Strategies for Molecular Imprinting and the Evolution of MIP Nanoparticles as Plastic Antibodies—Synthesis and Applications
Abstract
1. Introduction
2. Imprinting Challenges
- (i)
- Difficulty with imprinting of biological macromolecules, which are not soluble in organic solvents that are traditionally used in molecular imprinting. All bulk polymers, especially polymers imprinted with large templates such as proteins, also suffer from slow mass transfer kinetics. Protein recognition is the most important area of bioanalysis and drug development and for these reasons traditional MIPs are not considered as a viable alternative to antibodies.
- (ii)
- Template leakage (bleeding) which affects analytical applications of MIP particles. It is not feasible to use MIP as a biorecognition material in assays and sensors if there is a risk that leaked template can compromise clinical or forensic analysis.
- (iii)
- Heterogeneity of binding sites. Bulk MIPs always have large numbers of non-specific sites which contribute to the “polyclonal” nature of their binding profiles [12,21,22]. High levels of non-specific binding limit the utility of MIPs in diagnostic, pharmaceutical, and separation applications, except in a limited number of special cases where there is no alternative.
2.1. Imprinting of Proteins
2.2. Incomplete Template Removal and Template Leakage
2.3. Heterogeneous and Non-Specific Binding Sites
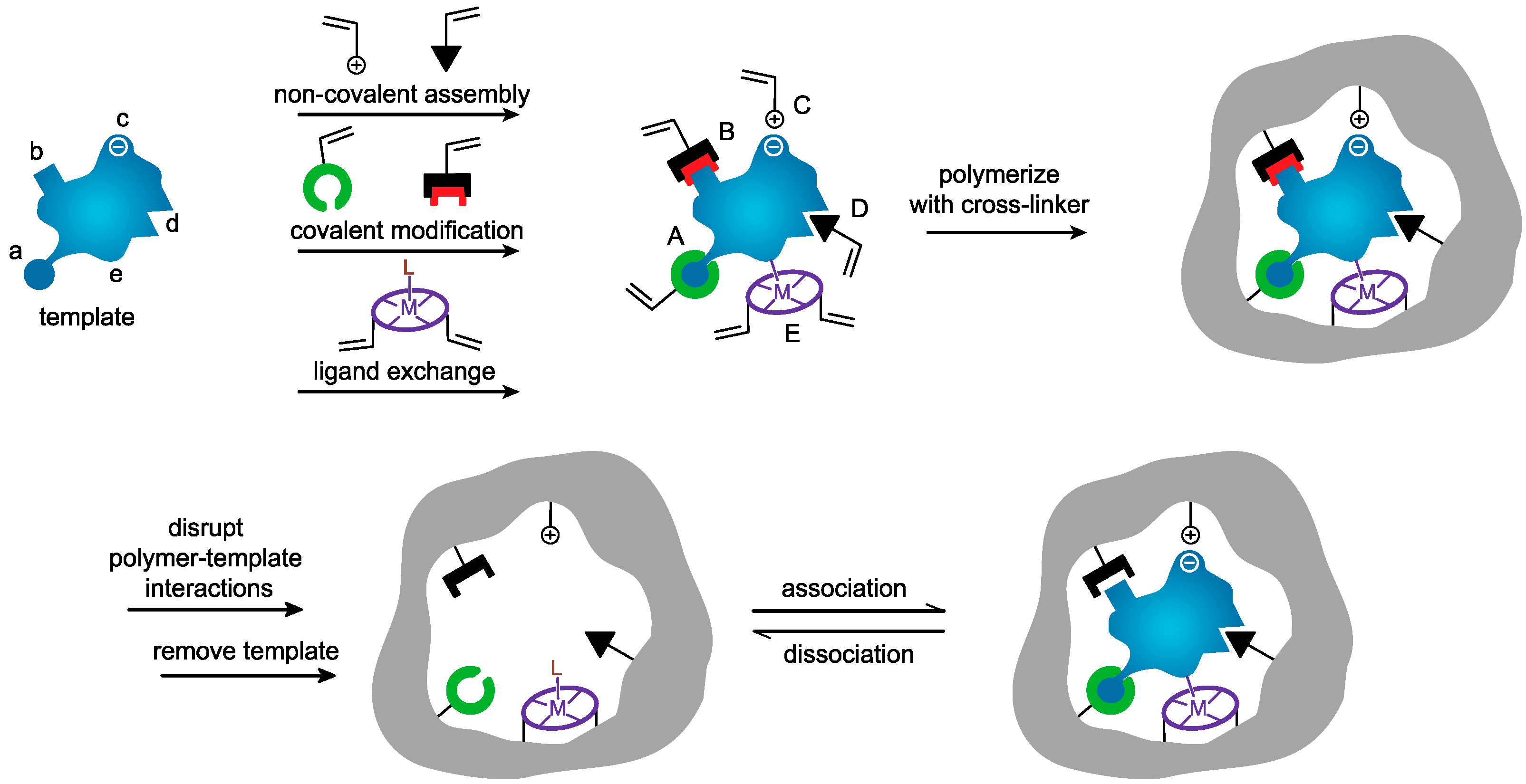
3. Synthesis of MIP Nanoparticles
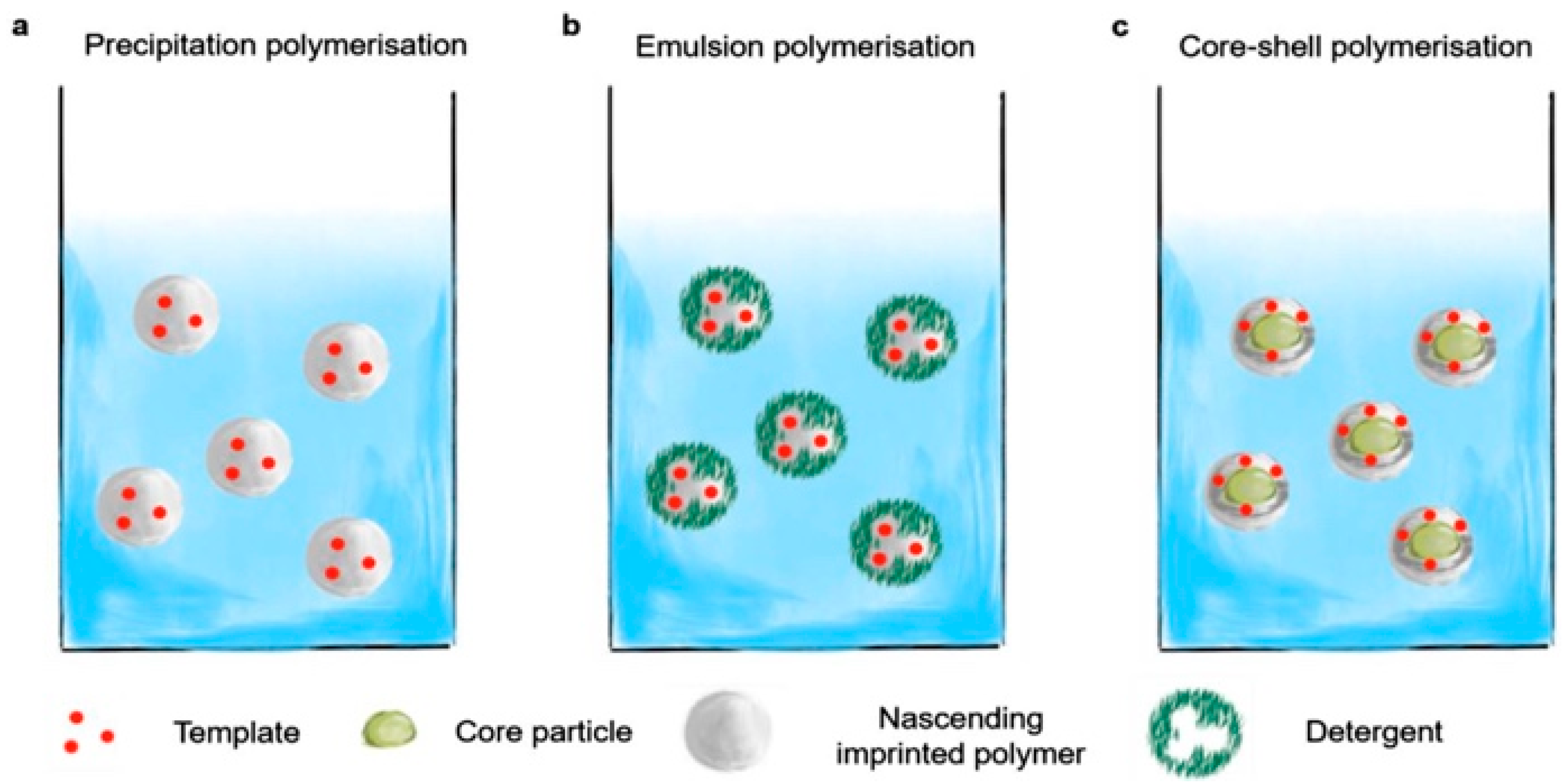
3.1. Precipitation Polymerization
3.2. Emulsion Polymerization
3.3. Core–Shell Grafting and Polymerization
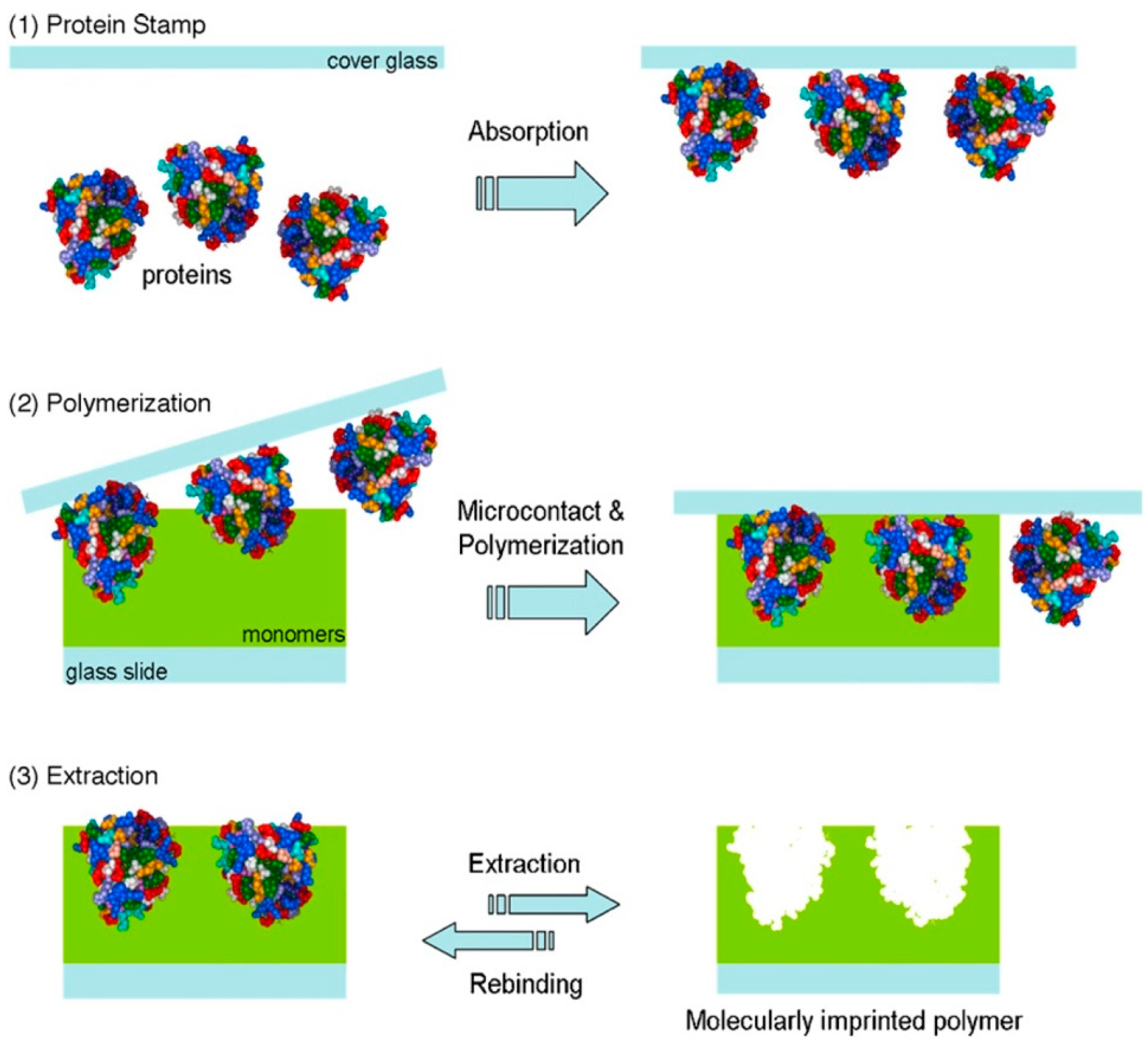
3.4. Solid Phase Imprinting
4. Applications of Nano MIPs
- Size of these nanoparticles is comparable to those of proteins and they have high apparent binding constants.
- NanoMIPs can be stored at room temperature for very long time [117].
- Synthesis of nanoMIPs requires weeks instead of months as in the case of antibodies [92].
- NanoMIPs can pass the cell membrane barrier and be delivered to cell targets orally [30].
- While it has not yet been demonstrated in practice, the production of nanoMIPs has the potential to be more economical than that of antibodies [119].
4.1. NanoMIPs in Separation
4.2. NanoMIPs in Catalysis
4.3. NanoMIPs in Assays and Sensors
4.4. NanoMIPs in Life Science and In Vivo Applications
5. Conclusions and Future Outlook
Funding
Conflicts of Interest
References
- Chen, B.; Piletsky, S.; Turner, A.P.F. Molecular Recognition: Design of “Keys”. Comb. Chem. High Throughput Screen. 2002, 5, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Wackerlig, J.; Schirhagl, R. Applications of molecularly imprinted polymer nanoparticles and their advances toward industrial use: A review. Anal. Chem. 2015, 88, 250–261. [Google Scholar] [CrossRef] [PubMed]
- MIPdatabase. Available online: https://www.mipdatabase.com (accessed on 10 December 2019).
- Ansell, R.J.; Ramstrom, O.; Mosbach, K. Towards artificial antibodies prepared by molecular imprinting. Clin. Chem. 1996, 42, 1506–1512. [Google Scholar] [CrossRef]
- Hussain, M.; Wackerlig, J.; Lieberzeit, P.A. Biomimetic Strategies for Sensing Biological Species. Biosensors 2013, 3, 89–107. [Google Scholar] [CrossRef]
- Svenson, J.; Nicholls, I.A. On the thermal and chemical stability of molecularly imprinted polymers. Anal. Chim. Acta 2001, 435, 19–24. [Google Scholar] [CrossRef]
- Polyakov, M. Adsorption properties and structure of silica gel. Zhurnal Fiz. Khimii 1931, 2, 799–805. [Google Scholar]
- Wulff, G.; Sarhan, A. Über die Anwendung von enzymanalog gebauten Polymeren zur Racemattrennung. Angew. Chem. 1972, 84, 364. [Google Scholar] [CrossRef]
- Arshady, R.; Mosbach, K. Synthesis of substrate-selective polymers by host-guest polymerization. Makromol. Chem. 1981, 182, 687–692. [Google Scholar] [CrossRef]
- Caro, E.; Masqué, N.; Marcé, R.M.; Borrull, F.; Cormack, P.A.; Sherrington, D.C. Non-covalent and semi-covalent molecularly imprinted polymers for selective on-line solid-phase extraction of 4-nitrophenol from water samples. J. Chromatogr. A 2002, 963, 169–178. [Google Scholar] [CrossRef]
- Shah, N.; Ul-Islam, M.; Haneef, M.; Park, J.K. A brief overview of molecularly imprinted polymers: From basics to applications. J. Pharm. Res. 2012, 3309–3317. [Google Scholar]
- Chen, L.; Xu, S.; Li, J. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef]
- Haupt, K. (Ed.) Molecular Imprinting; Springer: Berlin/Heidelberg, Germany, 2012; Volume 325. [Google Scholar]
- Karlsson, B.C.; O’Mahony, J.; Karlsson, J.G.; Bengtsson, H.; Eriksson, L.A.; Nicholls, I.A. Structure and dynamics of monomer-template complexation: An explanation for molecularly imprinted polymer recognition site heterogeneity. J. Am. Chem. Soc. 2009, 131, 13297–13304. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef]
- Alexander, C.; Andersson, H.S.; Andersson, L.I.; Ansell, R.J.; Kirsch, N.; Nicholls, I.A.; O’Mahony, J.; Whitcombe, M.J. Molecular imprinting science and technology: A survey of the literature for the years up to and including 2003. J. Mol. Recognit. 2006, 19, 106–180. [Google Scholar] [CrossRef]
- Ye, L. (Ed.) Molecular Imprinting: Principles and Applications of Micro- and Nanostructure Polymers; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Huckle, D. Point-of-care diagnostics: An advancing sector with nontechnical issues. Expert Rev. Mol. Diagn. 2008, 8, 679–688. [Google Scholar] [CrossRef]
- Huckle, D. Point-of-care diagnostics—Is this driven by supply or demand? Expert Opin. Med. Diagn. 2010, 4, 189–200. [Google Scholar] [CrossRef]
- Junker, R.; Petersmann, A.; Luppa, P.B. The relevance of POCT in healthcare. In Point-of-Care Testing: Principles and Clinical Applications; Luppa, P.B., Junker, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 9–17. [Google Scholar]
- Bossi, A.; Bonini, F.; Turner, A.; Piletsky, S. Molecularly imprinted polymers for the recognition of proteins: The state of the art. Biosens. Bioelectron. 2007, 22, 1131–1137. [Google Scholar] [CrossRef]
- Cheong, W.J.; Yang, S.H.; Ali, F. Molecular imprinted polymers for separation science: A review of reviews. J. Sep. Sci. 2013, 36, 609–628. [Google Scholar] [CrossRef]
- Baggiani, C.; Anfossi, L.; Giovannoli, C. MIP-based Immunoassays: State of the Art, Limitations and Perspectives. Mol. Impr. 2013, 1, 41–54. [Google Scholar] [CrossRef]
- Turner, N.W.; Jeans, C.W.; Brain, K.R.; Allender, C.J.; Hlady, V.; Britt, D.W. From 3D to 2D: A review of the molecular imprinting of proteins. Biotechnol. Prog. 2006, 22, 1474–1489. [Google Scholar] [CrossRef]
- Sellergren, B. Molecularly imprinted polymers: Shaping enzyme inhibitors. Nat. Chem. 2010, 2, 7–8. [Google Scholar] [CrossRef]
- Szenczi, A.; Kardos, J.; Medgyesi, G.A.; Zavodszky, P. The effect of solvent environment on the conformation and stability of human polyclonal IgG in solution. Biologicals 2006, 34, 5–14. [Google Scholar] [CrossRef]
- Vasapollo, G.; Sole, R.D.; Mergola, L.; Lazzoi, M.R.; Scardino, A.; Scorrano, S.; Mele, G. Molecularly imprinted polymers: Present and future prospective. Int. J. Mol. Sci. 2011, 12, 5908–5945. [Google Scholar] [CrossRef]
- Tan, C.J.; Tong, Y.W. Preparation of superparamagnetic ribonuclease A surface-imprinted submicrometer particles for protein recognition in aqueous media. Anal. Chem. 2007, 79, 299–306. [Google Scholar] [CrossRef]
- Awino, J.K.; Zhao, Y. Water-soluble molecularly imprinted nanoparticles (MINPs) with tailored, functionalized, modifiable binding pockets. Chem. Eur. J. 2015, 21, 655–661. [Google Scholar] [CrossRef]
- Paul, P.K.; Treetong, A.; Suedee, R. Biomimetic insulin-imprinted polymer nanoparticles as a potential oral drug delivery system. Acta Pharm. 2017, 67, 149–168. [Google Scholar] [CrossRef]
- Li, Y.; Yang, H.H.; You, Q.H.; Zhuang, Z.X.; Wang, X.R. Protein recognition via surface molecularly imprinted polymer nanowires. Anal. Chem. 2006, 78, 317–320. [Google Scholar] [CrossRef]
- Nicholls, I.A.; Rosengren, J.P. Molecular imprinting of surfaces. Bioseparation 2001, 10, 301–305. [Google Scholar] [CrossRef]
- Nishino, H.; Huang, C.S.; Shea, K.J. Selective Protein Capture by Epitope Imprinting. Angew. Chem. Int. Ed. 2006, 45, 2392–2396. [Google Scholar] [CrossRef]
- Perçin, I.; Idil, N.; Bakhshpour, M.; Yılmaz, E.; Mattiasson, B.; Denizli, A. Microcontact Imprinted Plasmonic Nanosensors: Powerful Tools in the Detection of Salmonella paratyphi. Sensors 2017, 17, 1375. [Google Scholar] [CrossRef]
- Shiomi, T.; Matsui, M.; Mizukami, F.; Sakaguchi, K. A method for the molecular imprinting of hemoglobin on silica surfaces using silanes. Biomaterials 2005, 26, 5564–5571. [Google Scholar] [CrossRef]
- Li, F.; Li, J.; Zhang, S. Molecularly imprinted polymer grafted on polysaccharide microsphere surface by the sol–gel process for protein recognition. Talanta 2008, 74, 1247–1255. [Google Scholar] [CrossRef]
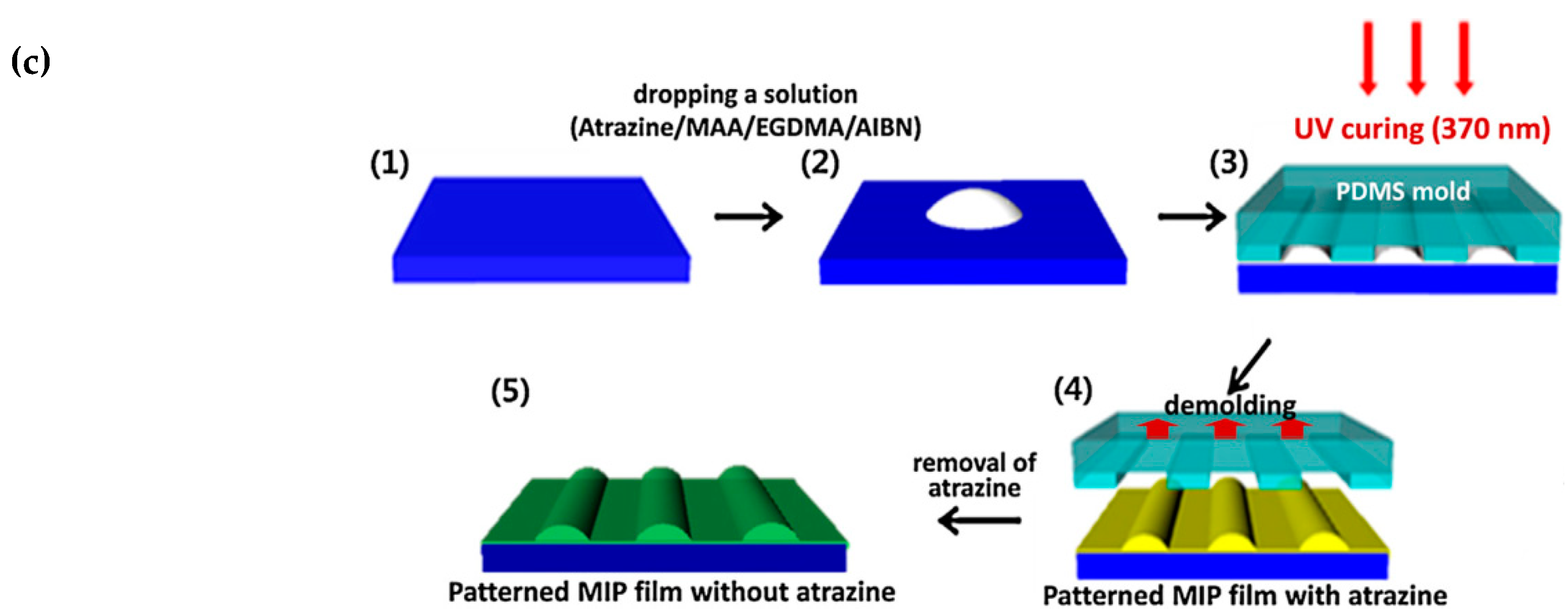
- Yang, J.C.; Shin, H.K.; Hong, S.W.; Park, J.Y. Lithographically patterned molecularly imprinted polymer for gravimetric detection of trace atrazine. Sens. Actuators B Chem. 2015, 216, 476–481. [Google Scholar] [CrossRef]
- Yang, H.H.; Zhang, S.Q.; Tan, F.; Zhuang, Z.X.; Wang, X.R. Surface molecularly imprinted nanowires for biorecognition. J. Am. Chem. Soc. 2005, 127, 1378–1379. [Google Scholar] [CrossRef]
- Ansari, S.; Karimi, M. Recent configurations and progressive uses of magnetic molecularly imprinted polymers for drug analysis. Talanta 2017, 167, 470–485. [Google Scholar] [CrossRef]
- Hu, Y.; Li, Y.; Liu, R.; Tan, W.; Li, G. Magnetic molecularly imprinted polymer beads prepared by microwave heating for selective enrichment of β-agonists in pork and pig liver samples. Talanta 2011, 84, 462–470. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, J.; Fang, G.; Xie, W. Improvement of the homogeneity of protein-imprinted polymer films by orientated immobilization of the template. Anal. Chim. Acta 2012, 726, 85–92. [Google Scholar] [CrossRef]
- Tan, C.J.; Tong, Y.W. Molecularly imprinted beads by surface imprinting. Anal. Bioanal. Chem. 2007, 389, 369–376. [Google Scholar] [CrossRef]
- Cumbo, A. Virus Imprinted Particles. Ph.D. Thesis, University of Basel, Basel, Switzerland, 2013. [Google Scholar]
- Yan, H.; Row, K.H. Characteristic and synthetic approach of molecularly imprinted polymer. Int. J. Mol. Sci. 2006, 7, 155–178. [Google Scholar] [CrossRef]
- Mandal, S.; Suriyanarayanan, S.; Nicholls, I.A.; Ramanujam, K. Selective Sensing of the Biotinyl Moiety Using Molecularly Imprinted Polyaniline Nanowires. J. Electrochem. Soc. 2018, 165, B669–B678. [Google Scholar] [CrossRef]
- Suriyanarayanan, S.; Mandal, S.; Ramanujam, K.; Nicholls, I.A. Electrochemically synthesized molecularly imprinted polythiophene nanostructures as recognition elements for an aspirin-chemosensor. Sens. Actuators B Chem. 2017, 253, 428–436. [Google Scholar] [CrossRef]
- Ndizeye, N.; Suriyanarayanan, S.; Nicholls, I.A. Hierarchical polymeric architectures through molecular imprinting in liquid crystalline environments. Eur. Polym. J. 2018, 106, 223–231. [Google Scholar] [CrossRef]
- Suriyanarayanan, S.; Petrone, L.; Ederth, T.; Nicholls, I.A. Biotinyl moiety-selective polymer films with highly ordered macropores. Chem. Commun. 2013, 49, 5274–5276. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qin, L.; He, X.W.; Li, W.Y.; Zhang, Y.K. Novel surface modified molecularly imprinted polymer using acryloyl-β-cyclodextrin and acrylamide as monomers for selective recognition of lysozyme in aqueous solution. J. Chromatogr. A 2009, 1216, 4560–4567. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; He, Y.; Ji, T.R.; Yan, X.P. Surface molecular imprinting on Mn-doped ZnS quantum dots for room-temperature phosphorescence optosensing of pentachlorophenol in water. Anal. Chem. 2009, 81, 1615–1621. [Google Scholar] [CrossRef]
- Tan, C.J.; Chua, H.G.; Ker, K.H.; Tong, Y.W. Preparation of bovine serum albumin surface-imprinted submicrometer particles with magnetic susceptibility through core-shell miniemulsion polymerization. Anal. Chem. 2008, 80, 683–692. [Google Scholar] [CrossRef]
- Jing, T.; Xia, H.; Guan, Q.; Lu, W.; Dai, Q.; Niu, J.; Lim, J.M.; Hao, Q.; Lee, Y.I.; Zhou, Y. Rapid and selective determination of urinary lysozyme based on magnetic molecularly imprinted polymers extraction followed by chemiluminescence detection. Anal. Chim. Acta 2011, 692, 73–79. [Google Scholar] [CrossRef]
- Li, Y.; Yin, X.F.; Chen, F.R.; Yang, H.H.; Zhuang, Z.X.; Wang, X.R. Synthesis of magnetic molecularly imprinted polymer nanowires using a nanoporous alumina template. Macromolecules 2006, 39, 4497–4499. [Google Scholar] [CrossRef]
- Xie, C.; Liu, B.; Wang, Z.; Gao, D.; Guan, G.; Zhang, Z. Molecular imprinting at walls of silica nanotubes for TNT recognition. Anal. Chem. 2008, 80, 437–443. [Google Scholar] [CrossRef]
- Rachkov, A.; Minoura, N. Recognition of oxytocin and oxytocin-related peptides in aqueous media using a molecularly imprinted polymer synthesized by the epitope approach. J. Chromatogr. A 2000, 889, 111–118. [Google Scholar] [CrossRef]
- Polevoda, B.; Sherman, F. Nα-terminal acetylation of eukaryotic proteins. J. Biol. Chem. 2000, 275, 36479–36482. [Google Scholar] [CrossRef]
- Whitcombe, M.J.; Chianella, I.; Larcombe, L.; Piletsky, S.A.; Noble, J.; Porter, R.; Horgan, A. The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem. Soc. Rev. 2011, 40, 1547–1571. [Google Scholar] [CrossRef] [PubMed]
- Kryscio, D.R.; Peppas, N.A. Critical review and perspective of macromolecularly imprinted polymers. Acta Biomater. 2012, 8, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Kolaskar, A.; Kulkarni-Kale, U. Prediction of three-dimensional structure and mapping of conformational epitopes of envelope glycoprotein of Japanese encephalitis virus. Virology 1999, 261, 31–42. [Google Scholar] [CrossRef] [PubMed]
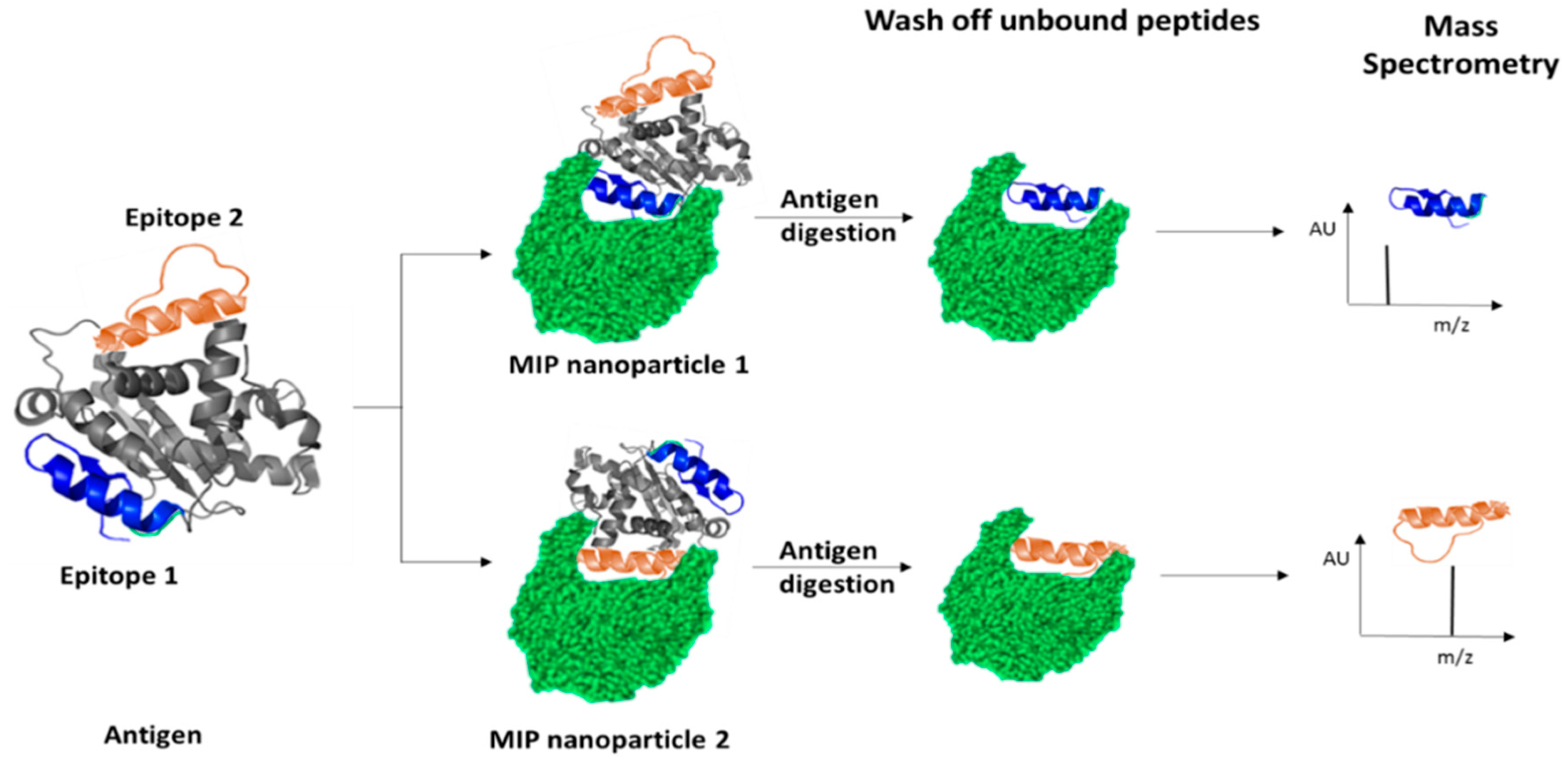
- Piletsky, S.; Piletska, E.; Canfarotta, F.; Karim, K.; Jones, D.; Norman, R.; Guerreiro, A. Methods and Kits for Determining Binding Sites. U.S. Patent WO/2018/178629, 19 March 2018. [Google Scholar]
- Lin, H.Y.; Hsu, C.Y.; Thomas, J.L.; Wang, S.E.; Chen, H.C.; Chou, T.C. The microcontact imprinting of proteins: The effect of cross-linking monomers for lysozyme, ribonuclease A and myoglobin. Biosens. Bioelectron. 2006, 22, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Sener, G.; Ozgur, E.; Rad, A.Y.; Uzun, L.; Say, R.; Denizli, A. Rapid real-time detection of procalcitonin using a microcontact imprinted surface plasmon resonance biosensor. Analyst 2013, 138, 6422–6428. [Google Scholar] [CrossRef] [PubMed]
- Osman, B.; Uzun, L.; Beşirli, N.; Denizli, A. Microcontact imprinted surface plasmon resonance sensor for myoglobin detection. Mater. Sci. Eng. C 2013, 33, 3609–3614. [Google Scholar] [CrossRef]
- Ertürk, G.; Mattiasson, B. Molecular imprinting techniques used for the preparation of biosensors. Sensors 2017, 17, 288. [Google Scholar] [CrossRef]
- Rosengren-Holmberg, J.P.; Andersson, J.; Smith, J.R.; Alexander, C.; Alexander, M.R.; Tovar, G.; Ekdahl, K.N.; Nicholls, I.A. Heparin molecularly imprinted surfaces for the attenuation of complement activation in blood. Biomater. Sci. 2015, 3, 1208–1217. [Google Scholar] [CrossRef]
- Bergmann, N.M.; Peppas, N.A. Molecularly imprinted polymers with specific recognition for macromolecules and proteins. Prog. Polym. Sci. 2008, 33, 271–288. [Google Scholar] [CrossRef]
- Dvorakova, G.; Haschick, R.; Chiad, K.; Klapper, M.; Müllen, K.; Biffis, A. Molecularly imprinted nanospheres by nonaqueous emulsion polymerization. Macromol. Rapid Commun. 2010, 31, 2035–2040. [Google Scholar] [CrossRef] [PubMed]
- Advincula, R.C. Engineering molecularly imprinted polymer (MIP) materials: Developments and challenges for sensing and separation technologies. Korean J. Chem. Eng. 2011, 28, 1313–1321. [Google Scholar] [CrossRef]
- Manesiotis, P.; Hall, A.J.; Courtois, J.; Irgum, K.; Sellergren, B. An artificial riboflavin receptor prepared by a template analogue imprinting strategy. Angew. Chem. 2005, 117, 3970–3974. [Google Scholar] [CrossRef]
- Ma, X.; Ji, W.; Chen, L.; Wang, X.; Liu, J.; Wang, X. Molecularly imprinted polymers with synthetic dummy templates for the preparation of capsaicin and dihydrocapsaicin from chili peppers. J. Sep. Sci. 2015, 38, 100–107. [Google Scholar] [CrossRef]
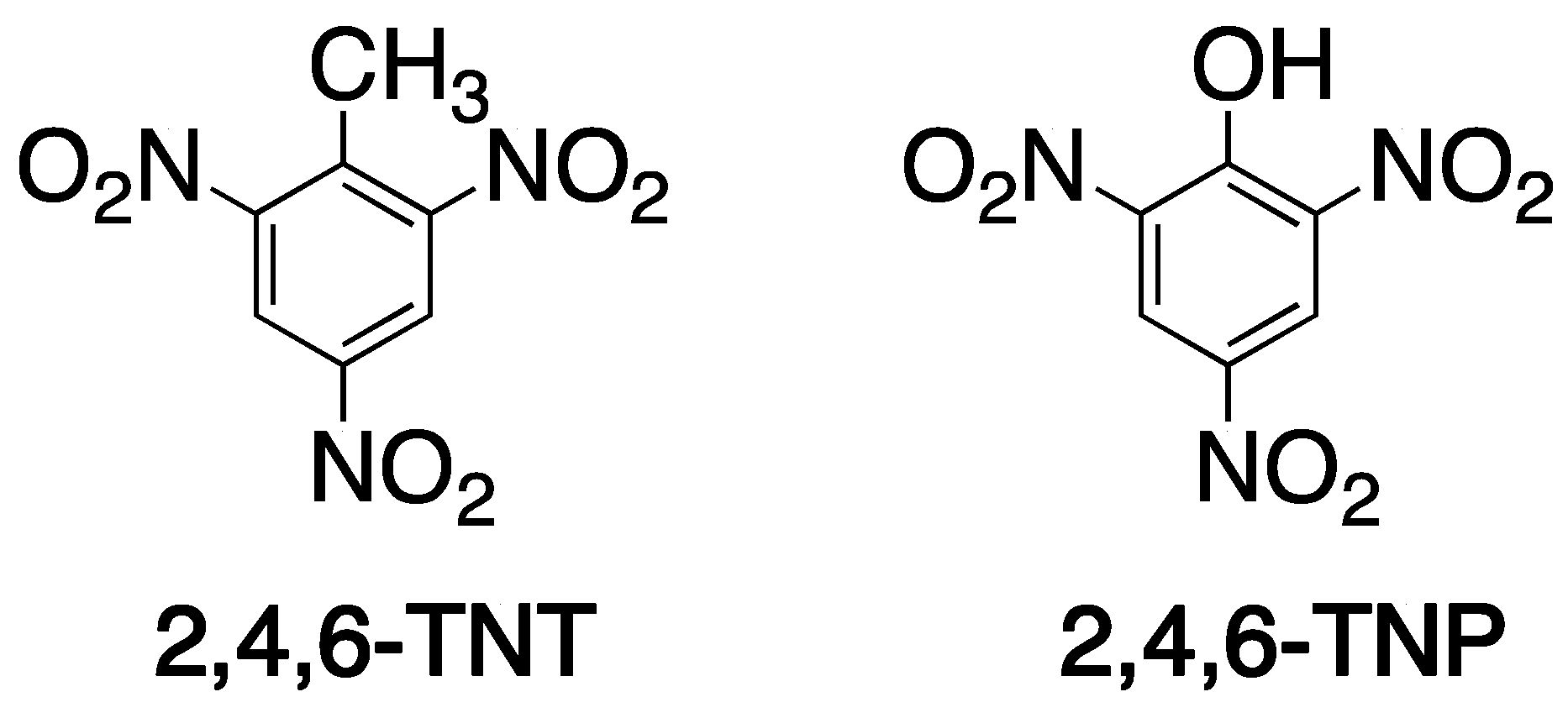
- Xu, S.; Lu, H.; Li, J.; Song, X.; Wang, A.; Chen, L.; Han, S. Dummy molecularly imprinted polymers-capped CdTe quantum dots for the fluorescent sensing of 2,4,6-trinitrotoluene. ACS Appl. Mater. Interfaces 2013, 5, 8146–8154. [Google Scholar] [CrossRef]
- Quaglia, M.; Chenon, K.; Hall, A.J.; De Lorenzi, E.; Sellergren, B. Target analogue imprinted polymers with affinity for folic acid and related compounds. J. Am. Chem. Soc. 2001, 123, 2146–2154. [Google Scholar] [CrossRef]
- Tokonami, S.; Shiigi, H.; Nagaoka, T. Review: Micro- and nanosized molecularly imprinted polymers for high-throughput analytical applications. Anal. Chim. Acta 2009, 641, 7–13. [Google Scholar] [CrossRef]
- Umpleby, R.J.; Baxter, S.C.; Rampey, A.M.; Rushton, G.T.; Chen, Y.; Shimizu, K.D. Characterization of the heterogeneous binding site affinity distributions in molecularly imprinted polymers. J. Chromatogr. B 2004, 804, 141–149. [Google Scholar] [CrossRef]
- Nicholls, I.A. Thermodynamic considerations for the design of and ligand recognition by non-covalent molecularly imprinted polymers. Chem. Lett. 1995, 24, 1035–1036. [Google Scholar] [CrossRef]
- Curcio, P.; Zandanel, C.; Wagner, A.; Mioskowski, C.; Baati, R. Semi-Covalent Surface Molecular Imprinting of Polymers by One-Stage Mini-emulsion Polymerization: Glucopyranoside as a Model Analyte. Macromol. Biosci. 2009, 9, 596–604. [Google Scholar] [CrossRef]
- Zimmerman, S.C.; Lemcoff, N.G. Synthetic hosts via molecular imprinting—Are universal synthetic antibodies realistically possible? Chem. Commun. 2004, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Spivak, D.A. Optimization, evaluation, and characterization of molecularly imprinted polymers. Adv. Drug. Deliv. Rev. 2005, 57, 1779–1794. [Google Scholar] [CrossRef] [PubMed]
- Piletska, E.V.; Pink, D.; Karim, K.; Piletsky, S.A. Development of a computationally-designed polymeric adsorbent specific for mycotoxin patulin. Analyst 2017, 142, 4678–4683. [Google Scholar] [CrossRef]
- Bates, F.; Busato, M.; Piletska, E.; Whitcombe, M.J.; Karim, K.; Guerreiro, A.; del Valle, M.; Giorgetti, A.; Piletsky, S. Computational design of molecularly imprinted polymer for direct detection of melamine in milk. Sep. Sci. Technol. 2017, 52, 1441–1453. [Google Scholar] [CrossRef]
- Cowen, T.; Karim, K.; Piletsky, S. Computational approaches in the design of synthetic receptors—A review. Anal. Chim. Acta 2016, 936, 62–74. [Google Scholar] [CrossRef]
- Guerreiro, A.R.; Chianella, I.; Piletska, E.; Whitcombe, M.J.; Piletsky, S.A. Selection of imprinted nanoparticles by affinity chromatography. Biosens. Bioelectron. 2009, 24, 2740–2743. [Google Scholar] [CrossRef]
- Poma, A.; Guerreiro, A.; Caygill, S.; Moczko, E.; Piletsky, S. Automatic reactor for solid-phase synthesis of molecularly imprinted polymeric nanoparticles (MIP NPs) in water. RSC Adv. 2014, 4, 4203–4206. [Google Scholar] [CrossRef]
- Canfarotta, F.; Poma, A.; Guerreiro, A.; Piletsky, S. Solid-phase synthesis of molecularly imprinted nanoparticles. Nat. Protoc. 2016, 11, 443–455. [Google Scholar] [CrossRef]
- Basozabal, I.; Guerreiro, A.; Gomez-Caballero, A.; Goicolea, M.A.; Barrio, R.J. Direct potentiometric quantification of histamine using solid-phase imprinted nanoparticles as recognition elements. Biosens. Bioelectron. 2014, 58, 138–144. [Google Scholar] [CrossRef]
- Otsu, T.; Matsunaga, T.; Kuriyama, A.; Yoshioka, M. Living radical polymerization through the use of iniferters: Controlled synthesis of polymers. Eur. Polym. J. 1989, 25, 643–650. [Google Scholar] [CrossRef]
- Cowen, T.; Busato, M.; Karim, K.; Piletsky, S.A. In Silico Synthesis of Synthetic Receptors: A Polymerization Algorithm. Macromol. Rapid Commun. 2016, 37, 2011–2016. [Google Scholar] [CrossRef] [PubMed]
- Cowen, T.; Karim, K.; Piletsky, S.A. Solubility and size of polymer nanoparticles. Polym. Chem. 2018, 9, 4566–4573. [Google Scholar] [CrossRef]
- Hoshino, Y.; Shea, K.J. The evolution of plastic antibodies. J. Mater. Chem. 2011, 21, 3517–3521. [Google Scholar] [CrossRef]
- Wulff, G. Fourty years of molecular imprinting in synthetic polymers: Origin, features and perspectives. Microchim. Acta 2013, 180, 1359–1370. [Google Scholar] [CrossRef]
- Poma, A.; Turner, A.P.; Piletsky, S.A. Advances in the manufacture of MIP nanoparticles. Trends Biotechnol. 2010, 28, 629–637. [Google Scholar] [CrossRef]
- Canfarotta, F.; Cecchini, A.; Piletsky, S. Nano-sized Molecularly Imprinted Polymers as Artificial Antibodies. In Molecularly Imprinted Polymers for Analytical Chemistry Applications; Kutner, W., Sharma, P.S., Eds.; Royal Society of Chemistry: London, UK, 2018; pp. 1–27. [Google Scholar]
- Ye, L.; Mosbach, K. Molecularly imprinted microspheres as antibody binding mimics. React. Funct. Polym. 2001, 48, 149–157. [Google Scholar] [CrossRef]
- Poma, A. Automatic Solid-Phase Synthesis of Molecularly Imprinted Nanoparticles (MIP NPs). Ph.D. Thesis, Cranfield University, Cranfield, UK, 2012. [Google Scholar]
- Ye, L.; Cormack, P.A.; Mosbach, K. Molecularly imprinted monodisperse microspheres for competitive radioassay. Anal. Commun. 1999, 36, 35–38. [Google Scholar] [CrossRef]
- Li, W.H.; Stöver, H.D. Monodisperse cross-linked core− shell polymer microspheres by precipitation polymerization. Macromolecules 2000, 33, 4354–4360. [Google Scholar] [CrossRef]
- Jing, T.; Gao, X.D.; Wang, P.; Wang, Y.; Lin, Y.F.; Hu, X.Z.; Hao, Q.L.; Zhou, Y.K.; Mei, S.R. Determination of trace tetracycline antibiotics in foodstuffs by liquid chromatography–tandem mass spectrometry coupled with selective molecular-imprinted solid-phase extraction. Anal. Bioanal. Chem. 2009, 393, 2009–2018. [Google Scholar] [CrossRef]
- Pérez-Moral, N.; Mayes, A. Comparative study of imprinted polymer particles prepared by different polymerisation methods. Anal. Chim. Acta 2004, 504, 15–21. [Google Scholar] [CrossRef]
- Downey, J.S.; McIsaac, G.; Frank, R.S.; Stöver, H.D. Poly (divinylbenzene) microspheres as an intermediate morphology between microgel, macrogel, and coagulum in cross-linking precipitation polymerization. Macromolecules 2001, 34, 4534–4541. [Google Scholar] [CrossRef]
- Beltran, A.; Marcé, R.; Cormack, P.; Borrull, F. Synthesis by precipitation polymerisation of molecularly imprinted polymer microspheres for the selective extraction of carbamazepine and oxcarbazepine from human urine. J. Chromatogr. A 2009, 1216, 2248–2253. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, T. Preparation of molecularly imprinted polymer containing selective cavities for urea molecule and its application for urea extraction. Anal. Chim. Acta 2010, 669, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hoshina, K.; Haginaka, J. Monodispersed, molecularly imprinted polymers for cinchonidine by precipitation polymerization. Talanta 2010, 80, 1713–1718. [Google Scholar] [CrossRef]
- Vaihinger, D.; Landfester, K.; Kräuter, I.; Brunner, H.; Tovar, G.E. Molecularly imprinted polymer nanospheres as synthetic affinity receptors obtained by miniemulsion polymerisation. Macromol. Chem. Phys. 2002, 203, 1965–1973. [Google Scholar] [CrossRef]
- Ambrosini, S.; Beyazit, S.; Haupt, K.; Bui, B.T.S. Solid-phase synthesis of molecularly imprinted nanoparticles for protein recognition. Chem. Commun. 2013, 49, 6746–6748. [Google Scholar] [CrossRef]
- Congur, G.; Senay, H.; Turkcan, C.; Canavar, E.; Erdem, A.; Akgol, S. Estrone specific molecularly imprinted polymeric nanospheres: Synthesis, characterization and applications for electrochemical sensor development. Comb. Chem. High Throughput Screen. 2013, 16, 503–510. [Google Scholar] [CrossRef]
- Van Herk, A.M.; Monteiro, M. Heterogeneous systems. In Handbook of Radical Polymerization; Matyjaszewski, K., Davis, T.P., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2003; pp. 301–332. [Google Scholar]
- Pérez, N.; Whitcombe, M.J.; Vulfson, E.N. Molecularly imprinted nanoparticles prepared by core-shell emulsion polymerization. J. Appl. Polym. Sci. 2000, 77, 1851–1859. [Google Scholar] [CrossRef]
- Mijangos, I.; Guerreiro, A.; Piletska, E.; Whitcombe, M.J.; Karim, K.; Chianella, I.; Piletsky, S. Macroradical initiated polymerisation of acrylic and methacrylic monomers. J. Sep. Sci. 2009, 32, 3340–3346. [Google Scholar] [CrossRef]
- Bossi, A.; Whitcombe, M.J.; Uludag, Y.; Fowler, S.; Chianella, I.; Subrahmanyam, S.; Sanchez, I.; Piletsky, S.A. Synthesis of controlled polymeric cross-linked coatings via iniferter polymerisation in the presence of tetraethyl thiuram disulphide chain terminator. Biosens. Bioelectron. 2010, 25, 2149–2155. [Google Scholar] [CrossRef]
- Carter, S.; Lu, S.Y.; Rimmer, S. Core-shell molecular imprinted polymer colloids. Supramol. Chem. 2003, 15, 213–220. [Google Scholar] [CrossRef]
- Li, L.; He, X.; Chen, L.; Zhang, Y. Preparation of core-shell magnetic molecularly imprinted polymer nanoparticles for recognition of bovine hemoglobin. Chem. Asian J. 2009, 4, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Madrakian, T.; Afkhami, A.; Mahmood-Kashani, H.; Ahmadi, M. Superparamagnetic surface molecularly imprinted nanoparticles for sensitive solid-phase extraction of tramadol from urine samples. Talanta 2013, 105, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dong, C.; Chu, J.; Qi, J.; Li, X. Surface molecular imprinting onto fluorescein-coated magnetic nanoparticles via reversible addition fragmentation chain transfer polymerization: A facile three-in-one system for recognition and separation of endocrine disrupting chemicals. Nanoscale 2011, 3, 280–287. [Google Scholar] [CrossRef]
- Gao, D.; Zhang, Z.; Wu, M.; Xie, C.; Guan, G.; Wang, D. A surface functional monomer-directing strategy for highly dense imprinting of TNT at surface of silica nanoparticles. J. Am. Chem. Soc. 2007, 129, 7859–7866. [Google Scholar] [CrossRef]
- Moczko, E.; Poma, A.; Guerreiro, A.; de Vargas Sansalvador, I.M.P.; Caygill, S.; Canfarotta, F.; Whitcombe, M.J.; Piletsky, S. Surface-modified multifunctional MIP nanoparticles. Nanoscale 2013, 5, 3733–3741. [Google Scholar] [CrossRef]
- Moczko, E.; Guerreiro, A.; Piletska, E.; Piletsky, S. PEG-stabilized core–shell surface-imprinted nanoparticles. Langmuir 2013, 29, 9891–9896. [Google Scholar] [CrossRef]
- Subrahmanyam, S.; Guerreiro, A.; Poma, A.; Moczko, E.; Piletska, E.; Piletsky, S. Optimisation of experimental conditions for synthesis of high affinity MIP nanoparticles. Eur. Polym. J. 2013, 49, 100–105. [Google Scholar] [CrossRef]
- Valero-Navarro, A.; Medina-Castillo, A.L.; Fernandez-Sanchez, J.F.; Fernández-Gutiérrez, A. Synthesis of a novel polyurethane-based-magnetic imprinted polymer for the selective optical detection of 1-naphthylamine in drinking water. Biosens. Bioelectron. 2011, 26, 4520–4525. [Google Scholar] [CrossRef]
- Piletsky, S.A.; Turner, N.W.; Laitenberger, P. Molecularly imprinted polymers in clinical diagnostics—Future potential and existing problems. Med. Eng. Phys. 2006, 28, 971–977. [Google Scholar] [CrossRef]
- Gutiérrez-Climente, R.; Gómez-Caballero, A.; Halhalli, M.; Sellergren, B.; Goicolea, M.A.; Barrio, R.J. Iniferter-mediated grafting of molecularly imprinted polymers on porous silica beads for the enantiomeric resolution of drugs. J. Mol. Recognit. 2016, 29, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, R.A.; Wu, M.; Zou, H. Recent progress of chiral monolithic stationary phases in CEC and capillary LC. Electrophoresis 2010, 31, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zong, H.Y.; Huang, Y.P.; Liu, Z.S. Liquid crystal-based molecularly imprinted nanoparticles with low crosslinking for capillary electrochromatography. J. Chromatogr. A 2013, 1309, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Maciel, E.V.S.; de Toffoli, A.L.; Neto, E.S.; Nazario, C.E.D.; Lancas, F.M. New materials in sample preparation: Recent advances and future trends. TrAC Trends Anal. Chem. 2019, 119, 115633. [Google Scholar] [CrossRef]
- Chen, L.; Liu, J.; Zeng, Q.; Wang, H.; Yu, A.; Zhang, H.; Ding, L. Preparation of magnetic molecularly imprinted polymer for the separation of tetracycline antibiotics from egg and tissue samples. J. Chromatogr. A 2009, 1216, 3710–3719. [Google Scholar] [CrossRef]
- Priego-Capote, F.; Ye, L.; Shakil, S.; Shamsi, S.A.; Nilsson, S. Monoclonal behavior of molecularly imprinted polymer nanoparticles in capillary electrochromatography. Anal. Chem. 2008, 80, 2881–2887. [Google Scholar] [CrossRef]
- Shi, X.X.; Xu, L.; Duan, H.Q.; Huang, Y.P.; Liu, Z.S. CEC separation of ofloxacin enantiomers using imprinted microparticles prepared in molecular crowding conditions. Electrophoresis 2011, 32, 1348–1356. [Google Scholar] [CrossRef]
- Li, S.; Li, W. Molecular imprinting: A versatile tool for separation, sensors and catalysis. Adv. Polym. Sci. 2007, 206, 191–210. [Google Scholar] [CrossRef]
- Wulff, G. Enzyme-like catalysis by molecularly imprinted polymers. Chem. Rev. 2002, 102, 1–28. [Google Scholar] [CrossRef]
- Berti, F.; Todros, S.; Lakshmi, D.; Whitcombe, M.J.; Chianella, I.; Ferroni, M.; Piletsky, S.A.; Turner, A.P.; Marrazza, G. Quasi-monodimensional polyaniline nanostructures for enhanced molecularly imprinted polymer-based sensing. Biosens. Bioelectron. 2010, 26, 497–503. [Google Scholar] [CrossRef]
- Wulff, G.; Liu, J. Design of biomimetic catalysts by molecular imprinting in synthetic polymers: The role of transition state stabilization. Acc. Chem. Res. 2011, 45, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Servant, A. Synthesis and Characterisation of Molecularly Imprinted Nanoparticles with Enzyme-Like Catalytic Activity for the Kemp Elimination. Ph.D. Thesis, Queen Mary University, London, UK, 2010. [Google Scholar]
- Anfossi, L.; Baggiani, C.; Giovannoli, C.; D’Arco, G.; Giraudi, G. Lateral-flow immunoassays for mycotoxins and phycotoxins: A review. Anal. Bioanal. Chem. 2013, 405, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Piletska, E.V.; Piletsky, S.S.; Whitcombe, M.J.; Chianella, I.; Piletsky, S.A. Development of a New Microtiter Plate Format for Clinically Relevant Assays. Anal. Chem. 2012, 84, 2038–2043. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Linares, E.M.; Pannuti, C.S.; Kubota, L.T.; Thalhammer, S. Immunospot assay based on fluorescent nanoparticles for Dengue fever detection. Biosens. Bioelectron. 2013, 41, 180–185. [Google Scholar] [CrossRef]
- Bedwell, T.S.; Whitcombe, M.J. Analytical applications of MIPs in diagnostic assays: Future perspectives. Anal. Bioanal. Chem. 2016, 408, 1735–1751. [Google Scholar] [CrossRef]
- Akbulut, M.; Lakshmi, D.; Whitcombe, M.J.; Piletska, E.V.; Chianella, I.; Güven, O.; Piletsky, S.A. Microplates with adaptive surfaces. ACS Comb. Sci. 2011, 13, 646–652. [Google Scholar] [CrossRef]
- Hoshino, Y.; Kodama, T.; Okahata, Y.; Shea, K.J. Peptide imprinted polymer nanoparticles: A plastic antibody. J. Am. Chem. Soc. 2008, 130, 15242–15243. [Google Scholar] [CrossRef]
- Hoshino, Y.; Koide, H.; Urakami, T.; Kanazawa, H.; Kodama, T.; Oku, N.; Shea, K.J. Recognition, neutralization, and clearance of target peptides in the bloodstream of living mice by molecularly imprinted polymer nanoparticles: A plastic antibody. J. Am. Chem. Soc. 2010, 132, 6644–6645. [Google Scholar] [CrossRef]
- Haupt, K.; Dzgoev, A.; Mosbach, K. Assay system for the herbicide 2, 4-dichlorophenoxyacetic acid using a molecularly imprinted polymer as an artificial recognition element. Anal. Chem. 1998, 70, 628–631. [Google Scholar] [CrossRef]
- Lin, C.I.; Joseph, A.K.; Chang, C.K.; Der Lee, Y. Synthesis and photoluminescence study of molecularly imprinted polymers appended onto CdSe/ZnS core-shells. Biosens. Bioelectron. 2004, 20, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Hunt, C.E.; Pasetto, P.; Ansell, R.J.; Haupt, K. A fluorescence polarisation molecular imprint sorbent assay for 2, 4-D: A non-separation pseudo-immunoassay. Chem. Commun. 2006, 1754–1756. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Villoslada, F.; Urraca, J.L.; Moreno-Bondi, M.C.; Orellana, G. Zearalenone sensing with molecularly imprinted polymers and tailored fluorescent probes. Sens. Actuators B Chem. 2007, 121, 67–73. [Google Scholar] [CrossRef]
- Lu, C.H.; Zhou, W.H.; Han, B.; Yang, H.H.; Chen, X.; Wang, X.R. Surface-imprinted core-shell nanoparticles for sorbent assays. Anal. Chem. 2007, 79, 5457–5461. [Google Scholar] [CrossRef] [PubMed]
- Poma, A. Solid-Phase Synthesis of Molecularly Imprinted Polymer Nanoparticles with a Reusable Template—“Plastic Antibodies”. Adv. Funct. Mater. 2013, 23, 2821–2827. [Google Scholar] [CrossRef]
- Chianella, I.; Guerreiro, A.; Moczko, E.; Caygill, J.S.; Piletska, E.V.; de Vargas Sansalvador, I.M.P.; Whitcombe, M.J.; Piletsky, S.A. Direct Replacement of Antibodies with Molecularly Imprinted Polymer Nanoparticles in ELISA—Development of a Novel Assay for Vancomycin. Anal. Chem. 2013, 85, 8462–8468. [Google Scholar] [CrossRef]
- Garcia, Y.; Smolinska-Kempisty, K.; Pereira, E.; Piletska, E.; Piletsky, S. Development of competitive ‘pseudo’-ELISA assay for measurement of cocaine and its metabolites using molecularly imprinted polymer nanoparticles. Anal. Methods 2017, 9, 4592–4598. [Google Scholar] [CrossRef]
- Tang, S.P.; Canfarotta, F.; Smolinska-Kempisty, K.; Piletska, E.; Guerreiro, A.; Piletsky, S. A pseudo-ELISA based on molecularly imprinted nanoparticles for detection of gentamicin in real samples. Anal. Methods 2017, 9, 2853–2858. [Google Scholar] [CrossRef]
- Piletska, E.V.; Piletsky, S.S.; Guerreiro, A.; Karim, K.; Whitcombe, M.J.; Piletsky, S.A. Microplates with enhanced immobilization capabilities controlled by a magnetic field. J. Chin. Adv. Mater. Soc. 2014, 2, 118–129. [Google Scholar] [CrossRef]
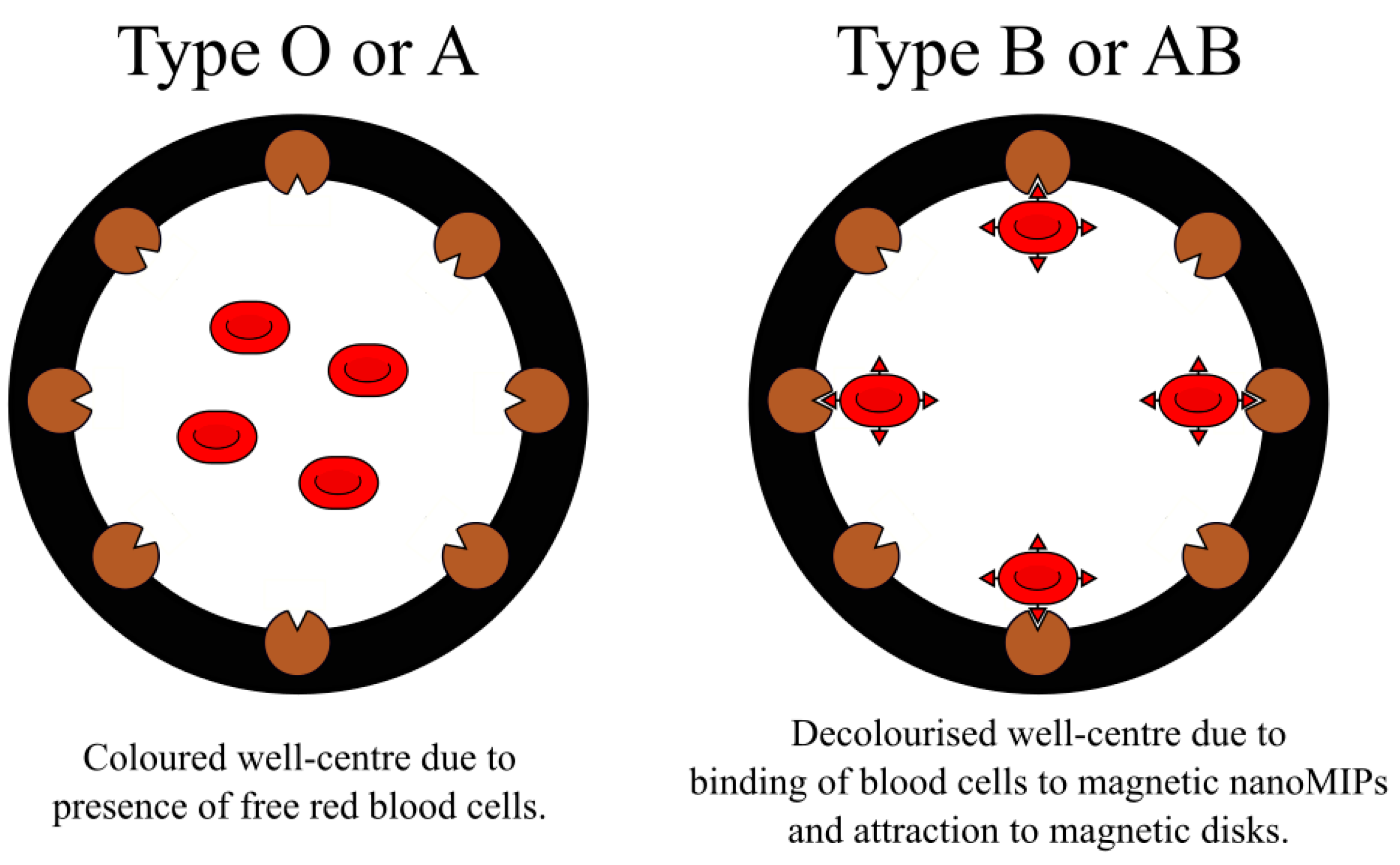
- Piletsky, S.; Rabinowicz, S.; Yang, Z.; Zagar, C.; Piletska, E.V.; Guerreiro, A.; Piletsky, S.A. Development of molecularly imprinted polymers specific for blood antigens for application in antibody-free blood typing. Chem. Commun. 2017, 53, 1793–1796. [Google Scholar] [CrossRef]
- Esen, C.; Czulak, J.; Cowen, T.; Piletska, E.; Piletsky, S.A. Highly Efficient Abiotic Assay Formats for Methyl Parathion–MINA as Alternative to ELISA. Anal. Chem. 2018, 91, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Piletsky, S.S.; Cass, A.E.; Piletska, E.V.; Czulak, J.; Piletsky, S.A. A Novel Assay Format as an Alternative to ELISA: MINA Test for Biotin. ChemNanoMat 2018, 4, 1214–1222. [Google Scholar] [CrossRef]
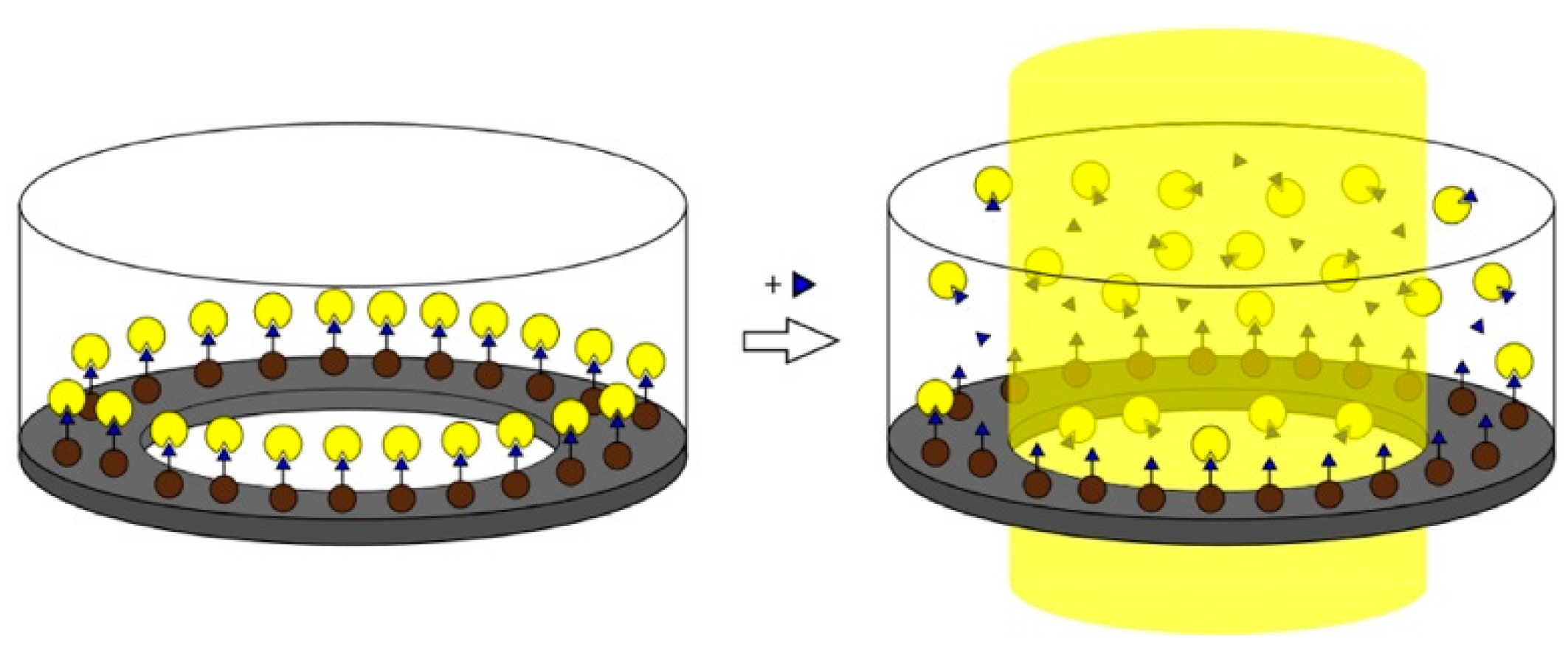
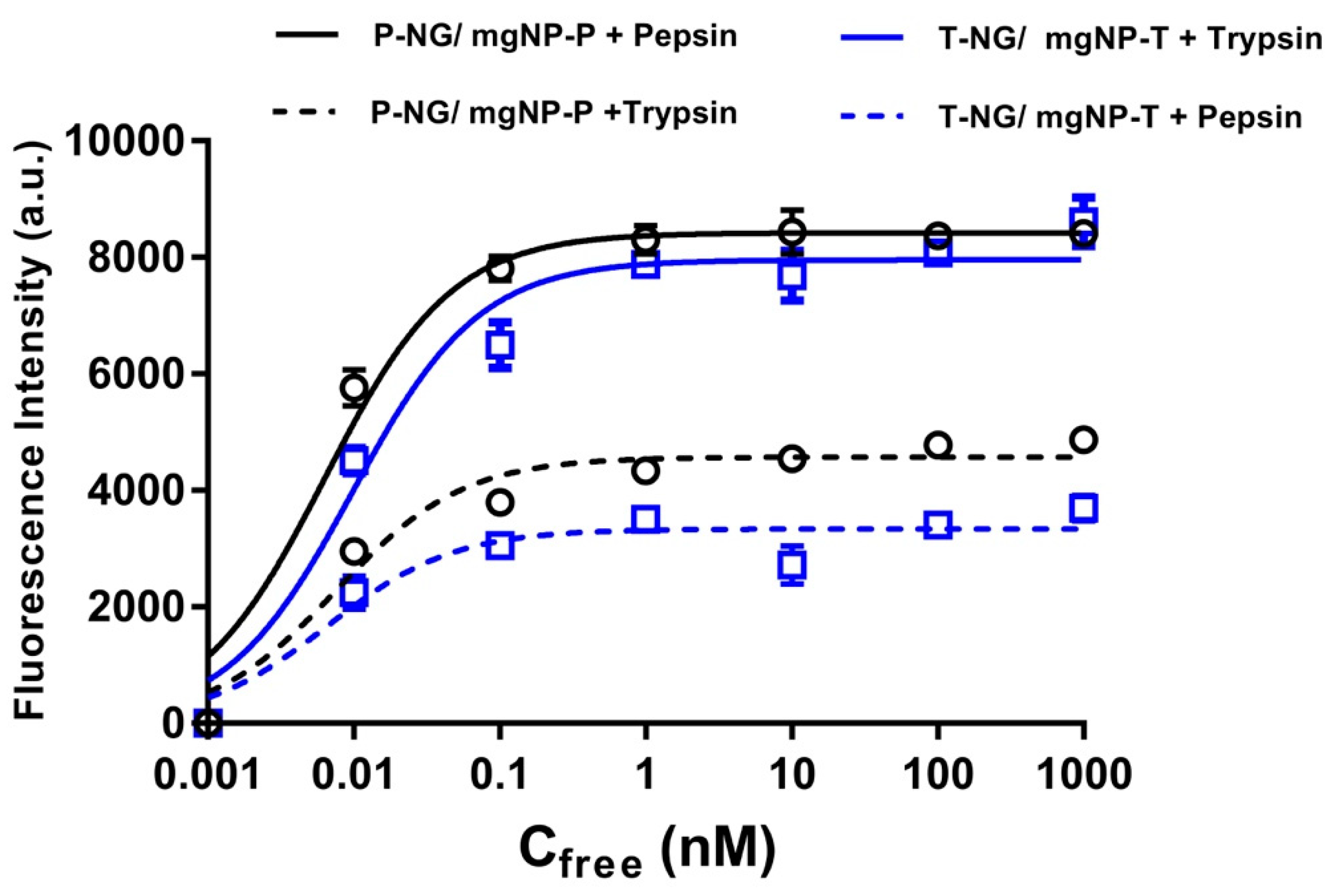
- Mahajan, R.; Rouhi, M.; Shinde, S.; Bedwell, T.; Incel, A.; Mavliutova, L.; Piletsky, S.; Nicholls, I.A.; Sellergren, B. Highly Efficient Synthesis and Assay of Protein-Imprinted Nanogels by Using Magnetic Templates. Angew. Chem. Int. Ed. 2019, 58, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Schirhagl, R.; Lieberzeit, P.A.; Dickert, F.L. Chemosensors for viruses based on artificial immunoglobulin copies. Adv. Mater. 2010, 22, 2078–2081. [Google Scholar] [CrossRef]
- Dickert, F.L.; Hayden, O.; Bindeus, R.; Mann, K.J.; Blaas, D.; Waigmann, E. Bioimprinted QCM sensors for virus detection—Screening of plant sap. Anal. Bioanal. Chem. 2004, 378, 1929–1934. [Google Scholar] [CrossRef]
- Piletsky, S.A.; Piletska, E.V.; Chen, B.; Karim, K.; Weston, D.; Barrett, G.; Lowe, P.; Turner, A.P. Chemical grafting of molecularly imprinted homopolymers to the surface of microplates. Application of artificial adrenergic receptor in enzyme-linked assay for β-agonists determination. Anal. Chem. 2000, 72, 4381–4385. [Google Scholar] [CrossRef]
- Titirici, M.M.; Sellergren, B. Thin molecularly imprinted polymer films via reversible addition− fragmentation chain transfer polymerization. Chem. Mater. 2006, 18, 1773–1779. [Google Scholar] [CrossRef]
- Cai, D.; Ren, L.; Zhao, H.; Xu, C.; Zhang, L.; Yu, Y.; Wang, H.; Lan, Y.; Roberts, M.F.; Chuang, J.H. A molecular-imprint nanosensor for ultrasensitive detection of proteins. Nat. Nanotechnol. 2010, 5, 597–601. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Liu, J.; Liu, J.; Tang, H.; Cao, C.; Zhao, D.; Ding, Y. Molecularly imprinted polymer decorated nanoporous gold for highly selective and sensitive electrochemical sensors. Sci. Rep. 2015, 5, 7699. [Google Scholar] [CrossRef]
- Reimhult, K.; Yoshimatsu, K.; Risveden, K.; Chen, S.; Ye, L.; Krozer, A. Characterization of QCM sensor surfaces coated with molecularly imprinted nanoparticles. Biosens. Bioelectron. 2008, 23, 1908–1914. [Google Scholar] [CrossRef]
- Fang, Y.; Yan, S.; Ning, B.; Liu, N.; Gao, Z.; Chao, F. Flow injection chemiluminescence sensor using molecularly imprinted polymers as recognition element for determination of maleic hydrazide. Biosens. Bioelectron. 2009, 24, 2323–2327. [Google Scholar] [CrossRef] [PubMed]
- Uchida, A.; Kitayama, Y.; Takano, E.; Ooya, T.; Takeuchi, T. Supraparticles comprised of molecularly imprinted nanoparticles and modified gold nanoparticles as a nanosensor platform. RSC Adv. 2013, 3, 25306–25311. [Google Scholar] [CrossRef]
- Dechtrirat, D.; Jetzschmann, K.J.; Stöcklein, W.F.; Scheller, F.W.; Gajovic-Eichelmann, N. Protein rebinding to a surface-confined imprint. Adv. Funct. Mater. 2012, 22, 5231–5237. [Google Scholar] [CrossRef]
- Canfarotta, F.; Waters, A.; Sadler, R.; McGill, P.; Guerreiro, A.; Papkovsky, D.; Haupt, K.; Piletsky, S. Biocompatibility and internalization of molecularly imprinted nanoparticles. Nano Res. 2016, 9, 3463–3477. [Google Scholar] [CrossRef]
- Curcio, M.; Cirillo, G.; Parisi, O.I.; Iemma, F.; Picci, N.; Puoci, F. Quercetin-imprinted nanospheres as novel drug delivery devices. J. Funct. Biomater. 2012, 3, 269–282. [Google Scholar] [CrossRef]
- Suksuwan, A.; Lomlim, L.; Rungrotmongkol, T.; Nakpheng, T.; Dickert, F.L.; Suedee, R. The composite nanomaterials containing (R)-thalidomide-molecularly imprinted polymers as a recognition system for enantioselective-controlled release and targeted drug delivery. J. Appl. Polym. Sci. 2015, 132. [Google Scholar] [CrossRef]
- Ishkuh, F.A.; Javanbakht, M.; Esfandyari-Manesh, M.; Dinarvand, R.; Atyabi, F. Synthesis and characterization of paclitaxel-imprinted nanoparticles for recognition and controlled release of an anticancer drug. J. Mater. Sci. 2014, 49, 6343–6352. [Google Scholar] [CrossRef]
- Parisi, O.I.; Morelli, C.; Puoci, F.; Saturnino, C.; Caruso, A.; Sisci, D.; Trombino, G.E.; Picci, N.; Sinicropi, M.S. Magnetic molecularly imprinted polymers (MMIPs) for carbazole derivative release in targeted cancer therapy. J. Mater. Chem. B 2014, 2, 6619–6625. [Google Scholar] [CrossRef]
- Puoci, F.; Iemma, F.; Picci, N. Stimuli-responsive molecularly imprinted polymers for drug delivery: A review. Curr. Drug Deliv. 2008, 5, 85–96. [Google Scholar] [CrossRef]
- Suedee, R.; Jantarat, C.; Lindner, W.; Viernstein, H.; Songkro, S.; Srichana, T. Development of a pH-responsive drug delivery system for enantioselective-controlled delivery of racemic drugs. J. Control. Release 2010, 142, 122–131. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Li, D.Y.; He, X.W.; Li, W.Y.; Zhang, Y.K. Epitope imprinted polymer nanoparticles containing fluorescent quantum dots for specific recognition of human serum albumin. Microchim. Acta 2015, 182, 1465–1472. [Google Scholar] [CrossRef]
- Li, C.E.; Zhong, S.A.; Li, X.J.; Guo, M. Silica particles coated with azobenzene-containing photoresponsive molecule-imprinted skin layer. Colloid. Polym. Sci. 2013, 291, 2049–2059. [Google Scholar] [CrossRef]
- Pan, G.; Guo, Q.; Cao, C.; Yang, H.; Li, B. Thermo-responsive molecularly imprinted nanogels for specific recognition and controlled release of proteins. Soft Matter 2013, 9, 3840–3850. [Google Scholar] [CrossRef]
- Lin, C.I.; Joseph, A.K.; Chang, C.K.; Der Lee, Y. Molecularly imprinted polymeric film on semiconductor nanoparticles: Analyte detection by quantum dot photoluminescence. J. Chromatogr. A 2004, 1027, 259–262. [Google Scholar] [CrossRef]
- Liu, R.; Guan, G.; Wang, S.; Zhang, Z. Core-shell nanostructured molecular imprinting fluorescent chemosensor for selective detection of atrazine herbicide. Analyst 2011, 136, 184–190. [Google Scholar] [CrossRef]
- Kunath, S.; Panagiotopoulou, M.; Maximilien, J.; Marchyk, N.; Sänger, J.; Haupt, K. Cell and tissue imaging with molecularly imprinted polymers as plastic antibody mimics. Adv. Healthc. Mater. 2015, 4, 1322–1326. [Google Scholar] [CrossRef]
- Dong, Y.; Li, W.; Gu, Z.; Xing, R.; Ma, Y.; Zhang, Q.; Liu, Z. Inhibition of HER2-Positive Breast Cancer Growth by Blocking the HER2 Signaling Pathway with HER2-Glycan-Imprinted Nanoparticles. Angew. Chem. Int. Ed. 2019, 58, 10621–10625. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Refaat, D.; Aggour, M.G.; Farghali, A.A.; Mahajan, R.; Wiklander, J.G.; Nicholls, I.A.; Piletsky, S.A. Strategies for Molecular Imprinting and the Evolution of MIP Nanoparticles as Plastic Antibodies—Synthesis and Applications. Int. J. Mol. Sci. 2019, 20, 6304. https://doi.org/10.3390/ijms20246304
Refaat D, Aggour MG, Farghali AA, Mahajan R, Wiklander JG, Nicholls IA, Piletsky SA. Strategies for Molecular Imprinting and the Evolution of MIP Nanoparticles as Plastic Antibodies—Synthesis and Applications. International Journal of Molecular Sciences. 2019; 20(24):6304. https://doi.org/10.3390/ijms20246304
Chicago/Turabian StyleRefaat, Doaa, Mohamed G. Aggour, Ahmed A. Farghali, Rashmi Mahajan, Jesper G. Wiklander, Ian A. Nicholls, and Sergey A. Piletsky. 2019. "Strategies for Molecular Imprinting and the Evolution of MIP Nanoparticles as Plastic Antibodies—Synthesis and Applications" International Journal of Molecular Sciences 20, no. 24: 6304. https://doi.org/10.3390/ijms20246304
APA StyleRefaat, D., Aggour, M. G., Farghali, A. A., Mahajan, R., Wiklander, J. G., Nicholls, I. A., & Piletsky, S. A. (2019). Strategies for Molecular Imprinting and the Evolution of MIP Nanoparticles as Plastic Antibodies—Synthesis and Applications. International Journal of Molecular Sciences, 20(24), 6304. https://doi.org/10.3390/ijms20246304