Mitochondrial Genomes Provide Insights into the Phylogeny of Culicomorpha (Insecta: Diptera)
Abstract
1. Introduction
2. Results
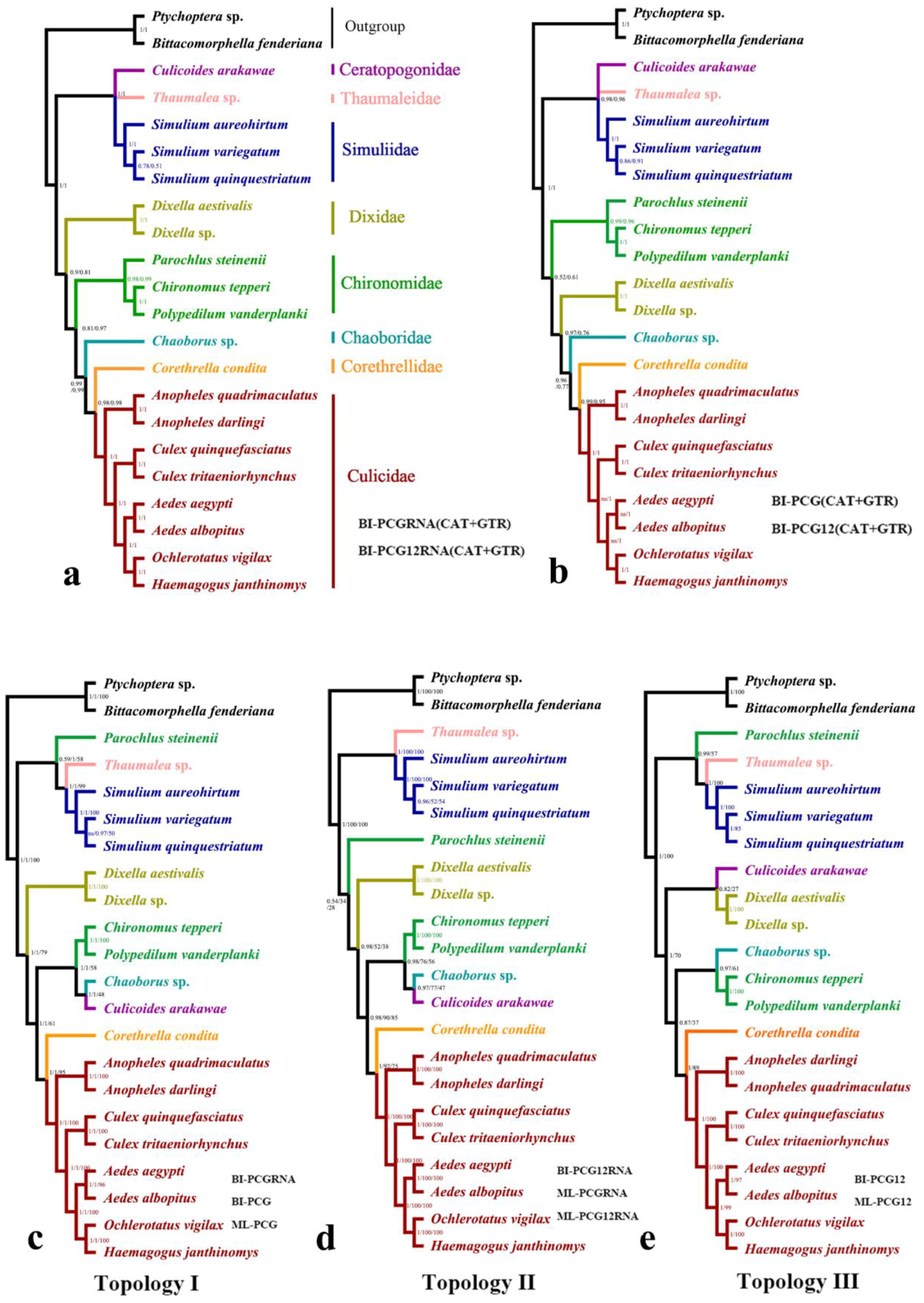
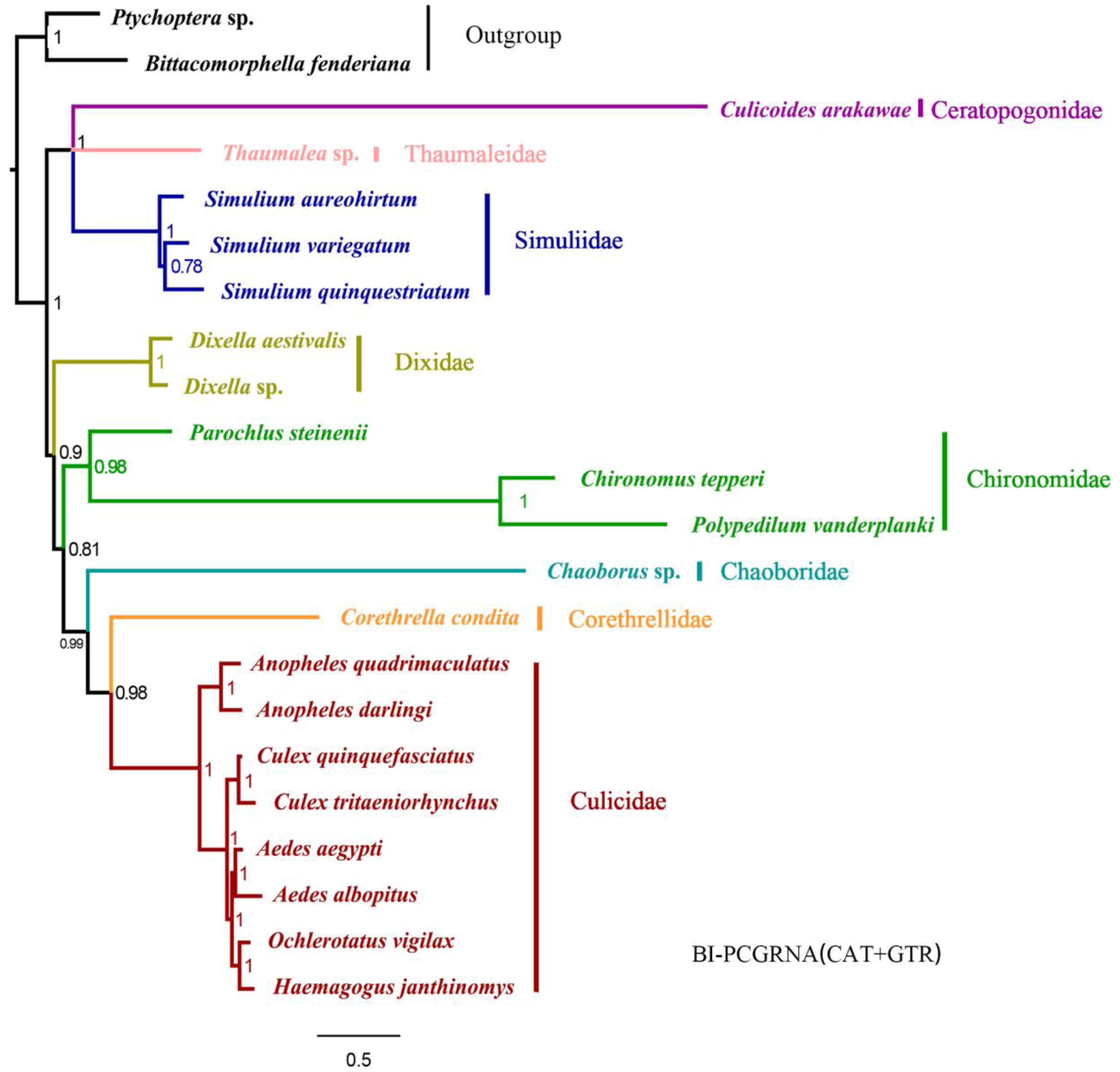
2.1. Phylogenetic Analyses
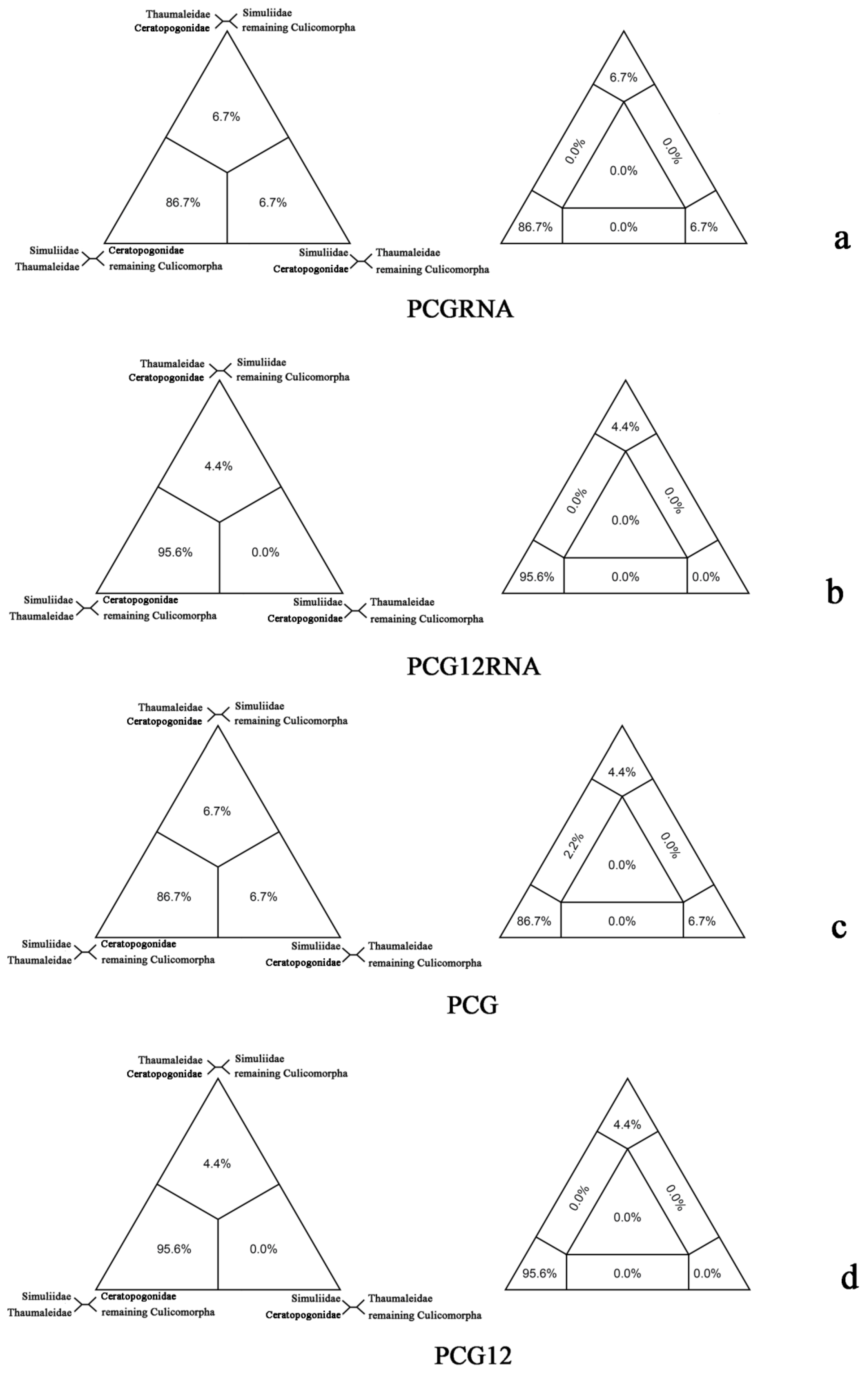
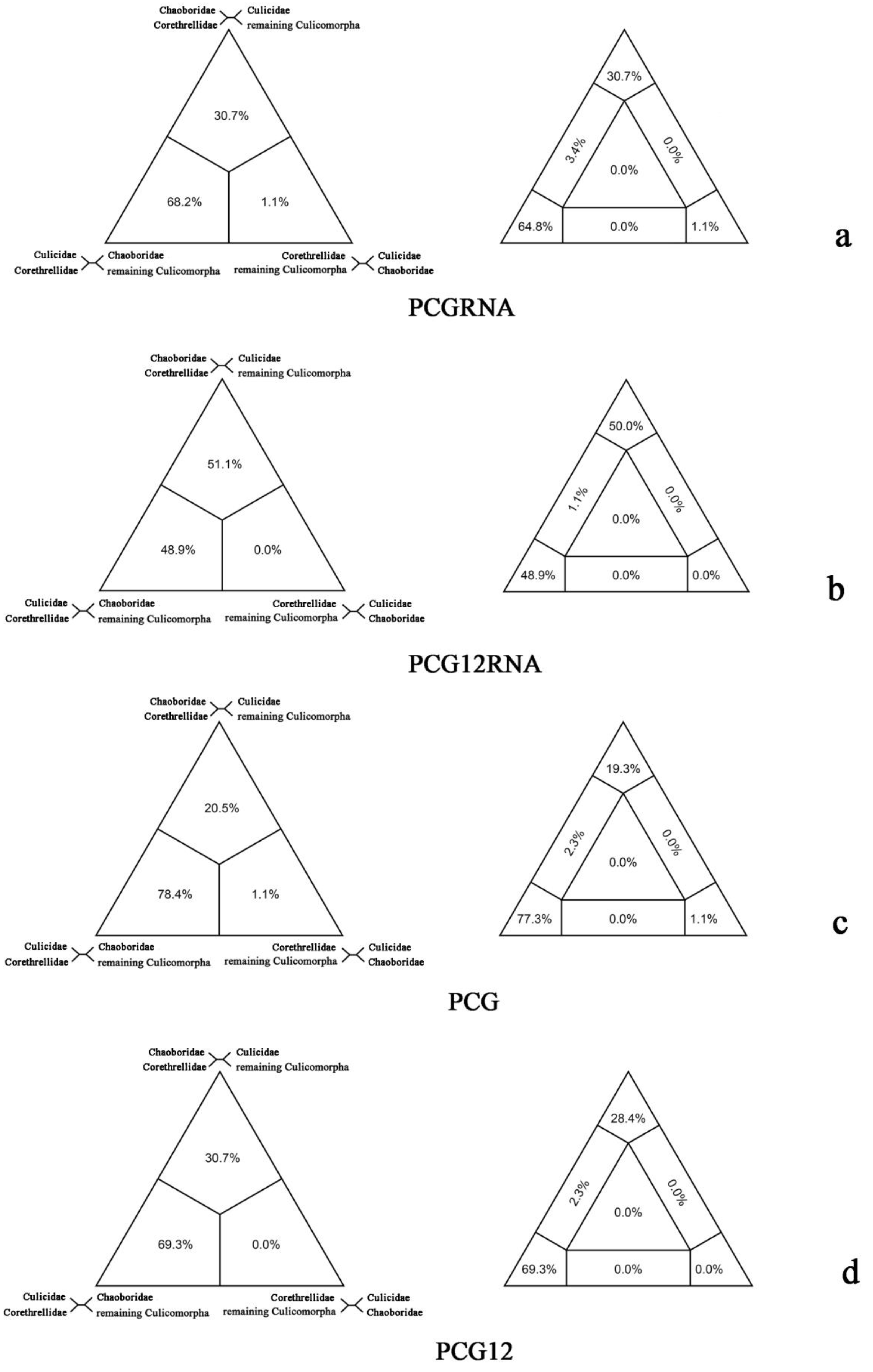
2.2. FcLM Analysis
3. Discussion
4. Materials and Methods
4.1. Taxon Sampling
4.2. DNA Extraction, PCR and Sequencing
4.3. Sequence Annotation
4.4. Dataset Concatenation and Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wood, D.M.; Borkent, A. Phylogeny and classification of the Nematocera; Research Branch Agriculture Canada: Ottawa, ON, Canada, 1989; Volume 3, pp. 1333–1370. [Google Scholar]
- Lane, R.P.; Crosskey, R.W. Medical Insects and Arachnids; Chapman & Hall: London, UK, 1993; p. 723. [Google Scholar]
- Gullan, P.J.; Cranston, P.S. Insects: An Outline of Entomology, 5th ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2014. [Google Scholar]
- Mullen, G.; Durden, L. Medical and veterinary entomology; Academic Press: London, UK, 2002. [Google Scholar]
- Hennig, W. Diptera (Zweiflügler). Handbuch der Zoologie (Berlin) 1973, 4, 1–200. [Google Scholar]
- Oosterbroek, P.; Courtney, G.W. Phylogeny of the nematocerous families of Diptera (Insecta). Zool. J. Linn. Soc. 1995, 115, 267–311. [Google Scholar] [CrossRef]
- Hackman, W.; Väisänen, R. Different classification systems in the Diptera. Ann. Zool. Fenn. 1982, 19, 209–219. [Google Scholar]
- Govalev, K.V. Geological history and the systematic position of the family Thaumaleidae (Diptera). Ent. Obozr. 1989, 69, 798–808. [Google Scholar]
- Kovalev, V.G. Flies. Muscidae; Trudy Paleontologicheskogo Instituta, 1990; Volume 239, pp. 123–177. [Google Scholar]
- Krzeminska, E.; Blagoderov, V.; Krzeminski, W. Elliidae, a new fossil family of the infraorder Axymyiomorpha (Diptera). Acta Zool. Cracov. 1993, 35, 581–591. [Google Scholar]
- Sæther, O.A. Phylogeny of Culicomorpha (Diptera). Syst. Entomol. 2000, 25, 223–234. [Google Scholar] [CrossRef]
- Borkent, A. The pupae of culicomorpha-morphology and a new phylogenetic tree. Zootaxa 2012, 3396, 1–98. [Google Scholar]
- Pawlowski, J.; Szadziewski, R.; Kmieciak, D.; Fahrni, J.; Bittar, G. Phylogeny of the infraorder Culicomorpha (Diptera: Nematocera) based on 28S RNA gene sequences. Syst. Entomol. 1996, 21, 167–178. [Google Scholar] [CrossRef]
- Miller, B.R.; Crabtree, M.B.; Savage, H.M. Phylogenetic relationships of the Culicomorpha inferred from 18S and 5.8S ribosomal DNA sequences (Diptera: Nematocera). Insect. Mol. Biol. 1997, 6, 105–114. [Google Scholar] [CrossRef]
- Wiegmann, B.M.; Trautwein, M.D.; Winkler, I.S.; Barr, N.B.; Kim, J.; Lambkin, C.; Bertone, M.A.; Cassel, B.K.; Bayless, K.M.; Heimberg, A.M.; et al. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. USA 2011, 108, 5690–5695. [Google Scholar] [CrossRef]
- Bertone, M.A.; Courtney, G.W.; Wiegmann, B.M. Phylogenetics and temporal diversification of the earliest true flies (Insecta: Diptera) based on multiple nuclear genes. Syst. Entomol. 2008, 33, 668–687. [Google Scholar] [CrossRef]
- Kutty, S.N.; Wong, W.H.; Meusemann, K.; Meier, R.; Cranston, P.S. A phylogenomic analysis of Culicomorpha (Diptera) resolves the relationships among the eight constituent families. Syst. Entomol. 2018, 43, 434–446. [Google Scholar] [CrossRef]
- Beckenbach, A.T.; Borkent, A. Molecular analysis of the biting midges (Diptera: Ceratopogonidae), based on mitochondrial cytochrome oxidase subunit 2. Mol. Phylogenet. Evol. 2003, 27, 21–35. [Google Scholar] [CrossRef]
- Beckenbach, A.T. Mitochondrial Genome Sequences of Nematocera (Lower Diptera): Evidence of Rearrangement following a Complete Genome Duplication in a Winter Crane Fly. Genome Biol. Evol. 2011, 4, 89–101. [Google Scholar] [CrossRef]
- Li, H.; Shao, R.; Song, N.; Song, F.; Jiang, P.; Li, Z.; Cai, W. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 2015, 5, 8527. [Google Scholar] [CrossRef] [PubMed]
- Caravas, J.; Friedrich, M. Shaking the Diptera tree of life: Performance analysis of nuclear and mitochondrial sequence data partitions. Syst. Entomol. 2013, 38, 93–103. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Winterton, S.L.; Yan, Y.; Aspöck, U.; Aspöck, H.; Yang, D. Mitochondrial phylogenomics illuminates the evolutionary history of Neuropterida. Cladistics 2017, 33, 617–636. [Google Scholar] [CrossRef]
- Li, H.; Shao, R.; Song, F.; Zhou, X.; Yang, Q.; Li, Z.; Cai, W. Mitochondrial genomes of two Barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): Contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLoS ONE 2013, 8, e61685. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Winterton, S.L.; Yang, D. The First Mitochondrial Genome for the Fishfly Subfamily Chauliodinae and Implications for the Higher Phylogeny of Megaloptera. PLoS ONE 2012, 7, e47302. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, M.J.; Vogler, A.P. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol. Phylogenet. Evol. 2012, 63, 299–304. [Google Scholar] [CrossRef]
- Siddall, M.E.; Whiting, M.F. Long-Branch Abstractions. Cladistics 1999, 15, 9–24. [Google Scholar] [CrossRef]
- Moulton, J.K. Molecular sequence data resolves basal divergences within Simuliidae (Diptera). Syst. Entomol. 2000, 25, 95–113. [Google Scholar] [CrossRef]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Cameron, S.L. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014, 39, 400–411. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic. Acids. Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.; Guindon, S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.; Wilgenbusch, J.C.; Warren, D.L.; Swofford, D.L. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 2008, 24, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Strimmer, K.; Haeseler, A.V. Likelihood-mapping: A simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 1997, 94, 6815–6819. [Google Scholar] [CrossRef]
- Schmidt, H.A.; Strimmer, K.; Vingron, M.; Haeseler, A.V. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 2002, 18, 502–504. [Google Scholar] [CrossRef] [PubMed]

| Infraorders | Family | Subfamily | Species | GenBank |
|---|---|---|---|---|
| Culicomorpha | Culicidae | Anophelinae | Anopheles darlingi | NC_014275 |
| Anopheles quadrimaculatus | NC_000875 | |||
| Culicinae | Aedes albopitus | NC_006817 | ||
| Aedes aegypti | NC_035159 | |||
| Culex quinquefasciatus | NC_014574 | |||
| Culex tritaeniorhynchus | NC_028616 | |||
| Haemagogus janthinomys | NC_028025.1 | |||
| Ochlerotatus vigilax | NC_027494 | |||
| Chironomidae | Chironominae | Chironomus tepperi | NC_016167 | |
| Polypedilum vanderplanki | NC_028015.1 | |||
| Podonominae | Parochlus steinenii | KT003702 | ||
| Simuliidae | Simuliinae | Simulium quinquestriatum * | MK281358 | |
| Simulium aureohirtum | KP793690.1 | |||
| Simulium variegatum | NC_033348.1 | |||
| Dixidae | – | Dixella aestivalis | NC_029354.1 | |
| – | Dixella sp. | KM245574 | ||
| Ceratopogonidae | Ceratopogoninae | Culicoides arakawae | NC_009809 | |
| Chaoboridae | Chaoborinae | Chaoborus sp. * | MK281356 | |
| Corethrellidae | – | Corethrella condita* | MK281357 | |
| Thaumaleidae | – | Thaumalea sp. * | MK281359 | |
| Psychodomorpha | Ptychopteridae | Ptychopterinae | Ptychoptera sp. | NC_016201 |
| Bittacomorphinae | Bittacomorphella fenderiana | JN_861745 |
| Phylogenetic Questions | Groups | Number of Species |
|---|---|---|
| What is the relationships of Ceratopogonidae, Thaumaleidae and Simuliidae? | G1(a): Ceratopogonidae | 1 |
| G2(b): Thaumaleidae | 1 | |
| G3(c): Simuliidae | 3 | |
| G4(d): remaining Culicomorpha | 15 | |
| Which family is the sister-group of the branch (Chaoboridae + (Corethrellidae + Culicidae)), Dixidae or Chironomidae? | G1(a): Dixidae | 2 |
| G2(b): Chironomidae | 3 | |
| G3(c): Chaoboridae + (Corethrellidae + Culicidae) | 10 | |
| G4(d): Thaumaleidae + Simuliidae | 4 | |
| What is the relationships of Corethrellidae, Chaoboridae and Culicidae? | G1(a): Chaoboridae | 1 |
| G2(b): Corethrellidae | 1 | |
| G3(c): Culicidae | 8 | |
| G4(d): remaining Culicomorpha | 10 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Kang, Z.; Ding, S.; Wang, Y.; Borkent, C.; Saigusa, T.; Yang, D. Mitochondrial Genomes Provide Insights into the Phylogeny of Culicomorpha (Insecta: Diptera). Int. J. Mol. Sci. 2019, 20, 747. https://doi.org/10.3390/ijms20030747
Zhang X, Kang Z, Ding S, Wang Y, Borkent C, Saigusa T, Yang D. Mitochondrial Genomes Provide Insights into the Phylogeny of Culicomorpha (Insecta: Diptera). International Journal of Molecular Sciences. 2019; 20(3):747. https://doi.org/10.3390/ijms20030747
Chicago/Turabian StyleZhang, Xiao, Zehui Kang, Shuangmei Ding, Yuyu Wang, Chris Borkent, Toyohei Saigusa, and Ding Yang. 2019. "Mitochondrial Genomes Provide Insights into the Phylogeny of Culicomorpha (Insecta: Diptera)" International Journal of Molecular Sciences 20, no. 3: 747. https://doi.org/10.3390/ijms20030747
APA StyleZhang, X., Kang, Z., Ding, S., Wang, Y., Borkent, C., Saigusa, T., & Yang, D. (2019). Mitochondrial Genomes Provide Insights into the Phylogeny of Culicomorpha (Insecta: Diptera). International Journal of Molecular Sciences, 20(3), 747. https://doi.org/10.3390/ijms20030747






