Transcriptome Analysis of Chinese Chestnut (Castanea mollissima Blume) in Response to Dryocosmus kuriphilus Yasumatsu Infestation
Abstract
:1. Introduction
2. Results
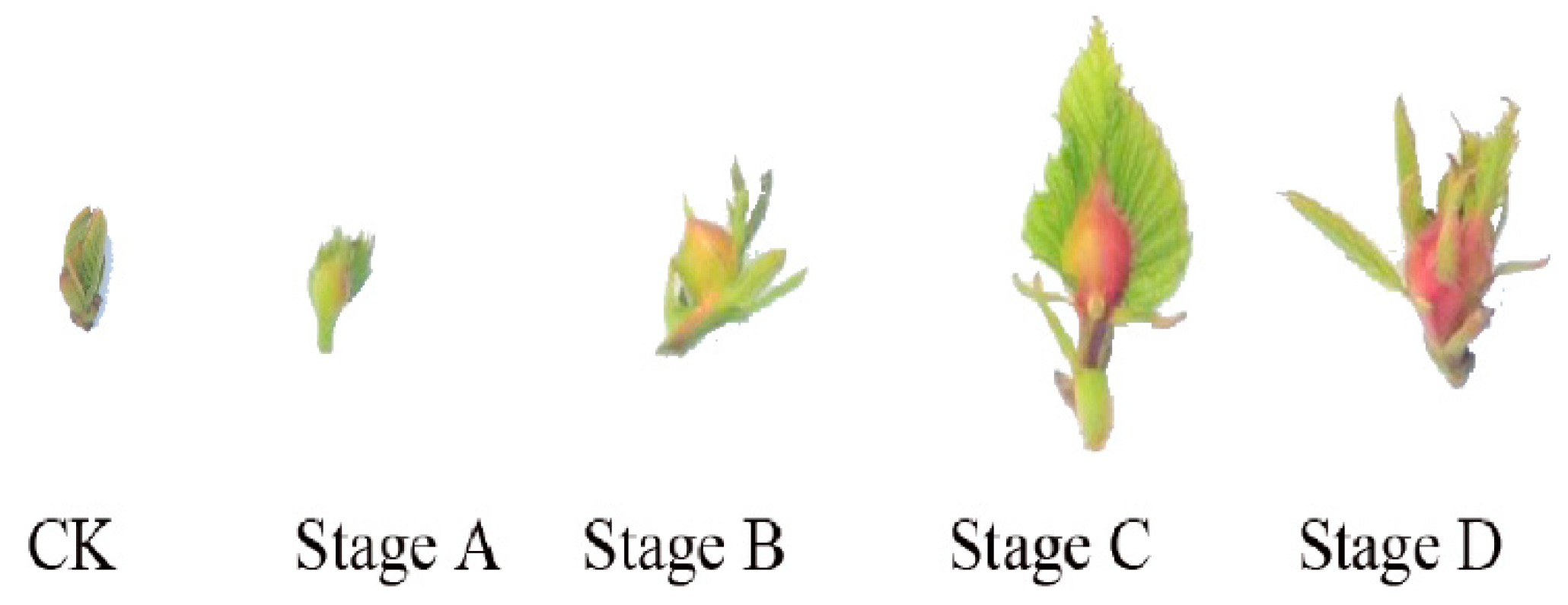
2.1. Gall Formation Process
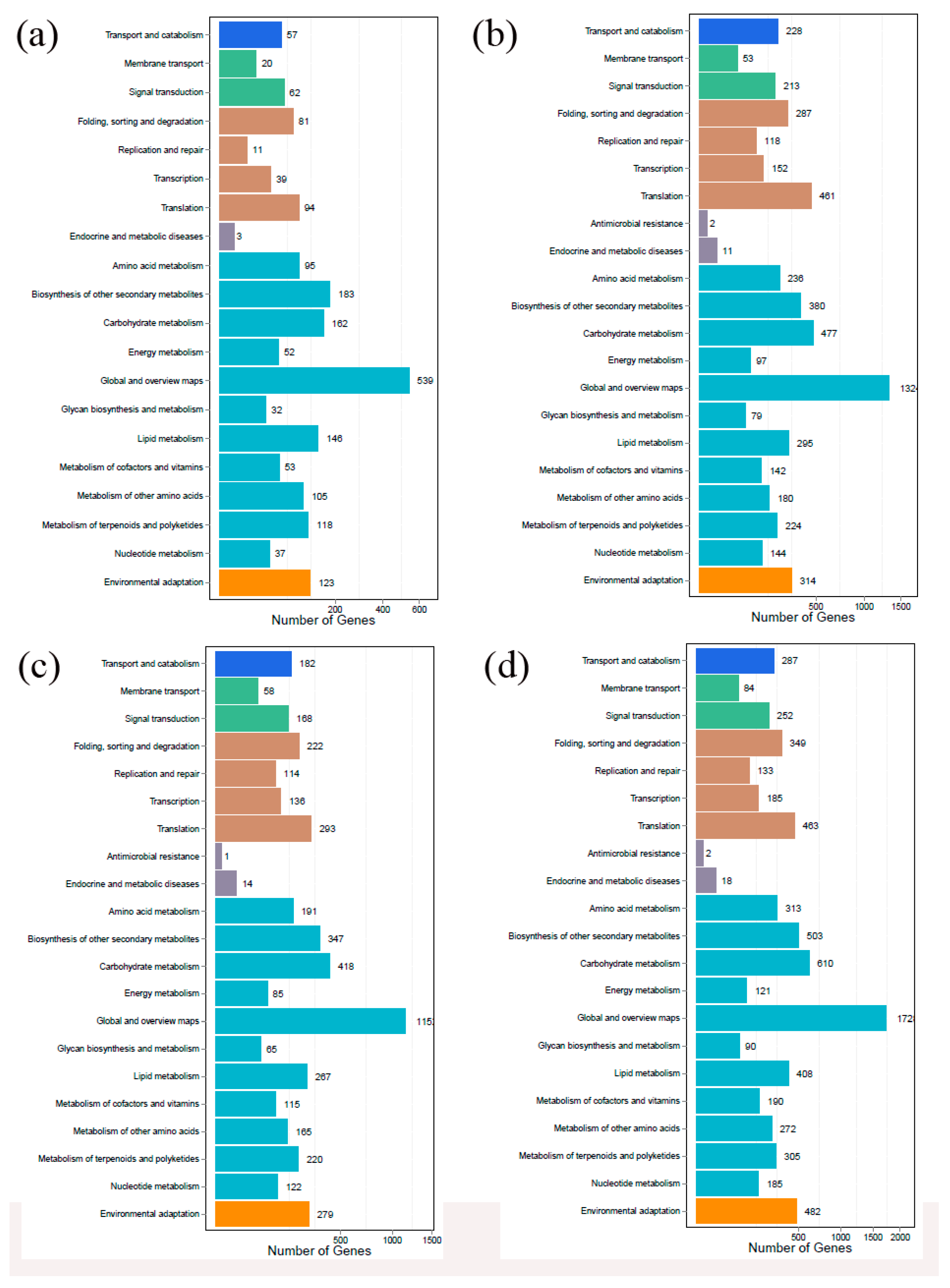
2.2. Sequencing, Assembly, and Annotation
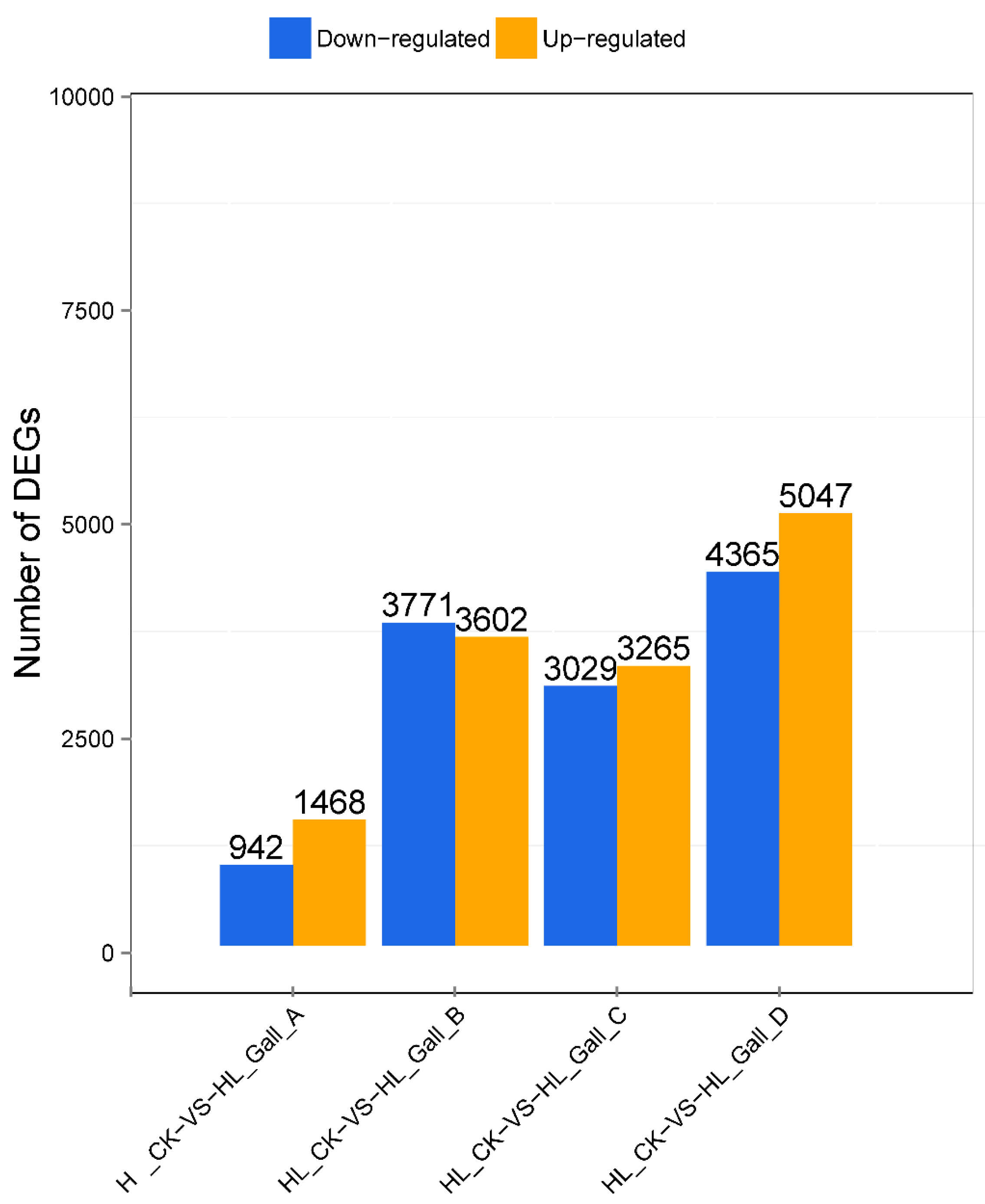
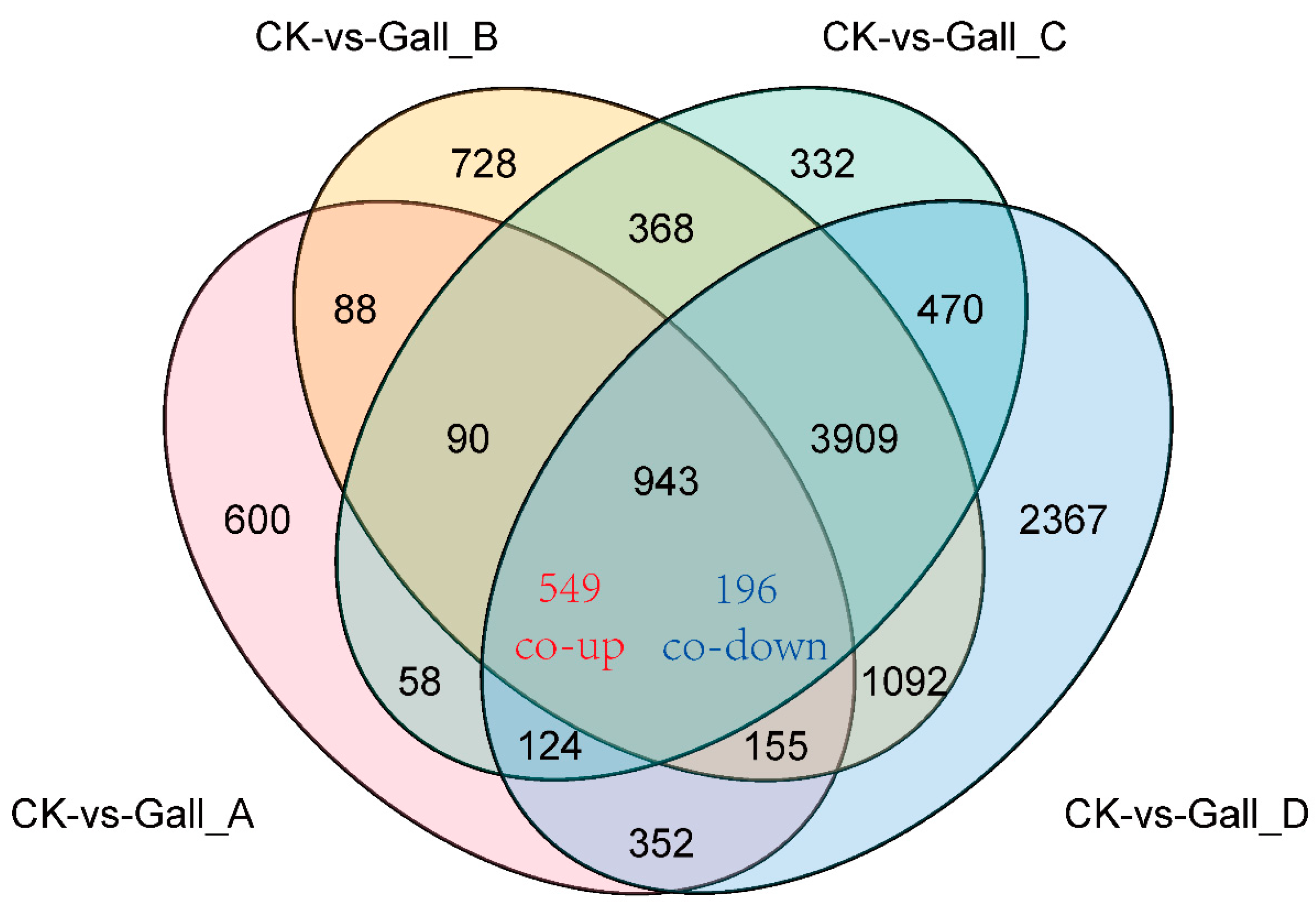
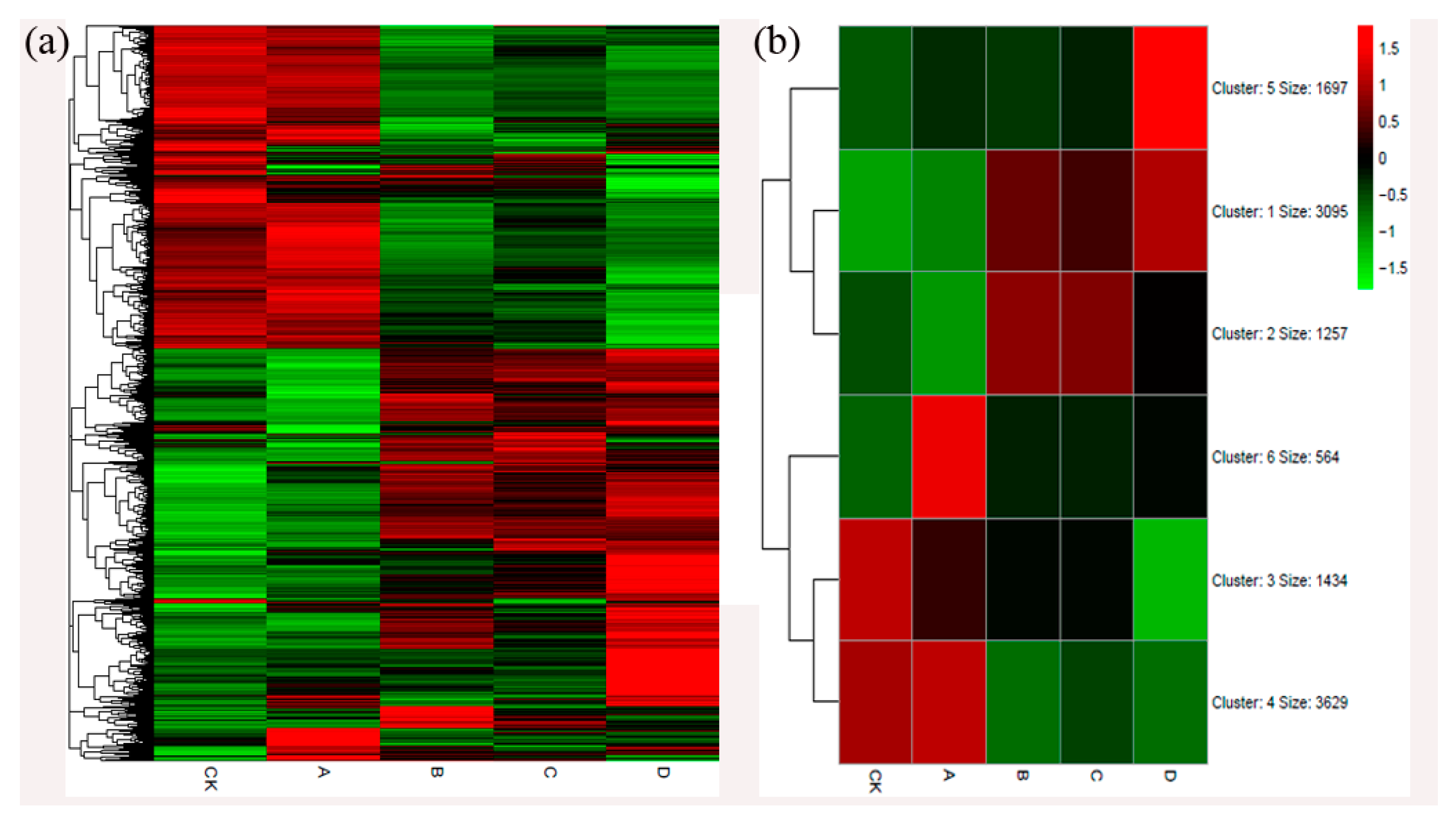
2.3. Analysis of Differentially Expressed Genes (DEGs) in Chinese Chestnut Leaf During the Leaf Gall Formation
2.4. DEGs Involved in Signaling and Transcription Were Altered during Leaf Gall Formation
2.5. DEGs Involved in Oxidation–Reduction Process Were Altered by D. kuriphilus Infestation
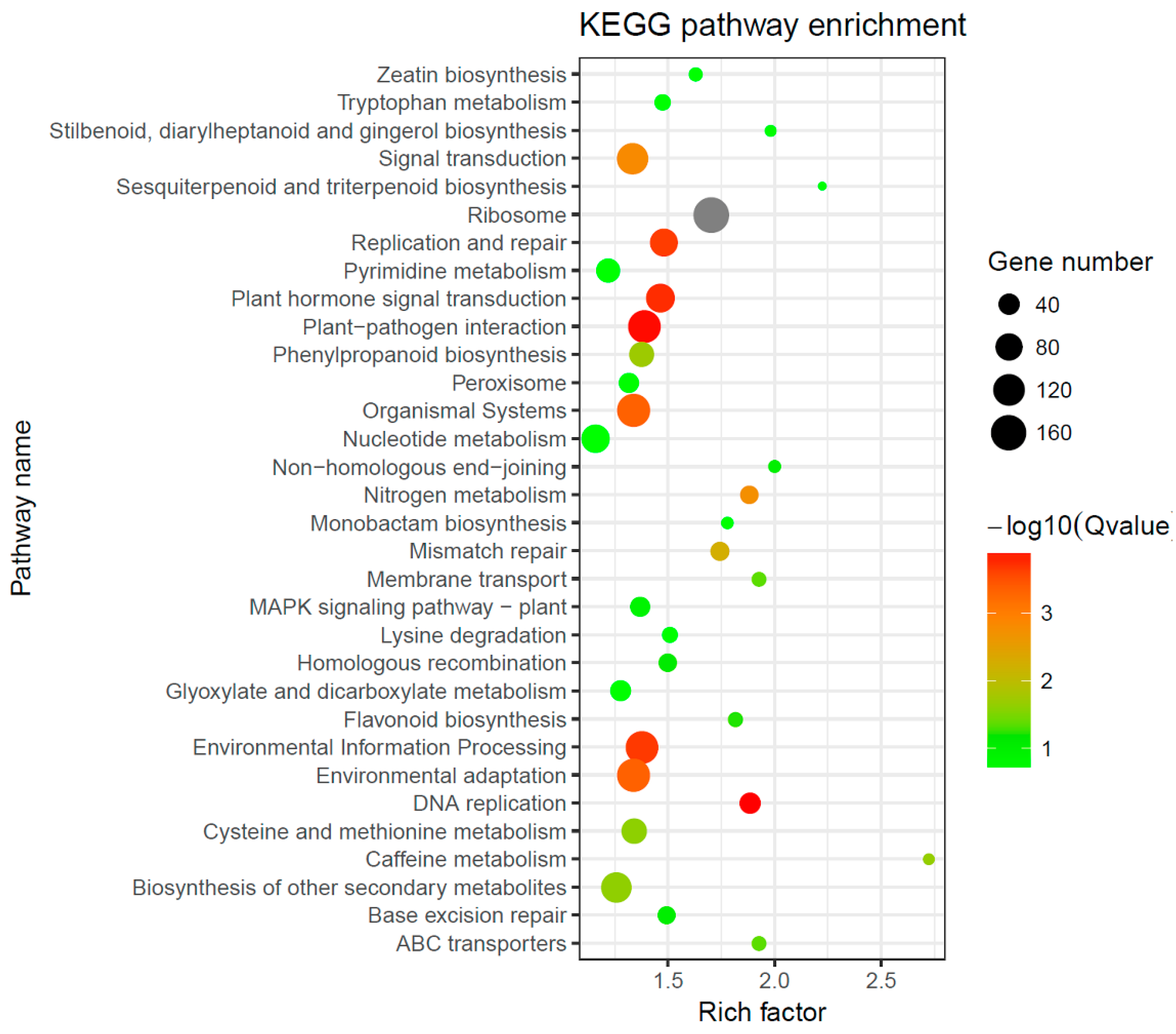
2.6. Regulation of Secondary Metabolism during Gall Formation by D. kuriphilus
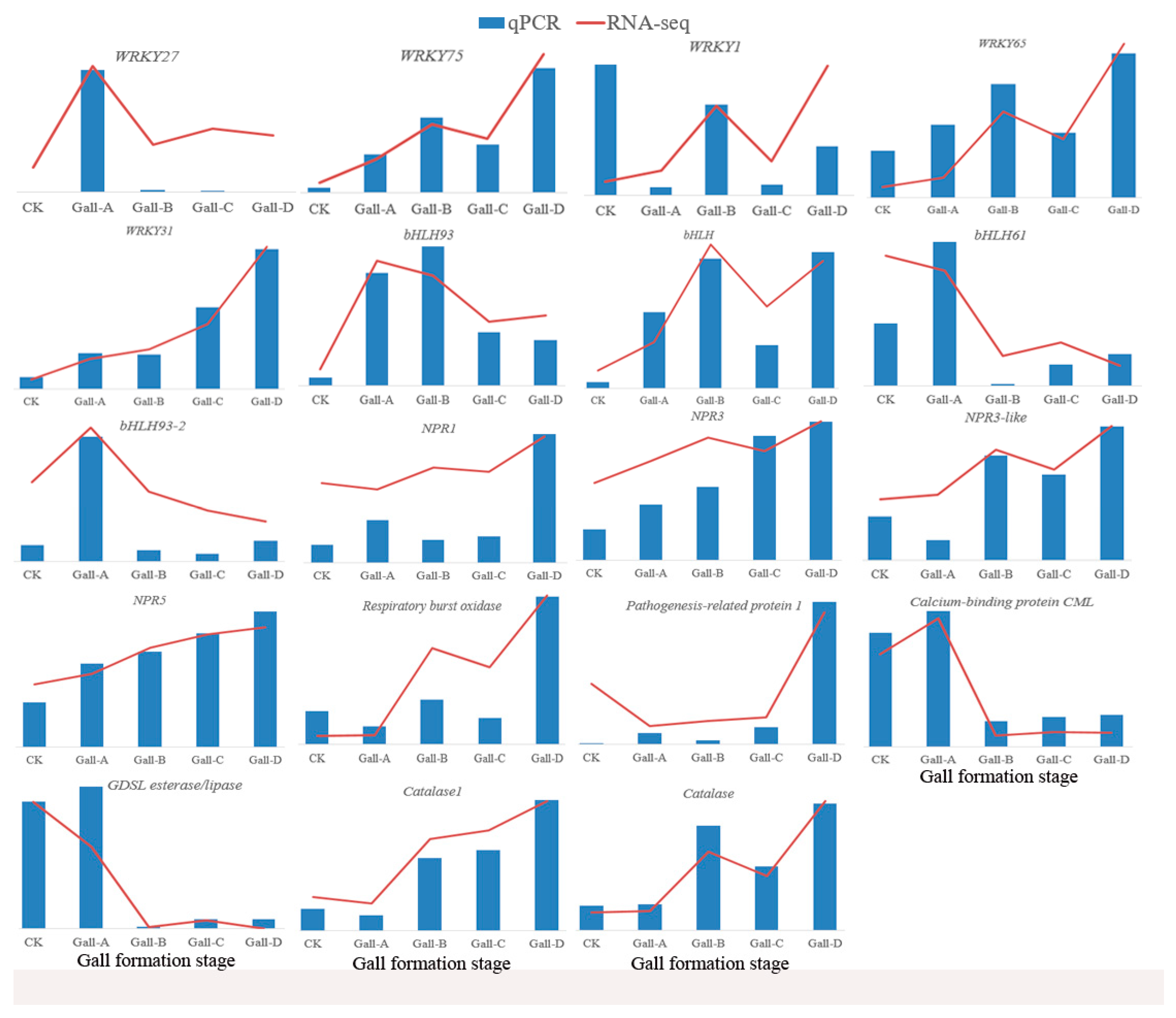
2.7. Validation of Some DEGs
3. Discussion
3.1. Signal Transduction and Transcription Factors Were Affected in Chestnuts during the Gall Formation by D. kuriphilus
3.2. Plant Secondary Metabolism Was Altered in Response to Gall Infestation
3.3. The Stress Response Ability of Chestnut Was Affected by Gall Infestation
4. Materials and Methods
4.1. Plant Material and Gall Collection
4.2. RNA Extraction, Library Construction, and Sequencing
4.3. De Novo Assembly and Annotation of the Castanea mollissima BL unigenes
4.4. Differential Expressed Genes (DEGs) Analysis
4.5. Validation of DEGs by Quantitative Real-Time PCR (qRT-PCR)
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DEGs | Differentially expressed genes |
| ROS | Reactive oxygen species |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene ontology |
| SOD | Superoxide dismutase |
| CAT | Catalase |
| BH | Benjamini and Hochberg |
| FDR | False discovery rate |
References
- Zhang, L.; Liu, T.; Hu, G.; Guo, K.; Wei, C. Comparison of physicochemical properties of starches from nine Chinese chestnut varieties. Molecules 2018, 23, 3248. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; DiLoreto, D.S.; Zhang, Y.; Smith, C.; Baier, K.; Powell, W.A.; Wheeler, N.; Sederoff, R.; Carlson, J.E. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol. 2009, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.R.; Rieske, L.K. Chestnut species and jasmonic acid treatment influence development and community interactions of galls produced by the Asian chestnut gall wasp, Dryocosmus kuriphilus. J. Insect Sci. 2011, 11, 140. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Hijii, N. Effects of gall formation by Dryocosmus kuriphilus Yasumatsu (Hym, Cynipidae) on the growth of chestnut trees. J. Appl. Entomol. 1997, 121, 9–15. [Google Scholar] [CrossRef]
- Stone, G.N.; Schönrogge, K. The adaptive significance of insect gall morphology. Trends Ecol. Evol. 2003, 18, 512–522. [Google Scholar] [CrossRef]
- Oliveira, D.C.; Isaias, R.M.S. Redifferentiation of leaflet tissues during midrib gall development in Copaifera langsdorffii (Fabaceae). S. Afr. J. Bot. 2010, 76, 239–248. [Google Scholar] [CrossRef]
- Pawlowski, T.A.; Staszak, A.M.; Karolewski, P.; Giertych, M.J. Plant development reprogramming by cynipid gall wasp: Proteomic analysis. Acta Physiol. Plant. 2017, 39, 114. [Google Scholar] [CrossRef]
- Inbar, M.; Izhaki, I.; Koplovich, A.; Lupo, I.; Silanikove, N.; Glasser, T.; Gerchman, Y.; Perevolotsky, A.; Levyadun, S. Why do many galls have conspicuous colors? A new hypothesis. Arthropod-Plant Interact. 2010, 4, 1–6. [Google Scholar] [CrossRef]
- Compson, Z.G.; Larson, K.C.; Zinkgraf, M.S.; Whitham, T.G. A genetic basis for the manipulation of sink–source relationships by the galling aphid Pemphigus batae. Oecologia 2011, 167, 711–721. [Google Scholar] [CrossRef]
- Oates, C.N.; Kulheim, C.; Myburg, A.A.; Slippers, B.; Naidoo, S. The transcriptome and terpene profile of eucalyptus grandis reveals mechanisms of defense against the insect pest, Leptocybe invasa. Plant Cell Physiol. 2015, 56, 1418–1428. [Google Scholar] [CrossRef]
- Ma, Z.C.; Zhu, L.; Song, T.Q.; Wang, Y.; Zhang, Q.; Xia, Y.Q.; Qiu, M.; Lin, Y.C.; Li, H.Y.; Kong, L.; et al. A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 2017, 355, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Wharton, P.S.; Schilder, A.C. Novel infection strategies of Colletotrichum acutatum on ripe blueberry fruit. Plant Pathol. 2008, 57, 122–134. [Google Scholar] [CrossRef]
- Tooker, J.F.; Rohr, J.R.; Abrahamson, W.G.; De Moraes, C.M. Gall insects can avoid and alter indirect plant defenses. New Phytol. 2008, 178, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Giron, D.; Huguet, E.; Stone, G.N.; Body, M. Insect-induced effects on plants and possible effectors used by galling and leaf-mining insects to manipulate their host-plant. J. Insect Physiol. 2016, 84, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.J.; Schonrogge, K.; Lim, K.Y.; Francis, P.; Lichtenstein, C.P. Cynipid galls: Insect-induced modifications of plant development create novel plant organs. Plant Cell Environ. 2004, 27, 327–335. [Google Scholar] [CrossRef]
- Allison, S.D.; Schultz, J.C. Biochemical responses of chestnut oak to a galling cynipid. J. Chem. Ecol. 2005, 31, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Tu, X.B.; Liu, Z.K.; Zhang, Z.H. Comparative transcriptomic analysis of resistant and susceptible alfalfa cultivars (Medicago sativa L.) after thrips infestation. BMC Genom. 2018, 19, 116. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Martinez, C.M.; Morris, J.A.; Hedley, P.E.; Bos, J.I.B. Barley transcriptome analyses upon interaction with different aphid species identify thionins contributing to resistance. Plant Cell Environ. 2017, 40, 2628–2643. [Google Scholar] [CrossRef]
- Wang, X.Y.; Lu, J.H.; Chen, H.F.; Shan, Z.H.; Shen, X.J.; Duan, B.B.; Zhang, C.J.; Yang, Z.L.; Zhang, X.J.; Qiu, D.Z.; et al. Comparative analyses of transcriptome and proteome in response to cotton bollworm between a resistant wild soybean and a susceptible soybean cultivar. Plant Cell Tissue Organ Cult. 2017, 129, 511–520. [Google Scholar] [CrossRef]
- Dini, F.; Sartor, C.; Marinoni, D.T.; Botta, R. Chestnut transcriptome NG sequencing: A new tool to investigate the response to gall wasp. II Eur. Congr. Chestnut 2014, 1043, 99–104. [Google Scholar] [CrossRef]
- Abe, Y.; Melika, G.; Stone, G.N. The diversity and phylogeography of cynipid gallwasps (Hymenoptera: Cynipidae) of the oriental and eastern Palearctic regions, and their associated communities. Orient Insects 2007, 41, 169–212. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. Lond. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Tooker, J.F.; Moraes, C.M.D. Feeding by Hessian fly [Mayetiola destructor (Say)] larvae does not induce plant indirect defences. Ecol. Entomol. 2010, 32, 153–161. [Google Scholar] [CrossRef]
- Tooker, J.F.; Moraes, C.M.D. Gall insects and indirect plant defenses: A case of active manipulation? Plant Signal Behav. 2008, 3, 503–504. [Google Scholar] [CrossRef] [PubMed]
- Nabity, P.D.; Haus, M.J.; Berenbaum, M.R.; DeLucia, E.H. Leaf-galling phylloxera on grapes reprograms host metabolism and morphology. Proc. Natl. Acad. Sci. USA 2013, 110, 16663–16668. [Google Scholar] [CrossRef] [PubMed]
- Agarrwal, R.; Padmakumari, A.P.; Bentur, J.S.; Nair, S. Metabolic and transcriptomic changes induced in host during hypersensitive response mediated resistance in rice against the Asian rice gall midge. Rice 2016, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef]
- Maffei, M.E.; Mithofer, A.; Arimura, G.I.; Uchtenhagen, H.; Bossi, S.; Bertea, C.M.; Cucuzza, L.S.; Novero, M.; Volpe, V.; Quadro, S.; et al. Effects of feeding Spodoptera littoralis on lima bean leaves. III. Membrane depolarization and involvement of hydrogen peroxide. Plant Physiol. 2006, 140, 1022–1035. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Occhipinti, A.; Zebelo, S.A.; Foti, M.; Fliegmann, J.; Bossi, S.; Maffei, M.E.; Bertea, C.M. Ginkgo biloba responds to herbivory by activating early signaling and direct defenses. PLoS ONE 2012, 7, e32822. [Google Scholar] [CrossRef]
- Zeng, W.Y.; Sun, Z.D.; Cai, Z.Y.; Chen, H.Z.; Lai, Z.G.; Yang, S.Z.; Tang, X.M. Comparative transcriptome analysis of soybean response to bean pyralid larvae. BMC Genom. 2017, 18, 871. [Google Scholar] [CrossRef]
- Van Verk, M.C.; Gatz, C.; Linthorst, H.J.M. Transcriptional regulation of plant defense responses. Adv. Bot. Res. 2009, 51, 397–438. [Google Scholar]
- Grunewald, W.; Karimi, M.; Wieczorek, K.; Van de Cappelle, E.; Wischnitzki, E.; Grundler, F.; Inze, D.; Beeckman, T.; Gheysen, G. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 2008, 148, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, J.; Huang, L.; Zhu, L.; Liu, X.; Weng, N.; Reese, J.C.; Harris, M.; Stuart, J.J.; Chen, M.S. Gene expression of different wheat genotypes during attack by virulent and avirulent Hessian fly (Mayetiola destructor) larvae. J. Chem. Ecol. 2007, 33, 2171–2194. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Matsuoka, M.; Yamanoto, N.; Ohashi, Y.; Kanomurakami, Y.; Ozeki, Y. Structure and characterization of a Cdna clone for phenylalanine ammonia-lyase from cut-injured roots of sweet-potato. Plant Physiol. 1989, 90, 1403–1407. [Google Scholar] [CrossRef]
- Ziv, C.; Zhao, Z.Z.; Gao, Y.G.; Xia, Y. Multifunctional roles of plant cuticle during plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1088. [Google Scholar] [CrossRef] [PubMed]
- Kosma, D.K.; Nemacheck, J.A.; Jenks, M.A.; Williams, C.E. Changes in properties of wheat leaf cuticle during interactions with Hessian fly. Plant J. 2010, 63, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, C.; Wang, H.Y.; Liu, X.M.; Wheeler, S.; Reese, J.C.; El Bouhssini, M.; Whitworth, R.J.; Chen, M.S. Mobilization of lipids and fortification of cell wall and cuticle are important in host defense against Hessian fly. BMC Genom. 2013, 14, 423. [Google Scholar] [CrossRef]
- Morant, M.; Bak, S.; Moller, B.L.; Werck-Reichhart, D. Plant cytochromes P450: Tools for pharmacology, plant protection and phytoremediation. Curr. Opin. Biotechnol. 2003, 14, 151–162. [Google Scholar] [CrossRef]
- Dini, F.; Sartor, C.; Botta, R. Detection of a hypersensitive reaction in the chestnut hybrid ‘Bouche de Betizac’ infested by Dryocosmus kuriphilus Yasumatsu. Plant Physiol. Biochem. 2012, 60, 67–73. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Zhang, Y.; Dong, J.; Ma, C.; Chen, W. NADPH oxidase RBOHD contributes to autophagy and hypersensitive cell death during the plant defense response in Arabidopsis thaliana. Biol. Plant. 2015, 59, 570–580. [Google Scholar] [CrossRef]
- Laloi, C.; Stachowiak, M.; Pers-Kamczyc, E.; Warzych, E.; Murgia, I.; Apel, K. Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Leszczynski, B. Changes in phenols content and metabolism in leaves of susceptible and resistant winter-wheat cultivars infested by Rhopalosiphum-Padi (L) (Hom, Aphididae). Z. Angew. Entomol. 1985, 100, 343–348. [Google Scholar] [CrossRef]
- Wei, Z.; Hu, W.; Lin, Q.S.; Cheng, X.Y.; Tong, M.J.; Zhu, L.L.; Chen, R.Z.; He, G.C. Understanding rice plant resistance to the Brown Planthopper (Nilaparvata lugens): A proteomic approach. Proteomics 2009, 9, 2798–2808. [Google Scholar] [CrossRef] [PubMed]
- Forslund, K.; Pettersson, J.; Bryngelsson, T.; Jonsson, L. Aphid infestation induces PR-proteins differently in barley susceptible or resistant to the birdcherry-oat aphid (Rhopalosiphum padi). Physiol. Plant 2000, 110, 496–502. [Google Scholar] [CrossRef]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.J.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.H.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Li, T.T.; Wu, Q.X.; Zhou, Y.J.; Yun, Z.; Duan, X.W.; Jiang, Y.M. L-Cysteine hydrochloride delays senescence of harvested longan fruit in relation to modification of redox status. Postharvest Biol. Technol. 2018, 143, 35–42. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Shi, F.; Chen, Y.; Wang, M.; Zhao, Y.; Geng, G. Transcriptome Analysis of Chinese Chestnut (Castanea mollissima Blume) in Response to Dryocosmus kuriphilus Yasumatsu Infestation. Int. J. Mol. Sci. 2019, 20, 855. https://doi.org/10.3390/ijms20040855
Zhu C, Shi F, Chen Y, Wang M, Zhao Y, Geng G. Transcriptome Analysis of Chinese Chestnut (Castanea mollissima Blume) in Response to Dryocosmus kuriphilus Yasumatsu Infestation. International Journal of Molecular Sciences. 2019; 20(4):855. https://doi.org/10.3390/ijms20040855
Chicago/Turabian StyleZhu, Cancan, Fenghou Shi, Yu Chen, Min Wang, Yuqiang Zhao, and Guomin Geng. 2019. "Transcriptome Analysis of Chinese Chestnut (Castanea mollissima Blume) in Response to Dryocosmus kuriphilus Yasumatsu Infestation" International Journal of Molecular Sciences 20, no. 4: 855. https://doi.org/10.3390/ijms20040855
APA StyleZhu, C., Shi, F., Chen, Y., Wang, M., Zhao, Y., & Geng, G. (2019). Transcriptome Analysis of Chinese Chestnut (Castanea mollissima Blume) in Response to Dryocosmus kuriphilus Yasumatsu Infestation. International Journal of Molecular Sciences, 20(4), 855. https://doi.org/10.3390/ijms20040855



