Factor Productivity (PFP)
2.5.3. Grain Yield (GY)
2.5.4. Biomass Yield (BY)
2.5.5. Percentage of Spikelet Fertility (PSPF)
2.5.6. 1000-Gwt
2.5.7. Filled Grain Number (FGN)
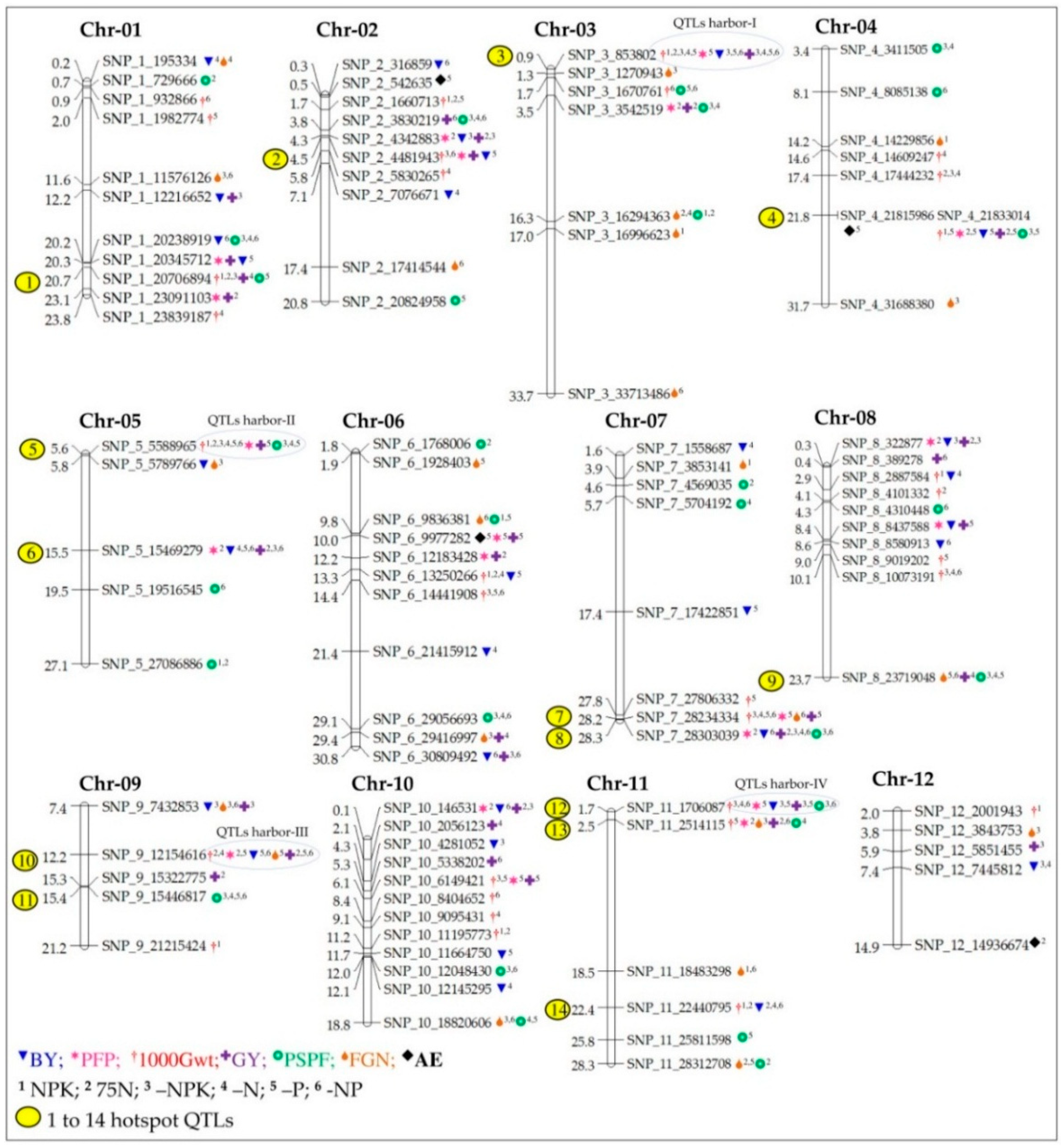
2.5.8. Hotspot QTLs for Multiple Traits
2.5.9. Fine-Tuning of QTLs Harbor-I to -IV
3. Discussion
3.1. Analysis of Critical NuUE Traits
3.2. Promising Traits of AE-Associated QTLs
3.3. Categorizing NuUE QTLs and Related Traits
3.4. Consistency and Comparisons of Major QTLs across Different Genetic Backgrounds
3.5. QTL Hotspots
4. Materials and Methods
4.1. Plant Materials
4.2. Measurements of Agronomic and Yield-Attributed Traits
4.3. Calculation of Agronomic Efficiency (AE) and Partial Factor Productivity (PFP)
- (i)
- AE (N) = (YNPK − Y0NPK)/FN
- (ii)
- AE (N) = (YNK − Y0NPK)/FN
- (iii)
- AE (75N)=(Y(75N) − Y0NPK)/F75N
- (iv)
- PFP(N) = Y(+NPK)/FN
- (v)
- PFP(N) = Y(−P)/FN
- (vi)
- PFP(N) = Y(75N)/FN
4.4. Genotyping via 6K SNP Array
4.5. Mapping of QTLs and Hotspot Regions for NuUE
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Godfray, H.C.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- FAO. The State of Food Insecurity in the World 2009. Available online: http://www.fao.org/tempref/docrep/fao/012/i0876e/i0876e_flyer.pdf (accessed on 30 October 2009).
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Edgerton, M.D. Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol. 2009, 149, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Naresh, R.K.; Gupta, R.K.; Ashok, K.; Singh, B.; Prakash, S.; Kumar, S.; Rathi, R.C. Direct-seeding and reduced-tillage options in the rice-wheat system of the Western Indo-Gangetic Plains. Int. J. Agric. Sci. 2011, 7, 197–208. [Google Scholar]
- Bindraban, P.S.; Dimkpa, C.; Nagarajan, L.; Roy, A.; Rabbinge, R. Revisiting fertilisers and fertilisation strategies for improved nutrient uptake by plants. Biol. Fertil. Soils 2015, 51, 897–911. [Google Scholar] [CrossRef]
- Mahender, A.; Anandan, A.; Pradhan, S.K.; Singh, O.N. Traits-related QTLs and genes and their potential applications in rice improvement under low phosphorus condition. Arch. Agron. Soil Sci. 2017, 64, 449–464. [Google Scholar] [CrossRef]
- Ali, J.; Xu, J.L.; Gao, Y.M.; Ma, X.F.; Meng, L.J.; Wang, Y.; Pang, Y.L.; Guan, Y.S.; Xu, M.R.; Revilleza, J.E.; et al. Harnessing the hidden genetic diversity for improving multiple abiotic stress tolerance in rice (Oryza sativa L.). PLoS ONE 2017, 12, e0172515. [Google Scholar] [CrossRef]
- Raun, W.R.; Johnson, G.V. Improving nitrogen use efficiency for cereal production. Agron. J. 1999, 91, 357–363. [Google Scholar] [CrossRef]
- Cassman, K.G.; Gines, G.C.; Dizon, M.A.; Samson, M.I.; Alcanter, J.M. Nitrogen use efficiency in tropical lowland rice systems: Contribution from indigenous and applied nitrogen. Field Crop. Res. 1996, 47, 1–12. [Google Scholar] [CrossRef]
- Giles, J. Nitrogen study fertilizes fears of pollution. Nature 2005, 433, 791. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Naylor, R.; Crews, T.; David, M.B.; Drinkwater, L.E.; Holland, E.; Johnes, P.J.; Katzenberger, J.; Martinelli, L.A.; Matson, P.A.; et al. Agriculture. Nutrient imbalances in agricultural development. Science 2009, 19, 324, 1519–1520. [Google Scholar] [CrossRef] [PubMed]
- Hirel, B.; Tétu, T.; Lea, P.J.; Dubois, F. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 2011, 3, 1452–1485. [Google Scholar] [CrossRef]
- Song, K.; Xue, Y.; Zheng, X.; Weiguang, L.; Qiao, H.; Qin, Q.; Yang, J. Effects of the continuous use of organic manure and chemical fertilizer on soil inorganic phosphorus fractions in calcareous soil. Sci. Rep. 2017, 7, 1164. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Bi, Y.; Rothstein, S.J. Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Fageria, N.K.; Baligar, V.C.; Li, Y.C. Nutrient uptake and use efficiency by tropical legume cover crops at varying pH of an Oxisol. J. Plant Nutr. 2014, 37, 294–311. [Google Scholar] [CrossRef]
- Das, G.; Rao, G.J.N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Anumalla, M.; Roychowdhury, R.; Geda, C.K.; Mazid, M.; Rathoure, A.K. Utilization of plant genetic resources and diversity analysis tools for sustainable crop improvement with special emphasis on rice. J. Adv. Res. 2015, 3, 1155–1175. [Google Scholar]
- Pang, Y.; Chen, K.; Wang, X.; Wang, W.; Xu, J.; Ali, J.; Li, Z. Simultaneous improvement and genetic dissection of salt tolerance of rice (Oryza sativa L.) by designed QTL pyramiding. Front. Plant Sci. 2017, 8, 1275. [Google Scholar] [CrossRef]
- Ali, J.; Jewel, Z.A.; Mahender, A.; Anandan, A.; Hernandez, J.; Li, Z. Molecular genetics and breeding for nutrient use efficiency in rice. Int. J. Mol. Sci. 2018, 19, 1762. [Google Scholar] [CrossRef]
- Jewel, Z.; Ali, J.; Pang, Y.; Anumalla, M.; Acero, B.; Hernandez, J.; Xu, J.; Li, Z. Developing Green Super Rice Varieties with High Nutrient Use Efficiency by Phenotypic Selection Under Varied Nutrient Conditions. Preprints 2018. [Google Scholar] [CrossRef]
- Novoa, R.; Loomis, R.S. Nitrogen and plant production. Plant Soil 1981, 58, 177–204. [Google Scholar] [CrossRef]
- Baligar, V.C.; Fageria, N.K.; He, Z.L. Nutrient use efficiency in plants. Commun. Soil Sci. Plant Anal. 2001, 32, 921–950. [Google Scholar] [CrossRef]
- McDonald, G.; William, B.; Chunyuan, H.; David, L. Nutrient use efficiency. In Genomics and Breeding for Climate-Resilient Crops; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2, pp. 333–393. [Google Scholar]
- Stahl, A.; Pfeifer, M.; Frisch, M.; Wittkop, B.; Snowdon, R.J. Recent genetic gains in nitrogen use efficiency in oilseed rape. Front. Plant Sci. 2017, 8, 963. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Xing, Y.; Yan, H.; Xu, C.; Li, X.; Zhang, Q. QTLs for low nitrogen tolerance at seedling stage identified using a recombinant inbred line population derived from an elite rice hybrid. Appl. Genet. 2005, 112, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Senthilvel, S.; Govindaraj, P.; Arumugachamy, S.; Latha, R.; Malarvizhi, P.; Gopalan, A.; Maheswaran, M. Mapping genetic loci associated with nitrogen use efficiency in rice (Oryza sativa ) L. In Proceedings of the 4th International Crop Science Congress, Brisbane, Australia, 26 September–1 October 2004. [Google Scholar]
- Srividya, A.; Vemireddy, L.R.; Hariprasad, A.S.; Jayaprada, M.; Sakile, S.; Puram, V.R.R.; Anuradha, G.; Siddiq, E.A. Identification and mapping of landrace derived QTL associated with yield and its components in rice under different nitrogen levels and environments. Int. J. Plant Breed. Genet. 2010, 4, 210–227. [Google Scholar] [CrossRef]
- Tong, H.H.; Chen, L.; Li, W.P.; Mei, H.; Xing, Y.; Yu, X.; Xu, X.; Zhang, S.; Luo, L. Identification and characterization of quantitative trait loci for grain yield and its components under different nitrogen fertilization levels in rice (Oryza sativa L.). Mol. Breed. 2011, 28, 495–509. [Google Scholar] [CrossRef]
- Vinod, K.K.; Heuer, S. Approaches towards nitrogen- and phosphorus-efficient rice. AoB Plants 2012, 28, 1–18. [Google Scholar] [CrossRef]
- Ni, J.J.; Wu, P.; Senadhira, D.; Huang, N. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 1998, 97, 1361–1369. [Google Scholar] [CrossRef]
- Wissuwa, M.; Ae, N. Further characterization of two QTLs that increase phosphorus uptake of rice (Oryza sativa L.) under phosphorus deficiency. Plant Soil 2001, 237, 275–286. [Google Scholar] [CrossRef]
- Lang, N.T.; Buu, B.C. Mapping QTLs for phosphorus deficiency tolerance in rice (Oryza sativa L.). Omonrice 2006, 14, 1–9. [Google Scholar]
- Cho, Y.I.; Jiang, W.Z.; Chin, J.H.; Piao, Z.; Cho, Y.G.; McCouch, S.; Koh, H.J. Identification of QTLs associated with physiological nitrogen use efficiency in rice. Mol. Cell 2007, 23, 72–79. [Google Scholar]
- Shimizu, A.; Kato, K.; Komatsu, A.; Motomura, K.; Ikehashi, H. Genetic analysis of root elongation induced by phosphorus deficiency in rice (Oryza sativa L.): Fine QTL mapping and multivariate analysis of related traits. Appl. Genet. 2008, 117, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Y.J.; Chen, D.Y.; Yu, S.B. Analysis of quantitative trait loci in response to nitrogen and phosphorus deficiency in rice using chromosomal segment substitution lines. Acta Agron. Sin. 2009, 35, 580–587. [Google Scholar] [CrossRef]
- Chin, J.H.; Gamuyao, R.; Dalid, C.; Bustamam, M.; Prasetiyono, J.; Moeljopawiro, S.; Wissuwa, M.; Heuer, S. Developing rice with high yield under phosphorus deficiency: Pup1 sequence to application. Plant Physiol. 2011, 156, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.D.; Liu, J.; Dai, L.F.; Zhang, F.T.; Wan, Y.; Xie, J.K. Linkage map construction and QTL identification of P-deficiency tolerance in Oryza rufipogon Griff. at early seedling stage. Euphytica 2017, 213, 96. [Google Scholar] [CrossRef]
- Wu, P.; Ni, J.J.; Luo, A.C. QTLs underlying rice tolerance to low-potassium stress in rice seedlings. Crop Sci. 1998, 38, 1458–1462. [Google Scholar] [CrossRef]
- Fageria, N.K.; Dos Santos, A.B.; De Moraes, M.F. Yield, potassium uptake, and use efficiency in upland rice genotypes. Commun. Soil Sci. Plant Anal. 2010, 41, 2676–2684. [Google Scholar] [CrossRef]
- Feltus, F.A.; Wan, J.; Schulze, S.R.; Estill, J.C.; Jiang, N.; Paterson, A.H. An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res. 2004, 14, 1812–1819. [Google Scholar] [CrossRef]
- Huang, Y.F.; Poland, J.A.; Wight, C.P.; Jackson, E.W.; Tinker, N.A. Using genotyping-by-sequencing (GBS) for genomic discovery in cultivated oat. PLoS ONE 2014, 9, e102448. [Google Scholar] [CrossRef]
- Yu, H.; Xie, W.; Li, J.; Zhou, F.; Zhang, Q. A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol. J. 2014, 12, 28–37. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- Kumar, S.; Banks, T.W.; Cloutier, S. SNP Discovery through next-generation sequencing and its applications. Int. J. Plant Genom. 2012, 2012, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wang, W.; Wang, P.; Jun, L.M.; Chung Sham, P.; Wang, J. A fast and accurate SNP detection algorithm for next-generation sequencing data. Nat. Commun. 2012, 3, 1258. [Google Scholar] [CrossRef] [PubMed]
- Thomson, M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Wang, K.; Cui, K.; Liu, G.; Xie, W.; Yu, H.; Pan, J.; Huang, J.; Nie, L.; Shah, F. Identification of quantitative trait loci for phosphorus use efficiency traits in rice using a high density SNP map. BMC Genet. 2014, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xie, W.; He, H.; Yu, H.; Chen, W.; Li, J.; Yu, R.; Yao, Y.; Zhang, W.; He, Y.; et al. A high-density SNP genotyping array for rice biology and molecular breeding. Mol. Plant 2014, 7, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Kurokawa, Y.; Noda, T.; Yamagata, Y.; Angeles-Shim, R.; Sunohara, H.; Uehara, K.; Furuta, T.; Nagai, K.; Jena, K.K.; Yasui, H.; et al. Construction of a versatile SNP array for pyramiding useful genes of rice. Plant Sci. 2016, 242, 131–139. [Google Scholar] [CrossRef]
- Feng, Y.; Zhai, R.-R.; Lin, Z.-C.; Cao, L.-Y.; Wei, X.-H.; Cheng, S.-H. Quantitative trait locus analysis for rice yield traits under two nitrogen levels. Rice Sci. 2015, 22, 108–115. [Google Scholar]
- Zhou, Y.; Tao, Y.; Tang, D.; Wang, J.; Zhong, J.; Wang, Y.; Yuan, Q.; Yu, X.; Zhang, Y.; Wang, Y.; et al. Identification of QTL associated with nitrogen uptake and nitrogen use efficiency using high throughput genotyped CSSLs in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1166. [Google Scholar] [CrossRef]
- Dai, G.J.; Cheng, S.H.; Hua, Z.T.; Zhang, M.L.; Jiang, H.B.; Feng, Y.; Shen, X.H.; Su, Y.A.; He, N.; Ma, Z.B.; et al. Mapping quantitative trait loci for nitrogen uptake and utilization efficiency in rice (Oryza sativa L.) at different nitrogen fertilizer levels. Genet. Mol. Res. 2015, 8, 10404–10414. [Google Scholar] [CrossRef] [PubMed]
- Wissuwa, M.; Yano, M.; Ae, N. Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa L.). Appl. Genet. 1998, 97, 777–783. [Google Scholar] [CrossRef]
- Feng, M.; Xianwu, Z.; Guohu, M.I.; Ping, H.E.; Lihuang, Z.; Fusuo, Z. Identification of quantitative trait loci affecting tolerance to low phosphorus in rice (Oryza sativa L.). Chin. Sci. Bull. 2000, 45, 520–525. [Google Scholar]
- Fang, P.; Wu, P. QTL × N-level interaction for plant height in rice (Oryza sativa L.). Plant Soil 2001, 236, 237–242. [Google Scholar] [CrossRef]
- Senthilvel, S.; Vinod, K.K.; Malarvizhi, P.; Maheswaran, M. QTL and QTL environment effects on agronomic and nitrogen acquisition traits in rice. J. Integr. Plant Biol. 2008, 50, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Ping, M.U.; Huang, C.; Li, J.X.; Liu, L.F.; Li, Z.C. Yield trait variation and QTL mapping in a DH population of rice under phosphorus deficiency. Acta Agron. Sin. 2008, 34, 1137–1142. [Google Scholar]
- Li, J.; Xie, Y.; Dai, A.; Liu, L.; Li, Z. Root and shoot traits responses to phosphorus deficiency and QTL analysis at seedling stage using introgression lines of rice. J. Genet. Genom. 2009, 36, 173–183. [Google Scholar] [CrossRef]
- Xiang, C.; Ren, J.; Zhao, X.-Q.; Ding, Z.-S.; Zhang, J.; Wang, C.; Zhang, J.-W.; Joseph, C.A.; Zhang, Q.; Pang, Y.-L.; et al. Genetic dissection of low phosphorus tolerance related traits using selected introgression lines in rice. Rice Sci. 2015, 22, 264–274. [Google Scholar]
- Yamaya, T.; Obara, M.; Nakajima, H.; Sasaki, S.; Hayakawa, T.; Sato, T. Genetic manipulation and quantitative trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 2002, 53, 917–925. [Google Scholar] [CrossRef]
- Wissuwa, M.; Wegner, J.; Ae, N.; Yano, M. Substitution mapping of Pup1: A major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Appl. Genet. 2002, 105, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.H.; Zhang, Y.L.; Ye, L.T.; Fan, X.R.; Xu, G.H.; Shen, Q.R. Responses of rice cultivars with different nitrogen use efficiency to partial nitrate nutrition. Ann. Bot. 2007, 99, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yuan, S.; Huang, L.; Sun, F.; Zhu, G.; Li, G.; Fahad, S.; Peng, S.; Wang, F. Physiological mechanisms underlying the high-grain yield and high-nitrogen use efficiency of elite rice varieties under a low rate of nitrogen application in China. Front. Plant Sci. 2016, 7, 1024. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.T.; Dang, D.T.; Pham, C.V.; Bertin, P. QTL mapping for nitrogen use efficiency and related physiological and agronomical traits during the vegetative phase in rice under hydroponics. Euphytica 2016, 212, 473–500. [Google Scholar] [CrossRef]
- Shimizu, A.; Yanagihara, S.; Kawasaki, S.; Ikehashi, H. Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Appl. Genet. 2004, 109, 1361–1368. [Google Scholar] [CrossRef]
- Rose, T.J.; Wissuwa, M. Rethinking internal phosphorus utilization efficiency: A new approach is needed to improve PUE in grain crops. Adv. Agron. 2012, 116, 185–217. [Google Scholar]
- Wissuwa, M.; Kretzschmar, T.; Rose, T.J. From promise to application: Root traits for enhanced nutrient capture in rice breeding. J. Exp. Bot. 2016, 67, 3605–3615. [Google Scholar] [CrossRef]
- Wiel van de, C.C.M.; van der Linden, C.G.; Scholten, O.E. Improving phosphorus use efficiency in agriculture: Opportunities for breeding. Euphytica 2016, 207, 1–22. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, W.H. Genetic approaches for improvement of the crop potassium acquisition and utilization efficiency. Curr. Opin. Plant Biol. 2015, 25, 46–52. [Google Scholar] [CrossRef]
- Xue, X.; Lu, J.; Ren, T.; Li, L.; Yousaf, M.; Cong, R.; Li, X. Positional difference in potassium concentration as diagnostic index relating to plant K status and yield level in rice (Oryza sativa L.). Soil Sci. Plant Nutr. 2016, 62, 31–38. [Google Scholar] [CrossRef]
- Carmeis Filho, A.C.A.; Crusciol, C.A.C.; Guimarães, T.M.; Calonego, J.C.; Mooney, S.J. Impact of amendments on the physical properties of soil under tropical long-term no till conditions. PLoS ONE 2016, 11, e0167564. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.H.; Mei, H.W.; Yu., X.Q.; Xu., X.Y.; Li, M.S.; Zhang, S.Q.; Luo, L.J. Identification of related QTLs at late developmental stage in rice (Oryza sativa L.) under two nitrogen levels. Acta Genet. Sin. 2006, 33, 458–467. [Google Scholar] [CrossRef]
- Piao, Z.; Li, M.; Li, P.; Zhang, C.; Wang, H.; Luo, Z.; Lee, J.; Yang, R. Bayesian dissection for genetic architecture of traits associated with nitrogen utilization efficiency in rice. Afr. J. Biotechnol. 2009, 8, 6834–6839. [Google Scholar]
- Wei, D.; Cui, K.; Ye, G.; Pan, J.; Xiang, J.; Huang, J.; Nie, L. QTL mapping for nitrogen-use efficiency and nitrogen deficiency tolerance traits in rice. Plant Soil 2012, 359, 281–295. [Google Scholar] [CrossRef]
- Tanksley, S.D. Mapping polygenes. Annu. Rev. Genet. 1993, 27, 205–233. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D.; Young, N.D.; Paterson, A.H.; Bonierbale, M.W. RFLP mapping in plant breeding: New tools for an old science. Nat. Biotechnol. 1989, 7, 257–264. [Google Scholar] [CrossRef]
- Li, Z.K.; Fu, B.Y.; Gao, Y.M.; Xu, J.L.; Ali, J.; Lafitte, H.; Jiang, Y.Z.; Rey, J.; Vijayakumar, C.; Maghirang, R.; et al. Genome-wide ILs and their use in genetic and molecular dissection of complex phenotypes in rice (Oryza sativa L.). Plant Mol. Biol. 2005, 59, 33–52. [Google Scholar] [CrossRef]
- Ali, J.; Xu, J.L.; Gao, Y.; Fontanilla, M.; Li, Z.K. Breeding for yield potential and enhanced productivity across different rice ecologies through green super rice (GSR) breeding strategy. In International Dialogue on Perceptions and Prospects of Designer Rice; Muralidharan, K., Siddiq, E.A., Eds.; Society for the Advancement of Rice Research, Directorate of Rice Research: Hyderabad, India, 2013; pp. 60–68. [Google Scholar]
- Rao, T.N. Improving Nutrient Use Efficiency: The Role of Beneficial Management Practices. In Better Crops-India; IPNI–India Program 133: Gurgaon, India, 2007; Volume 1, pp. 6–7. [Google Scholar]
- Prasad, B.; Patwary, A.K.; Biswas, P.S. Genetic variability and selection criteria in fine rice (Oryza sativa L.). Pak. J. Biol. Sci. 2001, 4, 1188–1190. [Google Scholar]
- Zahid, M.A.; Akhatar, M.; Sabar, N.; Zaheen, M.; Tahir, A. Correlation and path analysis studies of yield and grain traits in Basmati rice (Oryza sativa L.). Asian J. Plant Sci. 2006, 5, 643–645. [Google Scholar]
- Cruz, C.D.; Regazzi, A.J. Biometric Models Applied to Genetic Improvement, 2nd ed.; Universidade Federal de Viçosa: Viçosa, Brazil, 1997; 390p. [Google Scholar]
- Ashura, L. Inter-relationship between yield and some selected agronomic characters in rice. Afr. Crop Sci. J. 1998, 6, 83–88. [Google Scholar]
- Lebreton, C.; LazicJancic, V.; Steed, A.; Pekic, S.Q.S. Identification of QTL for drought responses in maize and their use in testing causal relationships between traits. J. Exp. Bot. 1995, 46, 853–865. [Google Scholar] [CrossRef]
- Cisse, L.; Amar, B. The importance of phosphatic fertilizer for increased crop production in developing countries. In Proceedings of the AFA 6th International Annual Conference, Cairo, Egypt, 31 January–2 February 2000. [Google Scholar]
- Yadav, R.L. Assessing on-farm efficiency and economics of fertilizer N, P and K in rice-wheat systems of India. Field Crop Res. 2003, 18, 39–51. [Google Scholar] [CrossRef]
- Yoshida, S. Fundamentals of Rice Crop Science; IRRI: Los Baños, Philippines, 1981; 269p. [Google Scholar]
- Xie, W.X.; Wang, G.H.; Zhang, Q.C.; Guo, H.C. Effects of nitrogen fertilization strategies on nitrogen use efficiency in physiology, recovery, and agronomy and redistribution of dry matter accumulation and nitrogen accumulation in two typical rice cultivars in Zhejiang. China J. Zhejiang Univ. Sci. B 2007, 8, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Bart, R.; Chern, M.; Canlas, P.E.; Bai, W.; Ronald, P.C. Over-expression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS ONE 2010, 5, e9262. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Yang, G.; Chen, Y.; Zhang, C.; Zhang, J.; Peng, X. Two hydroxyl pyruvate reductases encoded by OsHPR1 and OsHPR2 are involved in photo respiratory metabolism in rice. J. Integr. Plant Biol. 2014, 56, 170–180. [Google Scholar] [CrossRef]
- Yasuda, H.; Hirose, S.; Kawakatsu, T.; Wakasa, Y.; Takaiwa, F. Over-expression of BiPhas inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant Cell Physiol. 2009, 50, 1532–1543. [Google Scholar] [CrossRef]
- Wakasa, Y.; Yasuda, H.; Oono, Y.; Kawakatsu, T.; Hirose, S.; Takahashi, H.; Hayashi, S.; Yang, L.; Takaiwa, F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 2011, 65, 675–689. [Google Scholar] [CrossRef]
- Yang, Q.; He, H.; Li, H.; Tian, H.; Zhang, J.; Zhai, L.; Chen, J.; Wu, H.; Yi, G.; He, Z.H.; et al. NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice. PLoS ONE 2011, 6, e20015. [Google Scholar] [CrossRef]
- Qu, S.; Liu, G.; Zhou, B.; Bellizzi, M.; Zeng, L.; Dai, L.; Han, B.; Wang, G.L. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 2006, 172, 1901–1914. [Google Scholar] [CrossRef]
- Amarawathi, Y.; Singh, R.; Singh, A.K.; Singh, V.P.; Mohapatra, T.; Sharma, T.R.; Singh, N.K. Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol. Breed. 2008, 21, 49–65. [Google Scholar] [CrossRef]
- Cui, K.; Huang, J.; Xing, Y.; Yu, S.; Xu, C.; Peng, S. Mapping QTLs for seedling characteristics under different water supply conditions in rice (Oryza sativa). Physiol. Plant. 2008, 132, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Bernier, J.; Kumar, A.; Ramaiah, V.; Spaner, D.; Atlin, G. A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci. 2007, 47, 507–517. [Google Scholar] [CrossRef]
- Amanullah, M.A.; Almas, L.K.; Amanullaj, J.; Zahir, S.; Rahman, H.; Khalil, S.K. Agronomic efficiency and profitability of P-fertilizers applied at different planting densities of maize in Northwest. Pak. J. Plant Nutr. 2012, 35, 331–341. [Google Scholar] [CrossRef]
- Liu, X.; Bai, X.; Wang, X.; Chu, C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 2007, 164, 969–979. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Shen, J.; Li, L. Functional characterization of evolutionarily divergent 4-coumarate: Coenzyme a ligases in rice. Plant Physiol. 2011, 157, 574–586. [Google Scholar] [CrossRef]
- Tao, Z.; Liu, H.; Qiu, D.; Zhou, Y.; Li, X.; Xu, C.; Wang, S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009, 151, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Kou, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 2011, 62, 4863–4874. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Nguyen, B.D.; Sarkarung, S.; Martinez, C.; Paterson, A.H.; Nguyen, H.T. Mapping of genes controlling aluminum tolerance in rice: Comparison of different genetic backgrounds. Mol. Genet. Genom. 2002, 267, 772–780. [Google Scholar]
- Qi, D.; Guo, G.; Lee, M.C.; Zhang, J.; Cao, G.; Zhang, S.; Suh, S.C.; Zhou, Q.; Han, L. Identification of quantitative trait loci for the dead leaf rate and the seedling dead rate under alkaline stress in rice. J. Genet. Genom. 2008, 35, 299–305. [Google Scholar] [CrossRef]
- Sato, T.; Ueda, T.; Fukuta, Y.; Kumagai, T.; Yano, M. Mapping of quantitative trait loci associated with ultraviolet-B resistance in rice (Oryza sativa L.). Appl. Genet. 2003, 107, 1003–1008. [Google Scholar] [CrossRef]
- Xue, Y.; Wan, J.; Jiang, L.; Wang, C.; Liu, L.; Zhang, Y.M.; Zhai, H. Identification of quantitative trait loci associated with aluminum tolerance in rice (Oryza sativa L.). Euphytica 2006, 150, 37–45. [Google Scholar] [CrossRef]
- Mori, M.; Tomita, C.; Sugimoto, K.; Hasegawa, M.; Hayashi, N.; Dubouzet, J.G.; Ochiai, H.; Sekimoto, H.; Hirochika, H.; Kikuchi, S. Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol. Biol. 2007, 63, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Shimono, M.; Koga, H.; Akagi, A.; Hayashi, N.; Goto, S.; Sawada, M.; Kurihara, T.; Matsushita, A.; Sugano, S.; Jiang, C.J.; et al. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 2012, 13, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Cao, L.Y.; Wu, W.M.; Shen, X.H.; Zhan, X.D.; Zhai, R.R.; Wang, R.C.; Chen, D.B.; Cheng, S.H. Mapping QTLs for nitrogen-deficiency tolerance at seedling stage in rice (Oryza sativa L.). Plant Breed. 2010, 129, 652–656. [Google Scholar] [CrossRef]
- Heuer, S.; Lu, X.; Chin, J.H.; Tanaka, J.P.; Kanamori, H.; Matsumoto, T.; De Leon, T.; Ulat, V.J.; Ismail, A.M.; Yano, M.; et al. Comparative sequence analyses of the major quantitative trait locus phosphorus uptake 1 (Pup1) reveal a complex genetic structure. Plant Biotechnol. J. 2009, 7, 456–457. [Google Scholar] [CrossRef] [PubMed]
- Babu, R.C.; Nguyen, B.D.; Chamarerk, V.; Shanmugasundaram, P.; Chezhian, P.; Jeyaprakash, P.; Ganesh, S.K.; Palchamy, A.; Sadasivam, S.; Sarkarung, S.; et al. Genetic analysis of drought resistance in rice by molecular markers: Association between secondary traits and field performance. Crop Sci. 2003, 43, 1457–1469. [Google Scholar] [CrossRef]
- Andaya, V.C.; Mackill, D.J. Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. J. Exp. Bot. 2003, 54, 2579–2585. [Google Scholar] [CrossRef]
- Peng, J.W.; Liu, Q.; Rong, X.M.; Zhu, H.M.; Xie, G.X.; Tang, G.R. Effects of different rational ratio of N, P, K fertilizer and amount of N fertilizer on photosynthesis character and yield of rice. J. Hunan Agric. Univ. Nat. Sci. 2004, 30, 123–127. [Google Scholar]
- Wu, P.; Liao, C.Y.; Hu, B.; Yi, K.K.; Jin, W.Z.; Ni, J.J.; He, C. QTLs and epistasis for aluminum tolerance in rice (Oryza sativa L.) at different seedling stages. Appl. Genet. 2000, 100, 1295–1303. [Google Scholar] [CrossRef]
- Yi, K.; Wu, Z.; Zhou, J.; Du, L.; Guo, L.; Wu, Y.; Wu, P. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 2005, 138, 2087–2096. [Google Scholar] [CrossRef]
- Gamuyao, R.; Chin, J.H.; Pariasca-Tanaka, J.; Pesaresi, P.; Catausan, S.; Dalid, C.; Slamet-Loedin, I.; Tecson-Mendoza, E.M.; Wissuwa, M.; Heuer, S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 2012, 488, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Moncada, P.; Martinez, C.P.; Borrero, J.; Chatel, M.; Gauch, H.; Guimaraes, E.; Tohme, J.; McCouch, S.R. Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Appl. Genet. 2001, 102, 41–52. [Google Scholar] [CrossRef]
- Araki, R.; Hasegawa, H. Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breed. Sci. 2006, 56, 295–302. [Google Scholar] [CrossRef]
- Katayama, H.; Mori, M.; Kawamura, Y.; Tanaka, T.; Mori, M.; Hasegawa, H. Production and characterization of transgenic plants carrying a high-affinity nitrate transporter gene (OsNRT2.1). Breed. Sci. 2009, 59, 237–243. [Google Scholar] [CrossRef]
- Fu, Q.; Zhang, P.; Tan, L.; Zhu, Z.; Ma, D.; Fu, Y.; Zhan, X.; Cai, H.; Sun, C. Analysis of QTLs for yield-related traits in Yuanjiang common wild rice (Oryza rufipogon Griff.). J. Genet. Genom. 2010, 37, 147–157. [Google Scholar] [CrossRef]
- Ogawa, S.; Valencia, M.O.; Lorieux, M.; Arbelaez, J.D.; McCouch, M.; Ishitani, M.; Selvaraj, M.G. Identification of QTLs associated with agronomic performance under nitrogen-deficient conditions using chromosome segment substitution lines of a wild rice relative, Oryza rufipogon. Acta Physiol. Plant. 2016, 38, 103. [Google Scholar] [CrossRef]
- Hittalmani, S.; Shashidhar, H.E.; Bagali, P.G.; Ning, H.; Sidhu, J.S.; Singh, V.P.; Khush, G.S. Molecular mapping of quantitative trait loci for plant growth, yield and yield related traits across three diverse locations in a doubled haploid rice population. Euphytica 2002, 125, 207–214. [Google Scholar] [CrossRef]
- Pelgas, B.; Bousquet, J.; Meirmans, P.G.; Ritland, K.; Isabel, N. QTL mapping in white spruce: Gene maps and genomic regions underlying adaptive traits across pedigrees, years and environments. BMC Genom. 2011, 12, 145. [Google Scholar] [CrossRef]
- Koide, Y.; Pariasca-Tanaka, J.; Rose, T.; Fukuo, A.; Konisho, K.; Yanagihara, S.; Fukuta, Y.; Wissuwa, M. QTLs for phosphorus-deficiency tolerance detected in upland NERICA varieties. Plant Breed. 2013, 132, 259–265. [Google Scholar] [CrossRef]
- Mukherjee, A.; Sarkar, S.; Chakraborty, A.S.; Yelne, R.; Kavishetty, V.; Biswas, T.; Mandal, N.; Bhattacharyya, S. Phosphate acquisition efficiency and phosphate starvation tolerance locus (PSTOL1) in rice. J. Genet. 2014, 93, 683–688. [Google Scholar] [CrossRef]
- Yokoshom, K.; Yamaji, N.; Ueno, D.; Mitani, N.; Ma, J.F. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 2009, 149, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Ai, P.; Sun, S.; Zhao, J.; Fan, X.; Xin, W.; Guo, Q.; Yu, L.; Shen, Q.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009, 57, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Ying, Y.; Wang, L.; Wang, F.; Whelan, J.; Shou, H. Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 2010, 11, 166. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Jeong, H.J.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol. Biol. 2010, 73, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Kakei, Y.; Shimo, H.; Bashir, K.; Sato, Y.; Nishizawa, N.K. A rice phenolic efflux transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J. Biol. Chem. 2011, 286, 24649–24655. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C.; Radovanovic, D.; Geoffroy, J.; Zhang, Q.; Siegler, H.; Chiapello, M.; Casieri, L.; An, K.; An, G.; Guiderdoni, E.; et al. The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. Plant J. 2012, 69, 906–920. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fujii, Y.; Yamaji, N.; Masuda, S.; Takemoto, Y.; Kamiya, T.; Yusuyin, Y.; Iwasaki, K.; Kato, S.; Maeshima, M.; et al. Mn tolerance in rice is mediated by MTP8.1, a member of the cation diffusion facilitator family. J. Exp. Bot. 2013, 64, 4375–4387. [Google Scholar] [CrossRef]
- Funayama, K.; Kojima, S.; Tabuchi-Kobayashi, M.; Sawa, Y.; Nakayama, Y.; Hayakawa, T.; Yamaya, T. Cytosolic glutamine synthetase1;2 is responsible for the primary assimilation of ammonium in rice roots. Plant Cell Physiol. 2013, 54, 934–943. [Google Scholar] [CrossRef]
- Ogo, Y.; Kobayashi, T.; Nakanishi Itai, R.; Nakanishi, H.; Kakei, Y.; Takahashi, M.; Toki, S.; Mori, S.; Nishizawa, N.K. A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J. Biol. Chem. 2008, 19, 13407–13417. [Google Scholar] [CrossRef]
- Shim, D.; Hwang, J.U.; Lee, J.; Lee, S.; Choi, Y.; An, G.; Martinoia, E.; Lee, Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 2009, 21, 4031–4043. [Google Scholar] [CrossRef]
- Dong, Y.J.; Ogawa, T.; Kamiunten, H.; Lin, D.Z.; Cheng, S.H.; Terao, H.; Matsuo, M. Detection of QTLs for zinc toxicity tolerance in rice (Oryza sativa L.). Rice Genet. Newsl. 2004, 21, 33–36. [Google Scholar]
- Wan, J.L.; Zhai, H.Q.; Wan, J.M.; Yasui, H.; Yoshimura, A. Detection and analysis of QTLs for some traits associated with tolerance to ferrous iron toxicity in rice (Oryza sativa L.), using recombinant inbred lines. Rice Genet. Newsl. 2003, 20, 55–57. [Google Scholar]
- Xue, Y.; Jiang, L.; Su, N.; Wang, J.K.; Deng, P.; Ma, J.F.; Zhai, H.Q.; Wan, J.M. The genetic basis and fine-mapping of a stable quantitative-trait loci for aluminium tolerance in rice. Planta 2007, 227, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Hanh, N.T.T.; Cuòng, P.V.; Pierre, B. Rice nitrogen use efficiency: Genetic dissection. J. Sci. Dev. 2013, 11, 814–825. [Google Scholar]
- Jewel, Z.A.; Ali, J.; Pang, Y.; Mahender, A.; Acero, B.; Hernandez, J.; Xu, J.; Li, Z.K. Developing green super rice varieties with high nutrient use efficiency by phenotypic selection under varied nutrient conditions. Crop J. 2019, in press. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Dobermann, A.R. Nitrogen Use Efficiency-State of the Art; Agronomy-Faculty Publications: Lincoln, NE, USA, 2005; 316p. [Google Scholar]
- Murray, M.G.; Thampson, W.F. Rapid isolation of high molecular weight plant DNA. Nucl. Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Wang, J.; Li, H.; Zhang, L.; Meng, L. Users’ Manual of QTL IciMapping; The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS): Beijing, China; Genetic Resources Program, International Maize and Wheat Improvement Center (CIMMYT): Mexico City, Mexico, 2014. [Google Scholar]
- Churchill, G.A.; Doerge, R.W. Empirical threshold values for quantitative trait mapping. Genetics 1994, 138, 963–971. [Google Scholar]
- Chen, M.; Presting, G.; Barbazuk, W.B.; Goicoechea, J.L.; Blackmon, B.; Fang, G.; Kim, H.; Frisch, D.; Yu, Y.; Sun, S.; et al. An integrated physical and genetic map of the rice genome. Plant Cell 2002, 14, 537–545. [Google Scholar] [CrossRef]
- Tiwari, S.; Krishnamurthy, S.L.; Kumar, V.; Singh, B.; Rao, A.; Mithra, S.V.A.; Rai, V.; Singh, A.K.; Singh, N.K. Mapping QTLs for salt tolerance in rice (Oryza sativa L.) by bulked segregant analysis of recombinant inbred lines using 50K SNP chip. PLoS ONE 2016, 11, e0153610. [Google Scholar] [CrossRef]
- Warnes, G.; Leisch, F. Package genetics: Population Genetics. 2015. Available online: https://cran.r-project.org/web/packages/genetics/genetics.pdf (accessed on 17 May 2016).
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]


| Traits | NuUE Condition | Mean ± Std. Error of Mean | Range (Min–Max) | SD | Variance (S2) | CV% |
|---|---|---|---|---|---|---|
| GY | NPK | 32.48 ± 0.34 | 19.29–50.26 | 5.21 | 27.19 | 16.04 |
| 75N | 32.58 ± 0.35 | 20.82–48.83 | 5.44 | 29.69 | 16.70 | |
| –N | 21.61 ± 0.24 | 13.89–34.96 | 3.79 | 14.37 | 17.54 | |
| –P | 26.74 ± 0.31 | 17.57–42.19 | 4.78 | 22.86 | 17.88 | |
| –NP | 20.41 ± 0.29 | 11.86–40.43 | 4.49 | 20.20 | 22.00 | |
| –NPK | 20.49 ± 0.29 | 12.81–42.95 | 4.54 | 20.66 | 22.16 | |
| 1000 Gwt | NPK | 27.35 ± 0.13 | 19.20–31.75 | 2.05 | 4.21 | 7.50 |
| 75N | 27.35 ± 0.14 | 15.18–39.50 | 2.24 | 5.03 | 8.19 | |
| –N | 26.41 ± 0.11 | 20.65–30.60 | 1.82 | 3.31 | 6.89 | |
| –P | 27.13 ± 0.16 | 14.80–33.75 | 2.52 | 6.36 | 9.29 | |
| –NP | 26.88 ± 0.14 | 15.60–31.10 | 2.14 | 4.58 | 7.96 | |
| –NPK | 27.16 ± 0.12 | 21.50–33.35 | 1.83 | 3.37 | 6.76 | |
| PSPF | NPK | 87.02 ± 0.28 | 68.05–95.24 | 4.34 | 18.84 | 4.99 |
| 75N | 87.14 ± 0.24 | 72.39–94.35 | 3.76 | 14.15 | 4.32 | |
| –N | 89.24 ± 0.29 | 68.50–96.59 | 4.44 | 19.78 | 4.98 | |
| –P | 87.60 ± 0.27 | 72.95–95.95 | 4.21 | 17.75 | 4.81 | |
| –NP | 89.93 ± 0.28 | 70.52–97.37 | 4.30 | 18.54 | 4.78 | |
| –NPK | 89.88 ± 0.26 | 77.27–97.75 | 4.09 | 16.76 | 4.55 | |
| FGN | NPK | 1518.90 ± 17.95 | 956.83–2621.83 | 273.41 | 74,756.36 | 18.00 |
| 75N | 1402.42 ± 19.23 | 783.00–3177.50 | 293.04 | 85,876.01 | 20.90 | |
| –N | 1012.30 ± 11.88 | 686.50–1660.83 | 180.97 | 32,750.56 | 17.88 | |
| –P | 1318.12 ± 16.54 | 786.50–2251.17 | 252.01 | 63,512.83 | 19.12 | |
| –NP | 947.57 ± 13.26 | 445.33–1704.50 | 202.11 | 40,848.47 | 21.33 | |
| –NPK | 1040.31 ± 14.50 | 565.83–1675.83 | 220.94 | 48,814.58 | 21.24 | |
| BY | NPK | 79.72 ± 1.14 | 35.92–137.23 | 17.43 | 303.83 | 21.86 |
| 75N | 210.24 ± 1.29 | 41.52–30,066.06 | 1968.81 | 3,876,231.42 | 936.46 | |
| –N | 50.47 ± 1.48 | 23.46–336.63 | 22.54 | 508.41 | 44.66 | |
| –P | 69.26 ± 1.18 | 38.31–144.27 | 18.07 | 326.83 | 26.09 | |
| –NP | 45.01 ± 0.84 | 18.16–89.43 | 12.90 | 166.49 | 28.66 | |
| –NPK | 47.29 ± 0.91 | 18.09–116.50 | 13.95 | 194.736 | 29.50 |
| S. No. | Environment | Degrees of Freedom | Sum of Squares | Mean Squares | F-Value | Satterthwaite Denominator | Pr (>F) |
|---|---|---|---|---|---|---|---|
| 1 | NPK | 230 | 12,493.16 | 54.56 | 1.07 | 34.38 | 0.4293 |
| 2 | 75N | 230 | 12,736.71 | 55.62 | 1.20 | 451.78 | 0.0530 |
| 3 | –N | 230 | 6540.20 | 28.56 | 1.44 | 451.61 | 0.0005 *** |
| 4 | –P | 230 | 9273.85 | 40.50 | 1.38 | 62.36 | 0.0658 |
| 5 | –NP | 230 | 9288.00 | 40.56 | 1.76 | 229.00 | 0.0000 *** |
| 6 | –NPK | 230 | 9503.51 | 41.50 | 2.47 | 37.76 | 0.0007 *** |
| Effect | Model | AIC | BIC | Log-Likelihood | Chi Square | Degrees of Freedom | Pr (>Chisq) |
|---|---|---|---|---|---|---|---|
| Environment | 1 | 17,071.23 | 18,457.21 | −8301.62 | |||
| 2 | 17,058.00 | 18,449.91 | −8294.00 | 15.2284 | 1 | 0.0001 *** | |
| Genotype X Environment | 3 | 17,056.00 | 18,441.98 | −8294.00 | |||
| 4 | 17,058.00 | 18,449.91 | −8294.00 | 0 | 1 | 0.9992 |
| Agronomic Efficiency (AE) | |||
| AE applied nitrogen | AE formula | >15 kg grain kg−1 nitrogen applied | >Parents |
| AE(N) = grain yield (N fertilized–0NPK unfertilized) in kg ha−1/Fertilizer N in kg ha−1 | AE(N) = (YNPK − Y0NPK) ÷ FN | 117 | 74 |
| AE(N) = (YNK − Y0NPK) ÷ FN | 33 | 28 | |
| AE(N) = (Y75N − Y0NPK) ÷ F75N | 161 | 86 | |
| Partial Factor Productivity (PFP) | |||
| PFP Applied Nitrogen | PFP formula | >50 kg grain kg−1 nitrogen applied | >Parents |
| PFP(N) = grain yield N fertilized in kg ha−1/Fertilizer N in kg ha−1 | PFP(N) = Y(+NPK) ÷ FN | 16 | 25 |
| PFP(N) = Y(−P) ÷ FN | 4 | 61 | |
| PFP(N) = Y(75N) ÷ FN | 151 | 117 | |
| S. No. | Chr | Marker No. | Average Distance (Kb) | Genome Size (Kb) | Genome Size (Gramene) | Coverage Percentage | Genetic Distance (cM) | Physical Distance per (Kb) |
|---|---|---|---|---|---|---|---|---|
| 1 | Chr01 | 76 | 564.0 | 42,492.4 | 43,270.92 | 98.20 | 181.8 | 238.01 |
| 2 | Chr02 | 45 | 797.4 | 35,401.9 | 35,937.25 | 98.51 | 157.9 | 227.59 |
| 3 | Chr03 | 72 | 497.7 | 35,824.4 | 36,413.81 | 98.38 | 166.4 | 218.83 |
| 4 | Chr04 | 84 | 405.0 | 33,864.4 | 35,502.69 | 95.39 | 129.6 | 273.94 |
| 5 | Chr05 | 50 | 479.8 | 29,100.3 | 29,958.43 | 97.14 | 122.3 | 244.96 |
| 6 | Chr06 | 73 | 422.7 | 30,809.5 | 31,248.78 | 98.59 | 124.4 | 251.20 |
| 7 | Chr07 | 74 | 393.9 | 28,942.5 | 29,697.62 | 97.46 | 118.6 | 250.40 |
| 8 | Chr08 | 43 | 654.5 | 27,809.9 | 28,443.02 | 97.77 | 121.1 | 234.87 |
| 9 | Chr09 | 41 | 521.7 | 21,348.9 | 23,012.72 | 92.77 | 93.5 | 246.13 |
| 10 | Chr10 | 43 | 464.0 | 19,635.6 | 23,207.28 | 84.61 | 83.8 | 276.94 |
| 11 | Chr11 | 58 | 492.2 | 28,312.7 | 29,021.10 | 97.56 | 117.9 | 246.15 |
| 12 | Chr12 | 45 | 603.7 | 27,023.4 | 27,531.85 | 98.15 | 109.5 | 251.43 |
| S. No. | NuUE Condition a | Trait b | QTLs c | Chr | Position (bp) d | Peak Marker e | LOD Value | PVE% f | Additive Effect |
|---|---|---|---|---|---|---|---|---|---|
| 1 | –P | AE | qAE_2.1 | 2 | 542,635 | SNP_2_542635 | 2.77 | 6.43 | 3.16 |
| 2 | –P | AE | qAE_4.1 | 4 | 21,815,986 | SNP_4_21815986 | 4.01 | 9.17 | 2.13 |
| 3 | –P | AE | qAE_6.1 | 6 | 9,977,282 | SNP_6_9977282 | 4.52 | 10.27 | 2.28 |
| 4 | 75N | AE | qAE_12.1 | 12 | 14,936,674 | SNP_12_14936674 | 2.55 | 5.92 | −2.80 |
| 5 | –P | PFP | qPFP_1.1 | 1 | 20,345,712 | SNP_1_20345712 | 8.64 | 18.71 | 2.87 |
| 6 | –P | PFP | qPFP_2.2 | 2 | 4,481,943 | SNP_2_4481943 | 11.68 | 24.44 | −3.11 |
| 7 | –P | PFP | qPFP_3.1 | 3 | 853,802 | SNP_3_853802 | 8.93 | 19.28 | 3.05 |
| 8 | –P | PFP | qPFP_4.1 | 4 | 21,833,014 | SNP_4_21833014 | 10.59 | 22.44 | 3.03 |
| 9 | –P | PFP | qPFP_5.1 | 5 | 5,588,965 | SNP_5_5588965 | 3.39 | 7.81 | 2.10 |
| 10 | –P | PFP | qPFP_6.1 | 6 | 9,977,282 | SNP_6_9977282 | 8.07 | 17.60 | 2.71 |
| 11 | –P | PFP | qPFP_7.1 | 7 | 28,234,334 | SNP_7_28234334 | 7.35 | 16.16 | 2.82 |
| 12 | –P | PFP | qPFP_8.2 | 8 | 8,437,588 | SNP_8_8437588 | 9.90 | 21.14 | −2.89 |
| 13 | –P | PFP | qPFP_9.2 | 9 | 12,154,616 | SNP_9_12154616 | 8.73 | 18.89 | 3.15 |
| 14 | –P | PFP | qPFP_10.2 | 10 | 6,149,421 | SNP_10_6149421 | 12.15 | 25.28 | −3.16 |
| 15 | –P | PFP | qPFP_11.1 | 11 | 1,706,087 | SNP_11_1706087 | 5.93 | 13.25 | 2.29 |
| 16 | 75N | PFP | qPFP_1.2 | 1 | 23,091,103 | SNP_1_23091103 | 5.89 | 13.17 | 3.45 |
| 17 | 75N | PFP | qPFP_2.1 | 2 | 4,342,883 | SNP_2_4342883 | 9.44 | 20.25 | −3.99 |
| 18 | 75N | PFP | qPFP_3.2 | 3 | 3,542,519 | SNP_3_3542519 | 7.32 | 16.09 | 4.16 |
| 19 | 75N | PFP | qPFP_4.1 | 4 | 21,833,014 | SNP_4_21833014 | 7.60 | 16.66 | 3.68 |
| 20 | 75N | PFP | qPFP_5.2 | 5 | 15,469,279 | SNP_5_15469279 | 9.78 | 20.91 | −4.05 |
| 21 | 75N | PFP | qPFP_6.2 | 6 | 12,183,428 | SNP_6_12183428 | 4.46 | 10.14 | 2.92 |
| 22 | 75N | PFP | qPFP_7.2 | 7 | 28,303,039 | SNP_7_28303039 | 7.21 | 15.89 | 4.04 |
| 23 | 75N | PFP | qPFP_8.1 | 8 | 322,877 | SNP_8_322877 | 7.09 | 15.64 | −3.50 |
| 24 | 75N | PFP | qPFP_9.1 | 9 | 12,154,616 | SNP_9_12154616 | 7.87 | 17.19 | 4.23 |
| 25 | 75N | PFP | qPFP_10.1 | 10 | 146,531 | SNP_10_146531 | 9.13 | 19.68 | −3.92 |
| 26 | 75N | PFP | qPFP_11.2 | 11 | 2,514,115 | SNP_11_2514115 | 3.66 | 8.41 | 2.57 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jewel, Z.A.; Ali, J.; Mahender, A.; Hernandez, J.; Pang, Y.; Li, Z. Identification of Quantitative Trait Loci Associated with Nutrient Use Efficiency Traits, Using SNP Markers in an Early Backcross Population of Rice (Oryza sativa L.). Int. J. Mol. Sci. 2019, 20, 900. https://doi.org/10.3390/ijms20040900
Jewel ZA, Ali J, Mahender A, Hernandez J, Pang Y, Li Z. Identification of Quantitative Trait Loci Associated with Nutrient Use Efficiency Traits, Using SNP Markers in an Early Backcross Population of Rice (Oryza sativa L.). International Journal of Molecular Sciences. 2019; 20(4):900. https://doi.org/10.3390/ijms20040900
Chicago/Turabian StyleJewel, Zilhas Ahmed, Jauhar Ali, Anumalla Mahender, Jose Hernandez, Yunlong Pang, and Zhikang Li. 2019. "Identification of Quantitative Trait Loci Associated with Nutrient Use Efficiency Traits, Using SNP Markers in an Early Backcross Population of Rice (Oryza sativa L.)" International Journal of Molecular Sciences 20, no. 4: 900. https://doi.org/10.3390/ijms20040900
APA StyleJewel, Z. A., Ali, J., Mahender, A., Hernandez, J., Pang, Y., & Li, Z. (2019). Identification of Quantitative Trait Loci Associated with Nutrient Use Efficiency Traits, Using SNP Markers in an Early Backcross Population of Rice (Oryza sativa L.). International Journal of Molecular Sciences, 20(4), 900. https://doi.org/10.3390/ijms20040900