Comparative Proteomic Analysis of Pleurotus ostreatus Reveals Great Metabolic Differences in the Cap and Stipe Development and the Potential Role of Ca2+ in the Primordium Differentiation
Abstract
1. Introduction
2. Results
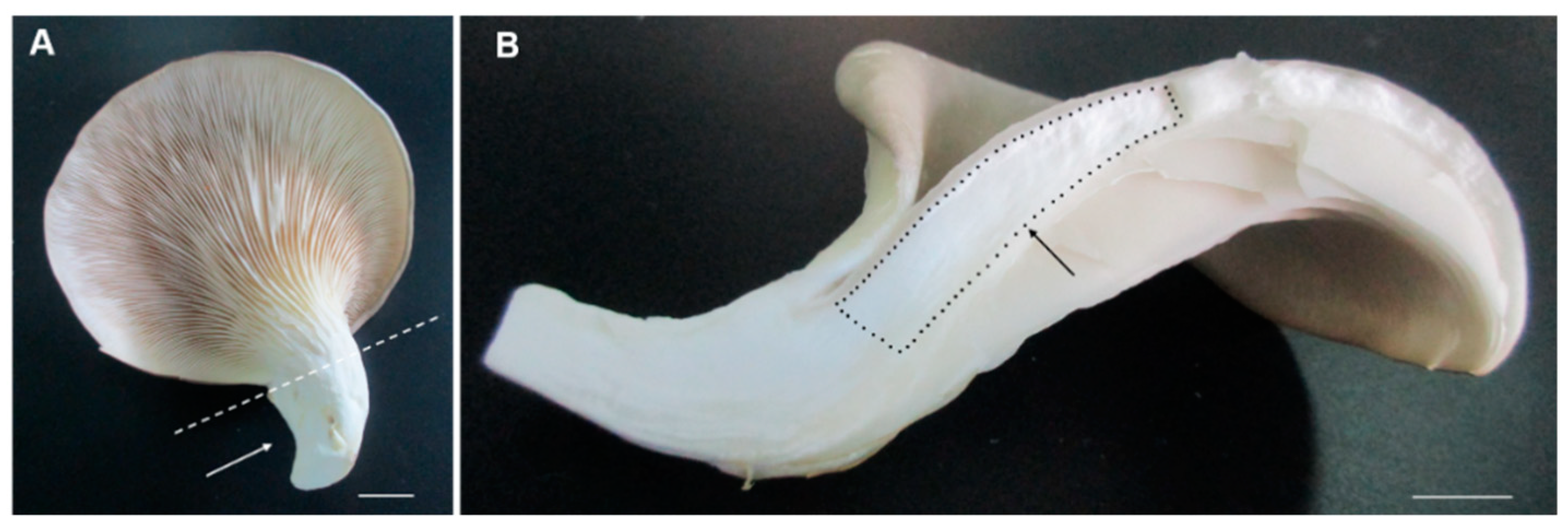
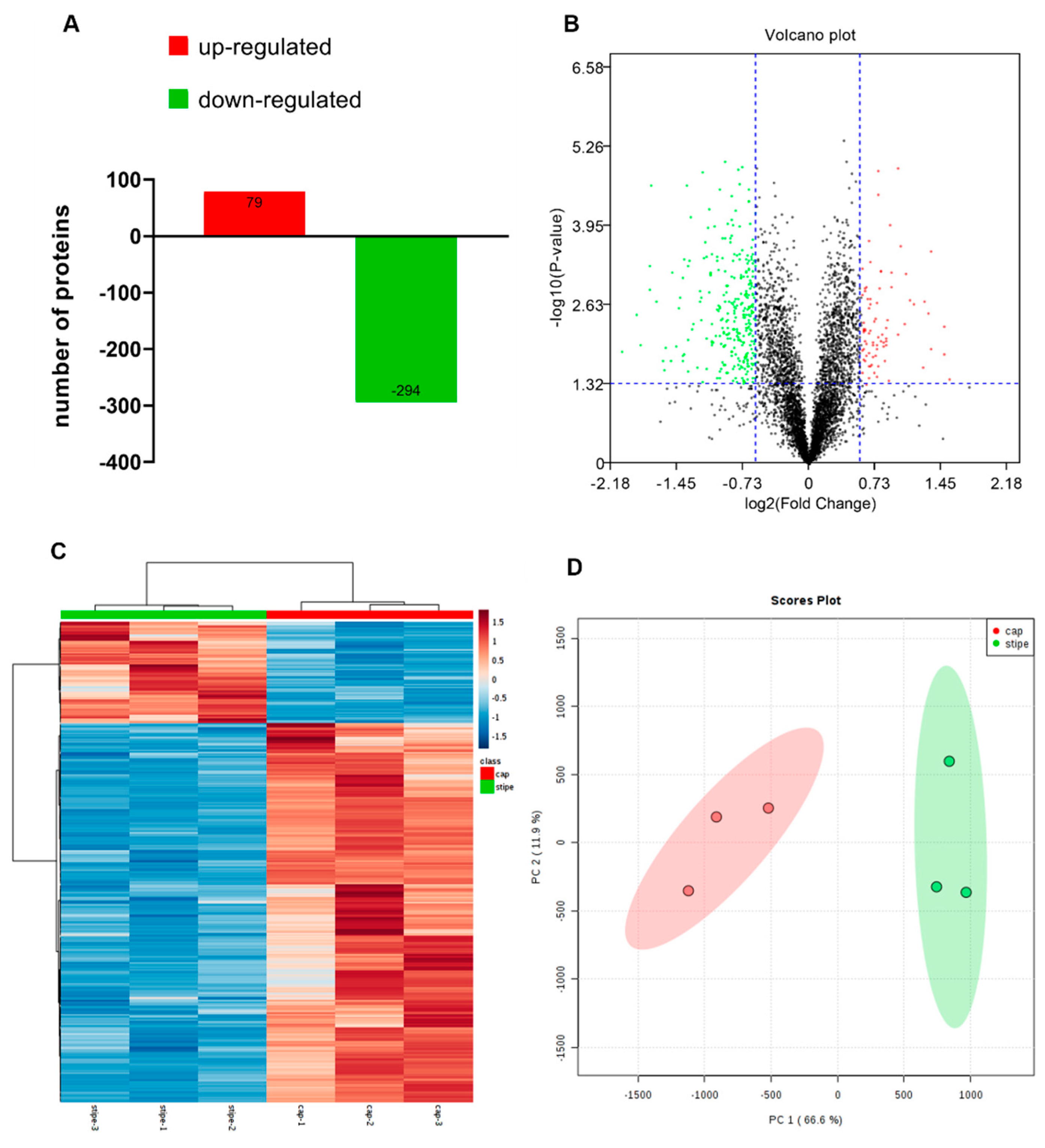
2.1. Isobaric Labeled Quantitative Proteomics Enabled the High-Throughput Proteomic Analysis of the Pleurotus ostreatus Fruiting Body
2.2. GO Analysis Revealed Great Differences in Membrane Part and Catalytic Activity
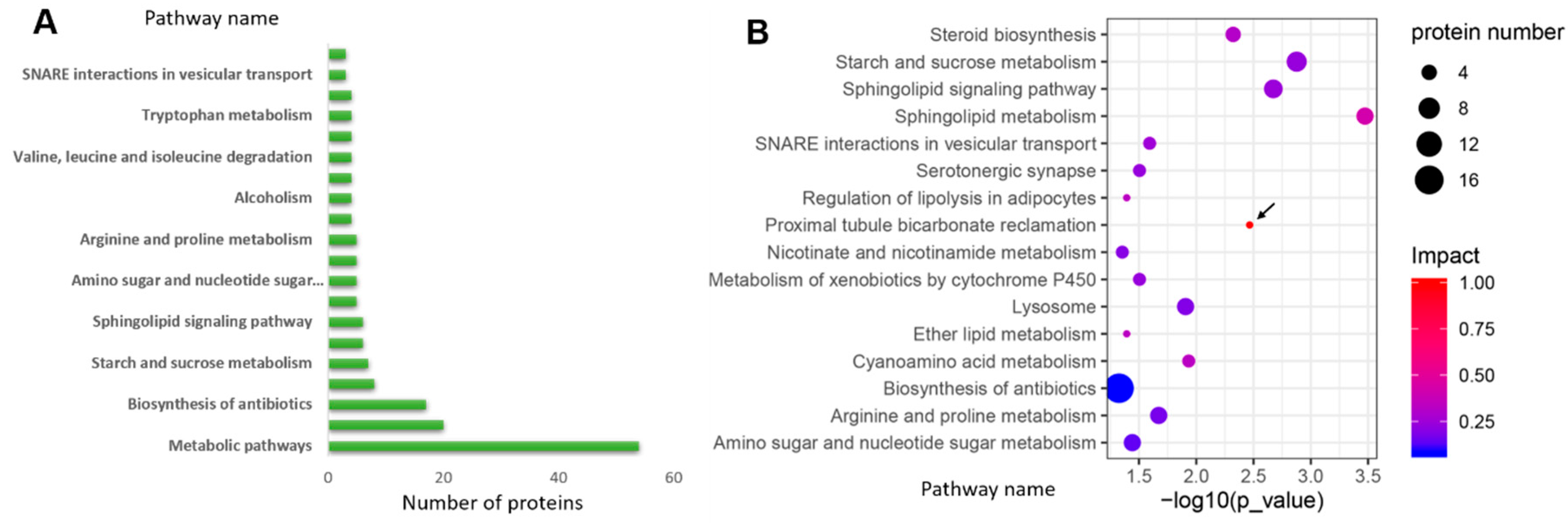
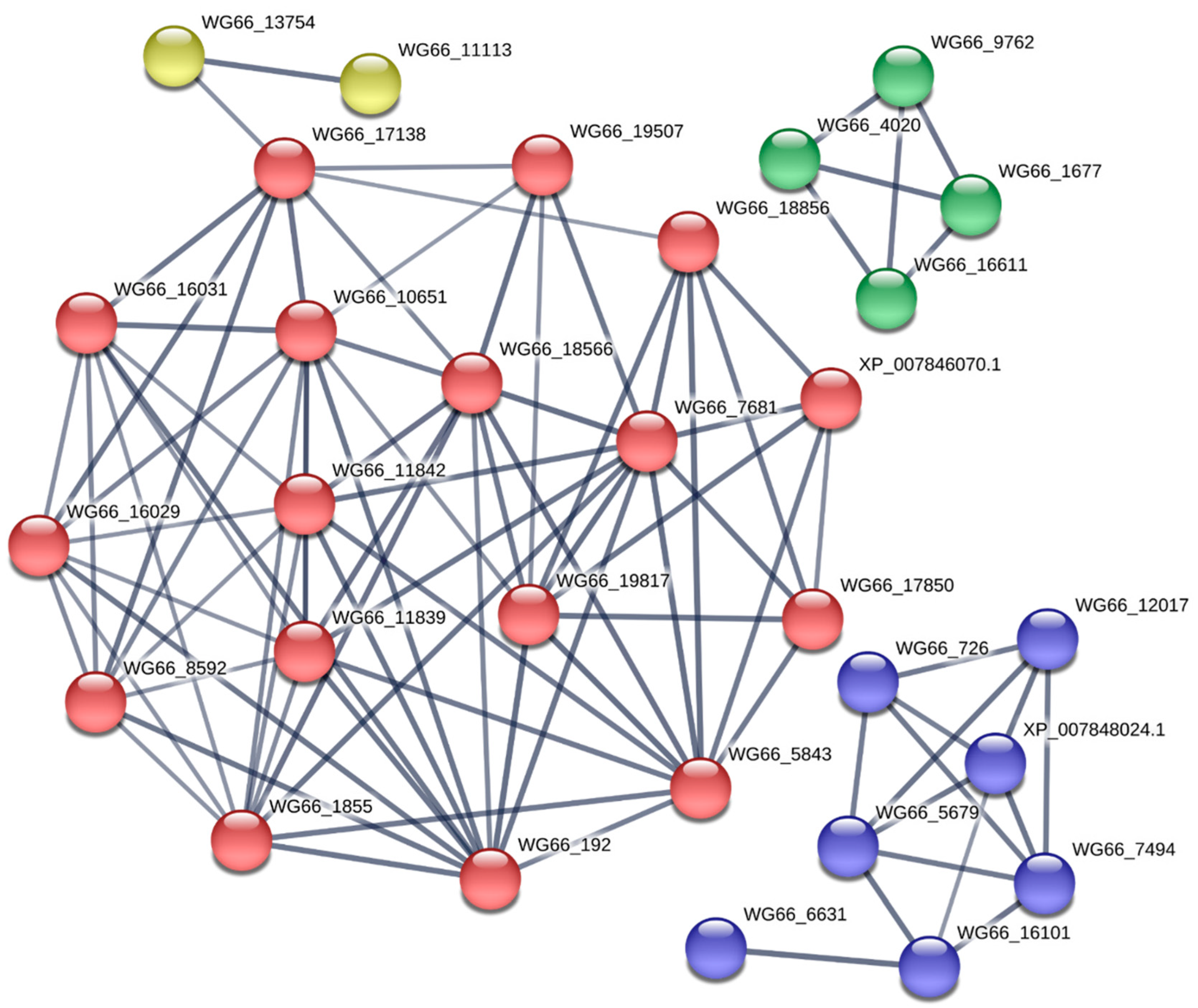
2.3. KEGG and Protein–Protein Interaction Analysis Revealed the Significant Metabolic Differences Between the Cap and Stipe
2.4. Quantitative Real-Time PCR Validation of the Expression of the DEPs
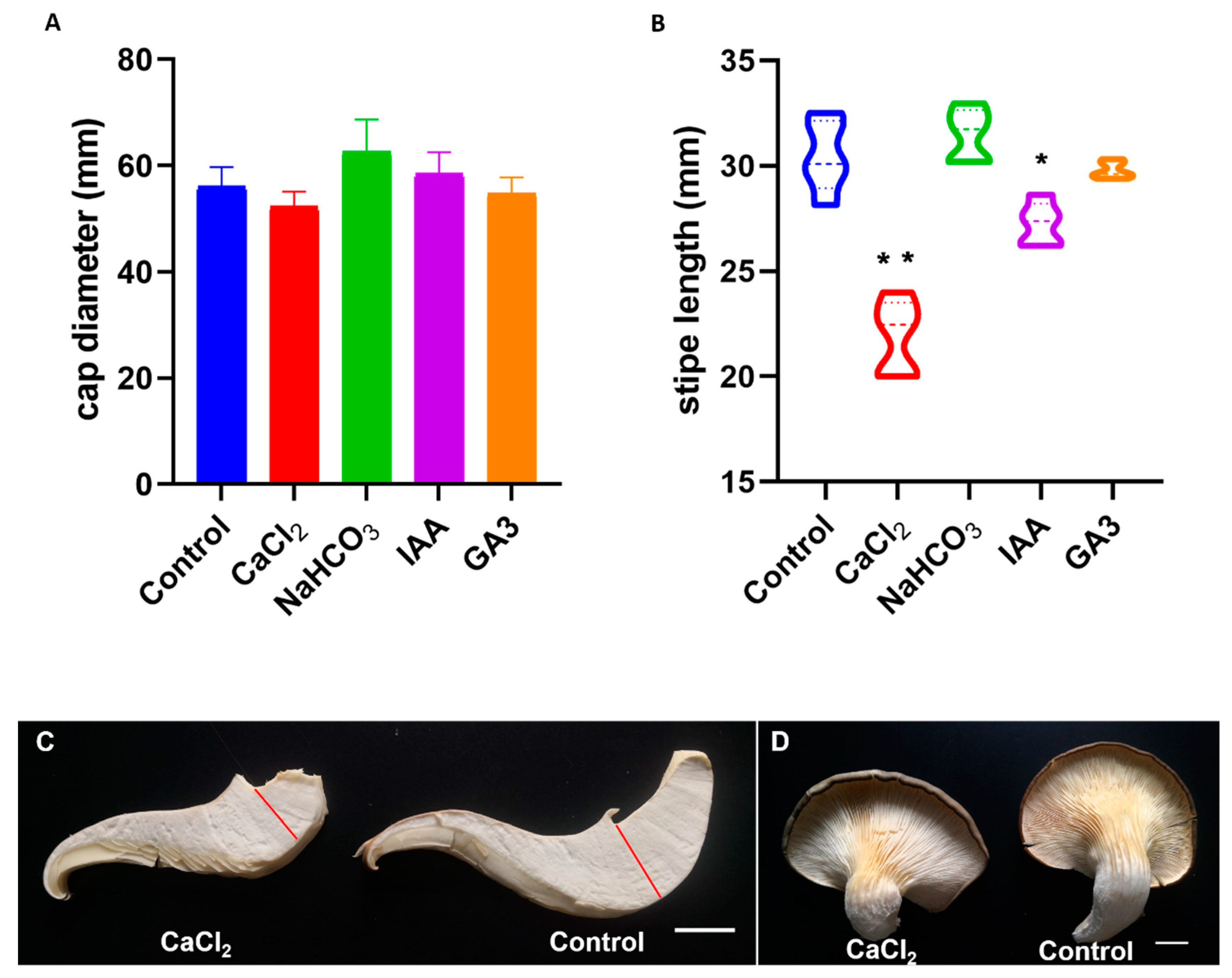
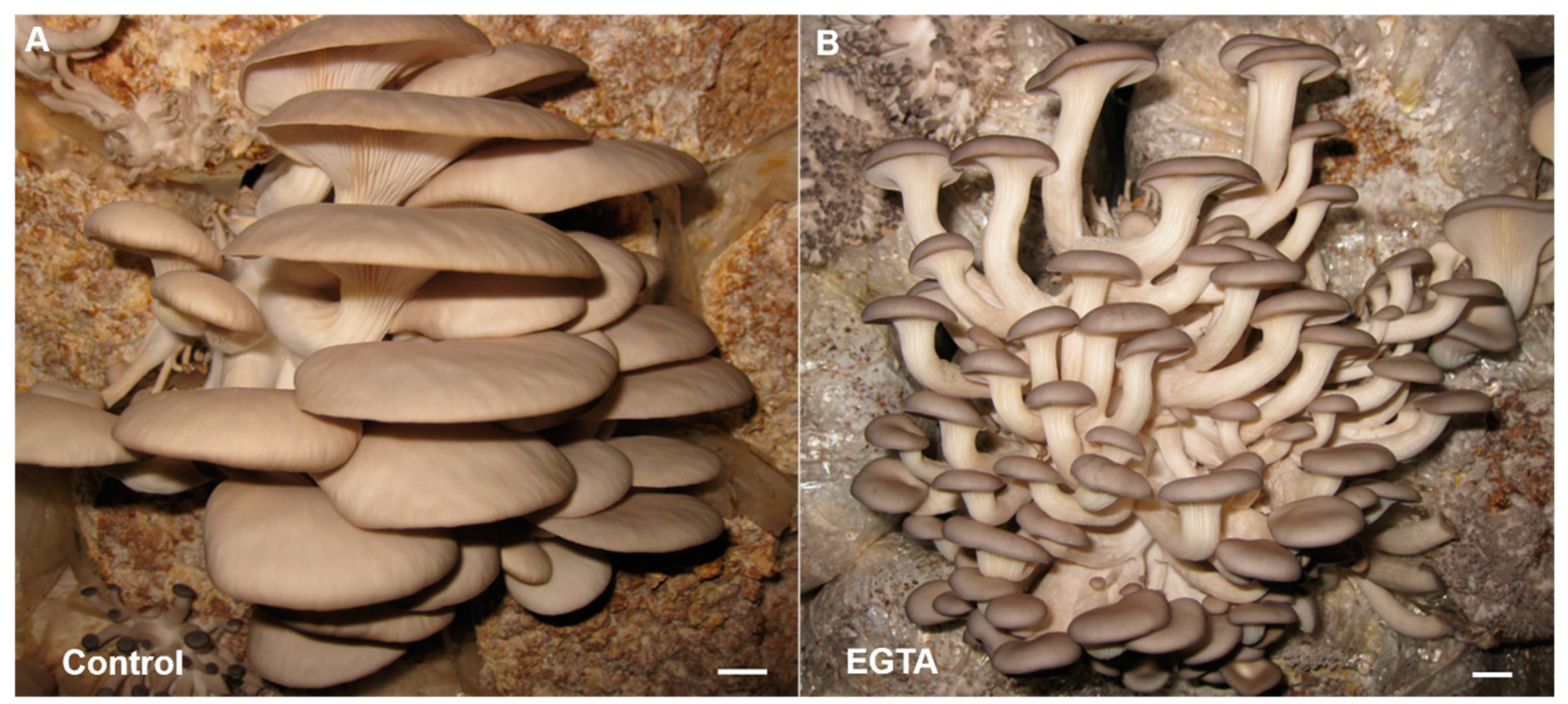
2.5. Ca2+ Plays a Regulatory Role in the Primordium Differentiation
3. Discussion
3.1. Potential Mechanism of Ca2+ on Inhibiting the Stipe Growth and Differentiation
3.2. Sphingolipids Are Indispensable Signaling Mediators in Regulating the Cap and Stipe Differentiation
4. Materials and Methods
4.1. Fungal and Culture Conditions
4.2. Chemical Inducers Application
4.3. Protein Extraction, Digestion, iTRAQ Labeling, and High pH Fractionation
4.4. LC–MS/MS Analysis
4.5. Data Analysis
4.6. Bioinformatics Analysis
4.7. RNA Extraction and Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sanchez, C. Cultivation of Pleurotus ostreatus and other edible mushrooms. Appl. Microbiol. Biotechnol. 2010, 85, 1321–1337. [Google Scholar] [CrossRef] [PubMed]
- Alam, N.; Amin, R.; Khan, A.; Ara, I.; Shim, M.J.; Lee, M.W.; Lee, T.S. Nutritional Analysis of Cultivated Mushrooms in Bangladesh-Pleurotus ostreatus, Pleurotus sajor-caju, Pleurotus floridaand Calocybe indica. Mycobiology 2008, 36, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y. Influences of environmental factors on fruiting body induction, development and maturation in mushroom-forming fungi. Fungal Biol. Rev. 2018, 32, 236–248. [Google Scholar] [CrossRef]
- Kues, U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 2000, 64, 316–353. [Google Scholar] [CrossRef]
- Kamada, T.; Sano, H.; Nakazawa, T.; Nakahori, K. Regulation of fruiting body photomorphogenesis in Coprinopsis cinerea. Fungal Genet. Biol. 2010, 47, 917–921. [Google Scholar] [CrossRef]
- Kuratani, M.; Tanaka, K.; Terashima, K.; Muraguchi, H.; Nakazawa, T.; Nakahori, K.; Kamada, T. The dst2 gene essential for photomorphogenesis of Coprinopsis cinerea encodes a protein with a putative FAD-binding-4 domain. Fungal Genet. Biol. 2010, 47, 152–158. [Google Scholar] [CrossRef]
- Fang, H.; Zhang, W.; Niu, X.; Liu, Z.; Lu, C.; Wei, H.; Yuan, S. Stipe wall extension of Flammulina velutipes could be induced by an expansin-like protein from Helix aspersa. Fungal Biol. 2014, 118, 1–11. [Google Scholar] [CrossRef]
- Kamada, T.; Takemaru, T.; Prosser, J.I.; Gooday, G.W. Right and left handed helicity of chitin microfibrils in stipe cells inCoprinus cinereus. Protoplasma 1991, 165, 64–70. [Google Scholar] [CrossRef]
- Niu, X.; Liu, Z.; Zhou, Y.; Wang, J.; Zhang, W.; Yuan, S. Stipe cell wall architecture varies with the stipe elongation of the mushroom Coprinopsis cinerea. Fungal Biol. 2015, 119, 946–956. [Google Scholar] [CrossRef]
- Money, N.P.; Ravishankar, J.P. Biomechanics of stipe elongation in the basidiomycete Coprinopsis cinerea. Mycol. Res. 2005, 109, 627–634. [Google Scholar] [CrossRef]
- Krizsan, K.; Almasi, E.; Merenyi, Z.; Sahu, N.; Viragh, M.; Koszo, T.; Mondo, S.; Kiss, B.; Balint, B.; Kues, U.; et al. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc. Natl. Acad. Sci. USA 2019, 116, 7409–7418. [Google Scholar] [CrossRef] [PubMed]
- Sipos, G.; Prasanna, A.N.; Walter, M.C.; O’Connor, E.; Balint, B.; Krizsan, K.; Kiss, B.; Hess, J.; Varga, T.; Slot, J.; et al. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat. Ecol. Evol. 2017, 1, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Almasi, E.; Sahu, N.; Krizsan, K.; Balint, B.; Kovacs, G.M.; Kiss, B.; Cseklye, J.; Drula, E.; Henrissat, B.; Nagy, I.; et al. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. N. Phytol. 2019, 224, 902–915. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Salamov, A.A.; Brown, D.W.; Nagy, L.G.; Floudas, D.; Held, B.W.; Levasseur, A.; Lombard, V.; Morin, E.; Otillar, R.; et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. USA 2014, 111, 9923–9928. [Google Scholar] [CrossRef]
- Floudas, D.; Held, B.W.; Riley, R.; Nagy, L.G.; Koehler, G.; Ransdell, A.S.; Younus, H.; Chow, J.; Chiniquy, J.; Lipzen, A.; et al. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet. Biol. 2015, 76, 78–92. [Google Scholar] [CrossRef]
- Kiss, E.; Hegedus, B.; Viragh, M.; Varga, T.; Merenyi, Z.; Koszo, T.; Balint, B.; Prasanna, A.N.; Krizsan, K.; Kocsube, S.; et al. Comparative genomics reveals the origin of fungal hyphae and multicellularity. Nat. Commun. 2019, 10, 4080. [Google Scholar] [CrossRef]
- Varga, T.; Krizsan, K.; Foldi, C.; Dima, B.; Sanchez-Garcia, M.; Sanchez-Ramirez, S.; Szollosi, G.J.; Szarkandi, J.G.; Papp, V.; Albert, L.; et al. Megaphylogeny resolves global patterns of mushroom evolution. Nat. Ecol. Evol. 2019, 3, 668–678. [Google Scholar] [CrossRef]
- Nagy, L.G.; Szollosi, G. Fungal Phylogeny in the Age of Genomics: Insights Into Phylogenetic Inference From Genome-Scale Datasets. Adv. Genet. 2017, 100, 49–72. [Google Scholar] [CrossRef]
- Chen, B.; van Peer, A.F.; Yan, J.; Li, X.; Xie, B.; Miao, J.; Huang, Q.; Zhang, L.; Wang, W.; Fu, J.; et al. Fruiting Body Formation in Volvariella volvacea Can Occur Independently of Its MAT-A-Controlled Bipolar Mating System, Enabling Homothallic and Heterothallic Life Cycles. G3 (Bethesda) 2016, 6, 2135–2146. [Google Scholar] [CrossRef]
- Gehrmann, T.; Pelkmans, J.F.; Ohm, R.A.; Vos, A.M.; Sonnenberg, A.S.M.; Baars, J.J.P.; Wosten, H.A.B.; Reinders, M.J.T.; Abeel, T. Nucleus-specific expression in the multinuclear mushroom-forming fungus Agaricus bisporus reveals different nuclear regulatory programs. Proc. Natl. Acad. Sci. USA 2018, 115, 4429–4434. [Google Scholar] [CrossRef]
- Ohm, R.A.; de Jong, J.F.; de Bekker, C.; Wosten, H.A.; Lugones, L.G. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol. Microbiol. 2011, 81, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; van Peer, A.F.; Chen, B.; Chen, Z.; Zhu, J.; Deng, Y.; Jiang, Y.; Li, S.; Wu, T.; Xie, B. Gene expression profiling reveals large regulatory switches between succeeding stipe stages in Volvariella volvacea. PLoS ONE 2014, 9, e97789. [Google Scholar] [CrossRef] [PubMed]
- Arima, T.; Yamamoto, M.; Hirata, A.; Kawano, S.; Kamada, T. The eln3 gene involved in fruiting body morphogenesis of Coprinus cinereus encodes a putative membrane protein with a general glycosyltransferase domain. Fungal Genet. Biol. 2004, 41, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, F.; Hon, C.C.; Zhang, Y.; Leung, F.C. The mitochondrial genome of the Basidiomycete fungus Pleurotus ostreatus (oyster mushroom). FEMS Microbiol. Lett. 2008, 280, 34–41. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qu, J.; Zhao, M.; Hsiang, T.; Feng, X.; Zhang, J.; Huang, C. Identification and Characterization of Small Noncoding RNAs in Genome Sequences of the Edible Fungus Pleurotus ostreatus. Biomed. Res. Int. 2016, 2016, 2503023. [Google Scholar] [CrossRef] [PubMed]
- Piscitelli, A.; Tarallo, V.; Guarino, L.; Sannia, G.; Birolo, L.; Pezzella, C. New lipases by mining of Pleurotus ostreatus genome. PLoS ONE 2017, 12, e0185377. [Google Scholar] [CrossRef]
- Lei, M.; Wu, X.; Zhang, J.; Wang, H.; Huang, C. Gene cloning, expression, and characterization of trehalose-6-phosphate synthase from Pleurotus ostreatus. J. Basic Microbiol. 2017, 57, 580–589. [Google Scholar] [CrossRef]
- Ohm, R.A.; de Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; de Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E.; et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 28, 957–963. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Nakade, K.; Sato, S.; Yoshida, K.; Miyazaki, K.; Natsume, S.; Konno, N. Lentinula edodes Genome Survey and Postharvest Transcriptome Analysis. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Lyons, J.M.; Karin, N.J. A role for G protein-coupled lysophospholipid receptors in sphingolipid-induced Ca2+ signaling in MC3T3-E1 osteoblastic cells. J. Bone Miner. Res. 2001, 16, 2035–2042. [Google Scholar] [CrossRef]
- Breslow, D.K. Sphingolipid homeostasis in the endoplasmic reticulum and beyond. Cold Spring Harb. Perspect Biol. 2013, 5, a013326. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, J.A.; Kim, T.H.; Li, H.Y.; Shin, K.O.; Lee, Y.M.; Oh, S.; Pewzner-Jung, Y.; Futerman, A.H.; Suh, S.H. Altering sphingolipid composition with aging induces contractile dysfunction of gastric smooth muscle via K(Ca) 1.1 upregulation. Aging Cell 2015, 14, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Kindman, L.A.; Kim, S.; McDonald, T.V.; Gardner, P. Characterization of a novel intracellular sphingolipid-gated Ca2+-permeable channel from rat basophilic leukemia cells. J. Biol. Chem. 1994, 269, 13088–13091. [Google Scholar] [PubMed]
- Foo, E.; Plett, J.M.; Lopez-Raez, J.A.; Reid, D. Editorial: The Role of Plant Hormones in Plant-Microbe Symbioses. Front. Plant. Sci. 2019, 10, 1391. [Google Scholar] [CrossRef]
- Sun, X.; Wang, N.; Li, P.; Jiang, Z.; Liu, X.; Wang, M.; Su, Z.; Zhang, C.; Lin, F.; Liang, Y. Endophytic fungus Falciphora oryzae promotes lateral root growth by producing indole derivatives after sensing plant signals. Plant. Cell Environ. 2019. [Google Scholar] [CrossRef]
- Bastias, D.A.; Alejandra Martinez-Ghersa, M.; Newman, J.A.; Card, S.D.; Mace, W.J.; Gundel, P.E. The plant hormone salicylic acid interacts with the mechanism of anti-herbivory conferred by fungal endophytes in grasses. Plant. Cell Environ. 2018, 41, 395–405. [Google Scholar] [CrossRef]
- Zuccaro, A.; Lahrmann, U.; Langen, G. Broad compatibility in fungal root symbioses. Curr. Opin. Plant. Biol. 2014, 20, 135–145. [Google Scholar] [CrossRef]
- Miller, A.J.; Vogg, G.; Sanders, D. Cytosolic calcium homeostasis in fungi: Roles of plasma membrane transport and intracellular sequestration of calcium. Proc. Natl. Acad. Sci. USA 1990, 87, 9348–9352. [Google Scholar] [CrossRef]
- Dunn, T.; Gable, K.; Beeler, T. Regulation of Cellular Ca2+ by Yeast Vacuoles. J. Biol. Chem. 1994, 269, 7273–7278. [Google Scholar]
- Campbell, A.K. Role of Intracellular Ca2+ in Plants and Fungi. John Wiley & Sons, Ltd. 2014. Available online: https://onlinelibrary.wiley.com/doi/10.1002/9781118675410.ch9 (accessed on 14 December 2019).
- Gururaj, C.; Federman, R.S.; Chang, A. Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J. Biol. Chem. 2013, 288, 20453–20463. [Google Scholar] [CrossRef]
- Liu, M.; Huang, C.; Polu, S.R.; Schneiter, R.; Chang, A. Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast. J. Cell Sci. 2012, 125, 2428–2435. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, L.L.; Toledo, M.S.; Ferreira, F.A.; Straus, A.H.; Takahashi, H.K. Structural diversity and biological significance of glycosphingolipids in pathogenic and opportunistic fungi. Front. Cell Infect. Microbiol. 2014, 4, 138. [Google Scholar] [CrossRef] [PubMed]
- Bothwell, J.H.F.; Ng, C.K.Y. The evolution of Ca2+ signalling in photosynthetic eukaryotes. N. Phytol. 2005, 166, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lakhani, V.; Costa, D.J.; Sharara, A.I.; Fitz, J.G.; Huang, L.W.; Peters, K.G.; Kindman, L.A. Sphingolipid-gated Ca2+ release from intracellular stores of endothelial cells is mediated by a novel Ca2+-permeable channel. J. Biol. Chem. 1995, 270, 5266–5269. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, F.A.; Nixon, G.F. Sphingolipids differentially regulate mitogen-activated protein kinases and intracellular Ca2+ in vascular smooth muscle: Effects on CREB activation. Br. J. Pharmacol. 2010, 147, 351–359. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef]
- Deng, Y.; Pakdel, M.; Blank, B.; Sundberg, E.L.; Burd, C.G.; von Blume, J. Activity of the SPCA1 Calcium Pump Couples Sphingomyelin Synthesis to Sorting of Secretory Proteins in the Trans-Golgi Network. Dev. Cell 2018, 47, 464–478. [Google Scholar] [CrossRef]
- Abbineni, P.S.; Coorssen, J.R. Sphingolipids modulate docking, Ca2+ sensitivity and membrane fusion of native cortical vesicles. Int. J. Biochem. Cell Biol. 2018, 104, 43–54. [Google Scholar] [CrossRef]
- Kajiwara, K.; Muneoka, T.; Watanabe, Y.; Karashima, T.; Kitagaki, H.; Funato, K. Perturbation of sphingolipid metabolism induces endoplasmic reticulum stress-mediated mitochondrial apoptosis in budding yeast. Mol. Microbiol. 2012, 86, 1246–1261. [Google Scholar] [CrossRef]
- Jia, D.; Wang, B.; Li, X.; Peng, W.; Zhou, J.; Tan, H.; Tang, J.; Huang, Z.; Tan, W.; Gan, B.; et al. Proteomic Analysis Revealed the Fruiting-Body Protein Profile of Auricularia polytricha. Curr. Microbiol. 2017, 74, 943–951. [Google Scholar] [CrossRef]
- Castanera, R.; Lopez-Varas, L.; Pisabarro, A.G.; Ramirez, L. Validation of Reference Genes for Transcriptional Analyses in Pleurotus ostreatus by Using Reverse Transcription-Quantitative PCR. Appl. Environ. Microbiol. 2015, 81, 4120–4129. [Google Scholar] [CrossRef] [PubMed]

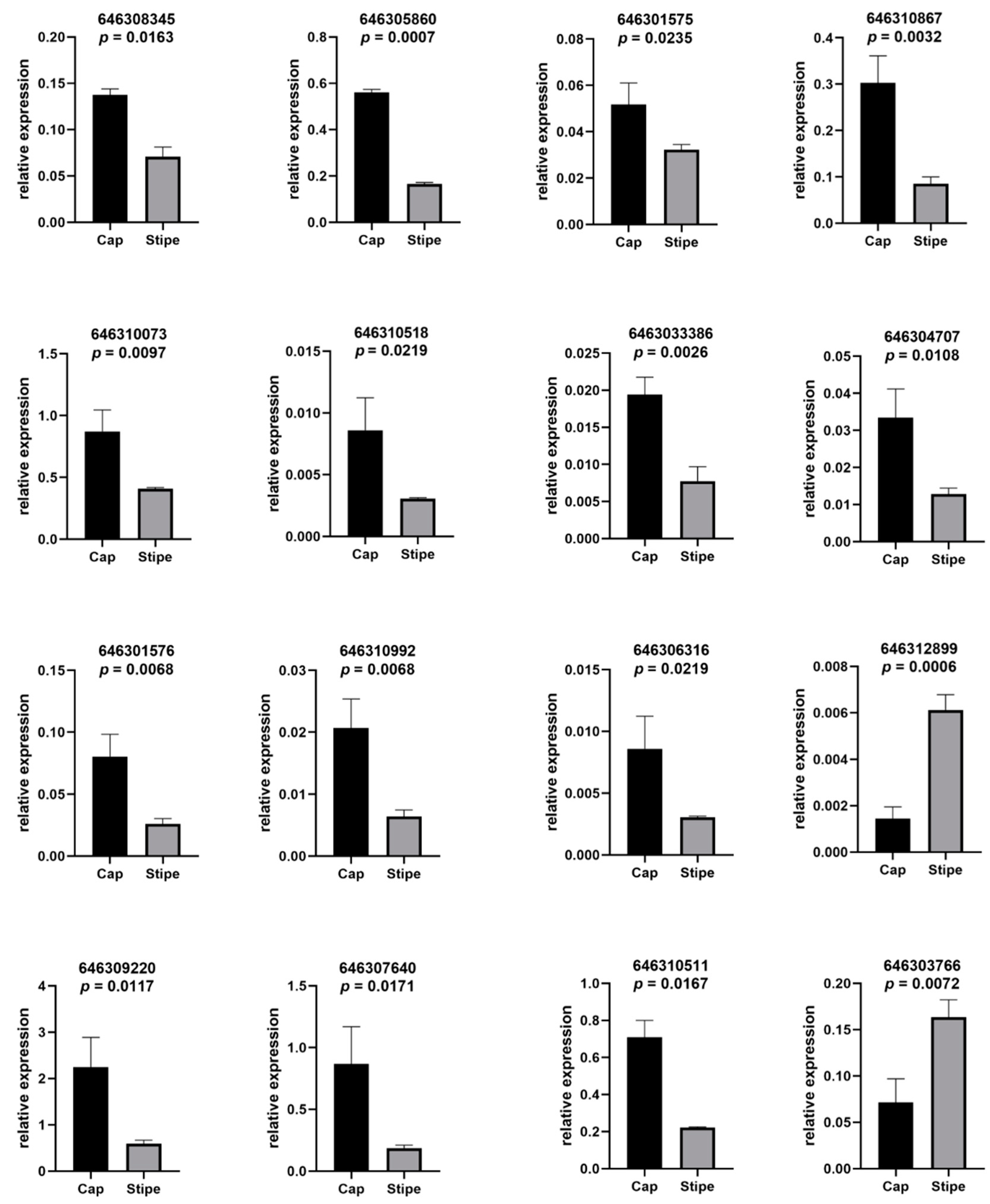
| Pathway Name | Accession | Annotation | Ratio Stipe/Cap | p-Value |
|---|---|---|---|---|
| Sphingolipid metabolism | 646310867 | KYQ37540.1 putative sphingomyelin phosphodiesterase asm-3 | 1.532292106 | 0.019700878 |
| 646310992 | KDQ32134.1 glycoside hydrolase family 27 protein | 0.381533502 | 0.020176311 | |
| 646306316 | KYQ40993.1 Sphingosine-1-phosphate lyase | 0.633097441 | 0.000435157 | |
| 646310518 | KDQ31660.1 glycoside hydrolase family 30 protein | 0.568217459 | 0.008990541 | |
| 646310073 | KYQ45887.1 Inositol phosphosphingolipids phospholipase C | 0.566318538 | 0.002894861 | |
| Starch and sucrose metabolism | 646305860 | KDQ27006.1 glycoside hydrolase family 13 protein | 0.602831197 | 0.039272074 |
| 646308345 | KDQ29489.1 glycoside hydrolase family 13 protein | 0.626457034 | 0.040795714 | |
| 646310601 | KDQ31743.1 glycosyltransferase family 20 protein | 0.524911032 | 0.022925933 | |
| 646302526 | KDQ23675.1 glycosyltransferase family 35 protein | 0.540832049 | 0.007134693 | |
| 646301575 | KDQ22726.1glycosyltransferase family 3 protein | 0.659292035 | 0.022073266 | |
| 646308401 | KDQ29545.1 glycoside hydrolase family 3 protein | 1.530577815 | 0.006173318 | |
| 646310511 | KDQ31653.1 glycoside hydrolase family 3 protein | 0.586462189 | 0.006815785 | |
| Sphingolipid signaling pathway | 646303386 | AAK15758.1 ras-like protein | 0.628664495 | 0.002764913 |
| 646310867 | KYQ37540.1 putative sphingomyelin phosphodiesterase asm-3 | 1.532292106 | 0.019700878 | |
| 646310073 | KYQ45887.1 Inositol phosphosphingolipids phospholipase C | 0.566318538 | 0.002894861 | |
| 646304707 | ESK89222.1 spo14 | 0.651073198 | 0.023107308 | |
| 646301576 | XP_001886453.1 heterotrimeric G-protein alpha subunit, GPA3-like protein | 0.573976915 | 0.003752769 | |
| 646306316 | KYQ40993.1 Sphingosine-1-phosphate lyase | 0.633097441 | 0.000435157 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Hu, J.; Li, Y.; Yang, B.; Guan, Y.; Xu, C.; Chen, F.; Chi, J.; Bao, Y. Comparative Proteomic Analysis of Pleurotus ostreatus Reveals Great Metabolic Differences in the Cap and Stipe Development and the Potential Role of Ca2+ in the Primordium Differentiation. Int. J. Mol. Sci. 2019, 20, 6317. https://doi.org/10.3390/ijms20246317
Zhu W, Hu J, Li Y, Yang B, Guan Y, Xu C, Chen F, Chi J, Bao Y. Comparative Proteomic Analysis of Pleurotus ostreatus Reveals Great Metabolic Differences in the Cap and Stipe Development and the Potential Role of Ca2+ in the Primordium Differentiation. International Journal of Molecular Sciences. 2019; 20(24):6317. https://doi.org/10.3390/ijms20246317
Chicago/Turabian StyleZhu, Weiwei, Jinbo Hu, Yang Li, Bing Yang, Yanli Guan, Chong Xu, Fei Chen, Jingliang Chi, and Yongming Bao. 2019. "Comparative Proteomic Analysis of Pleurotus ostreatus Reveals Great Metabolic Differences in the Cap and Stipe Development and the Potential Role of Ca2+ in the Primordium Differentiation" International Journal of Molecular Sciences 20, no. 24: 6317. https://doi.org/10.3390/ijms20246317
APA StyleZhu, W., Hu, J., Li, Y., Yang, B., Guan, Y., Xu, C., Chen, F., Chi, J., & Bao, Y. (2019). Comparative Proteomic Analysis of Pleurotus ostreatus Reveals Great Metabolic Differences in the Cap and Stipe Development and the Potential Role of Ca2+ in the Primordium Differentiation. International Journal of Molecular Sciences, 20(24), 6317. https://doi.org/10.3390/ijms20246317





