Metabolomic Profile of BALB/c Macrophages Infected with Leishmania amazonensis: Deciphering L-Arginine Metabolism
Abstract
1. Introduction
2. Results
2.1. L. amazonensis Infected Macrophages Show a Differential Metabolomic Profile than Uninfected Macrophages
2.2. L. amazonensis Infection of Macrophages Impact on the Modulation of Arginine and Proline Metabolism
2.3. Parasite Metabolism Intercross the Changes in L-Arginine and Proline Metabolism During Infection
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Parasite Culture
4.3. In Vitro Macrophage Infections
4.4. Metabolite Extraction
4.5. CE-MS Metabolic Fingerprinting
4.6. CE-MS Data Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NOS2 | nitric oxide synthase 2 |
| NO | nitric oxide |
| CAT2 | cationic amino acid transporters 2 |
| ARG1 | arginase 1 |
| AAP3 | amino acid permease 3 |
| La-WT | Leishmania amazonensis-wild type |
| La-arg− | Leishmania amazonensis-arginase knockout |
| CE-MS | Capillary Eletrophoresis-Mass Spectrometer |
| BMDMs | bone marrow-derived macrophages |
References
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Alvar, J.; Velez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; den Boer, M. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- Naderer, T.; McConville, M.J. The leishmania-macrophage interaction: A metabolic perspective. Cell. Microbiol. 2008, 10, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Singh, N.; Mishra, M.; Kumar, V.; Gour, J.K.; Bajpai, S.; Singh, S.; Pandey, H.P.; Singh, R.K. Identification of tlr inducing th1-responsive leishmania donovani amastigote-specific antigens. Mol. Cell. Biochem. 2012, 359, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Ben-Othman, R.; Dellagi, K.; Guizani-Tabbane, L. Leishmania major parasites induced macrophage tolerance: Implication of mapk and nf-kappab pathways. Mol. Immunol. 2009, 46, 3438–3444. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Mosser, D.M.; Zhang, X. Activation of the mapk, erk, following leishmania amazonensis infection of macrophages. J. Immunol. 2007, 178, 1077–1085. [Google Scholar] [CrossRef]
- Ghalib, H.W.; Whittle, J.A.; Kubin, M.; Hashim, F.A.; el-Hassan, A.M.; Grabstein, K.H.; Trinchieri, G.; Reed, S.G. Il-12 enhances th1-type responses in human leishmania donovani infections. J. Immunol. 1995, 154, 4623–4629. [Google Scholar]
- Wilhelm, P.; Ritter, U.; Labbow, S.; Donhauser, N.; Rollinghoff, M.; Bogdan, C.; Korner, H. Rapidly fatal leishmaniasis in resistant c57bl/6 mice lacking tnf. J. Immunol. 2001, 166, 4012–4019. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Aoki, J.I.; Acuña, S.M.; Zampieri, R.A.; Markus, R.P.; Floeter-Winter, L.M.; Muxel, S.M. Melatonin and leishmania amazonensis infection altered mir-294, mir-30e, and mir-302d impacting on tnf, mcp-1, and nos2 expression. Front. Cell. Infect. Microbiol. 2019, 9, 60. [Google Scholar] [CrossRef]
- Nasseri, M.; Modabber, F.Z. Generalized infection and lack of delayed hypersensitivity in balb/c mice infected with leishmania tropica major. Infect. Immun. 1979, 26, 611–614. [Google Scholar]
- Bacellar, O.; D´Oliveira, A., Jr.; Jeronimo, S.; Carvalho, E.M. Il-10 and il-12 are the main regulatory cytokines in visceral leishmaniasis. Cytokine 2000, 12, 1228–1231. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Ghosh, S.; Jhonson, P.L.; Bhattacharya, S.K.; Majumdar, S. Immunomodulatory role of interleukin-10 in visceral leishmaniasis: Defective activation of protein kinase c-mediated signal transduction events. Infect. Immun. 2001, 69, 1499–1507. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rath, M.; Muller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Gomes, F.L.; Roma, E.H.; Carneiro, M.B.; Doria, N.A.; Sacks, D.L.; Peters, N.C. Site-dependent recruitment of inflammatory cells determines the effective dose of leishmania major. Infect. Immun. 2014, 82, 2713–2727. [Google Scholar] [CrossRef] [PubMed]
- Closs, E.I.; Simon, A.; Vekony, N.; Rotmann, A. Plasma membrane transporters for arginine. J. Nutr. 2004, 134, 2752–2759. [Google Scholar] [CrossRef]
- Closs, E.I.; Boissel, J.P.; Habermeier, A.; Rotmann, A. Structure and function of cationic amino acid transporters (cats). J. Membr. Biol. 2006, 213, 67–77. [Google Scholar] [CrossRef]
- Yeramian, A.; Martin, L.; Arpa, L.; Bertran, J.; Soler, C.; McLeod, C.; Modolell, M.; Palacin, M.; Lloberas, J.; Celada, A.; et al. Macrophages require distinct arginine catabolism and transport systems for proliferation and for activation. Eur. J. Immunol. 2006, 36, 1516–1526. [Google Scholar] [CrossRef]
- Laranjeira-Silva, M.F.; Zampieri, R.A.; Muxel, S.M.; Floeter-Winter, L.M.; Markus, R.P. Melatonin attenuates leishmania (l.) amazonensis infection by modulating arginine metabolism. J. Pineal Res. 2015, 4, 478–487. [Google Scholar] [CrossRef]
- Iniesta, V.; Gomez-Nieto, L.C.; Corraliza, I. The inhibition of arginase by n(omega)-hydroxy-l-arginine controls the growth of leishmania inside macrophages. J. Exp. Med. 2001, 193, 777–784. [Google Scholar] [CrossRef]
- Wanasen, N.; Soong, L. L-arginine metabolism and its impact on host immunity against leishmania infection. Immunol. Res. 2008, 41, 15–25. [Google Scholar] [CrossRef]
- Acuña, S.M.; Aoki, J.I.; Laranjeira da Silva, M.F.; Zampieri, R.A.; Fernandes, J.C.; Muxel, S.M.; Floeter-Winter, L.M. Arginase expression modulates nitric oxide production in leishmania (leishmania) amazonensis. PLoS ONE 2017, 12, e0187186. [Google Scholar] [CrossRef] [PubMed]
- Aoki, J.I.; Muxel, S.M.; Zampieri, R.A.; Acuna, S.M.; Fernandes, J.C.R.; Vanderlinde, R.H.; Sales, M.; Floeter-Winter, L.M. L-arginine availability and arginase activity: Characterization of amino acid permease 3 in leishmania amazonensis. PLoS Negl. Trop. Dis. 2017, 11, 0006025. [Google Scholar] [CrossRef]
- Muxel, S.M.; Laranjeira-Silva, M.F.; Zampieri, R.A.; Floeter-Winter, L.M. Leishmania (leishmania) amazonensis induces macrophage mir-294 and mir-721 expression and modulates infection by targeting nos2 and l-arginine metabolism. Sci. Rep. 2017, 7, 44141. [Google Scholar] [CrossRef] [PubMed]
- McConville, M.J.; de Souza, D.; Saunders, E.; Likic, V.A.; Naderer, T. Living in a phagolysosome; metabolism of leishmania amastigotes. Trends Parasitol. 2007, 23, 368–375. [Google Scholar] [CrossRef]
- McConville, M.J.; Saunders, E.C.; Kloehn, J.; Dagley, M.J. Leishmania carbon metabolism in the macrophage phagolysosome- feast or famine? F1000Research 2015, 4, 938. [Google Scholar] [CrossRef]
- McConville, M.J. Metabolic crosstalk between leishmania and the macrophage host. Trends Parasitol. 2016, 32, 666–668. [Google Scholar] [CrossRef]
- Castilho-Martins, E.A.; Canuto, G.A.; Muxel, S.M.; da Silva, M.F.; Floeter-Winter, L.M.; Del Aguila, C.; Lopez-Gonzalvez, A.; Barbas, C. Capillary electrophoresis reveals polyamine metabolism modulation in leishmania (leishmania) amazonensis wild type and arginase knockout mutants under arginine starvation. Electrophoresis 2015, 18, 2314–2323. [Google Scholar] [CrossRef]
- Castilho-Martins, E.A.; Laranjeira da Silva, M.F.; dos Santos, M.G.; Muxel, S.M.; Floeter-Winter, L.M. Axenic leishmania amazonensis promastigotes sense both the external and internal arginine pool distinctly regulating the two transporter-coding genes. PLoS ONE 2011, 6, e27818. [Google Scholar] [CrossRef]
- Laranjeira-Silva, M.F.; Zampieri, R.A.; Muxel, S.M.; Beverley, S.M.; Floeter-Winter, L.M. Leishmania amazonensis arginase compartmentalization in the glycosome is important for parasite infectivity. PLoS ONE 2012, 7, e34022. [Google Scholar]
- Pinney, J.W.; Papp, B.; Hyland, C.; Wambua, L.; Westhead, D.R.; McConkey, G.A. Metabolic reconstruction and analysis for parasite genomes. Trends Parasitol. 2007, 23, 548–554. [Google Scholar] [CrossRef]
- Chavali, A.K.; Whittemore, J.D.; Eddy, J.A.; Williams, K.T.; Papin, J.A. Systems analysis of metabolism in the pathogenic trypanosomatid leishmania major. Mol. Syst. Biol. 2008, 4, 177. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zeng, A.P. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics 2003, 19, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Ma, H.W.; Zhao, X.M.; Goryanin, I. The reconstruction and analysis of tissue specific human metabolic networks. Mol. Biosyst. 2012, 8, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Cornish-Bowden, A.; Cardenas, M.L. Metabolic analysis in drug design. C. R. Biol. 2003, 326, 509–515. [Google Scholar] [CrossRef]
- Kafsack, B.F.; Llinas, M. Eating at the table of another: Metabolomics of host-parasite interactions. Cell Host Microbe 2010, 7, 90–99. [Google Scholar] [CrossRef]
- Gregory, D.J.; Olivier, M. Subversion of host cell signalling by the protozoan parasite leishmania. Parasitology 2005, 130, 27–35. [Google Scholar] [CrossRef]
- Aoki, J.I.; Muxel, S.M.; Zampieri, R.A.; Laranjeira-Silva, M.F.; Muller, K.E.; Nerland, A.H.; Floeter-Winter, L.M. Rna-seq transcriptional profiling of leishmania amazonensis reveals an arginase-dependent gene expression regulation. PLoS Negl. Trop. Dis. 2017, 11. [Google Scholar] [CrossRef]
- Fernandes, M.C.; Dillon, L.A.; Belew, A.T.; Bravo, H.C.; Mosser, D.M.; El-Sayed, N.M. Dual transcriptome profiling of leishmania-infected human macrophages reveals distinct reprogramming signatures. mBio 2016, 7. [Google Scholar] [CrossRef]
- Muxel, S.M.; Acuna, S.M.; Aoki, J.I.; Zampieri, R.A.; Floeter-Winter, L.M. Toll-like receptor and mirna-let-7e expression alter the inflammatory response in leishmania amazonensis-infected macrophages. Front. Immunol. 2018, 9, 2792. [Google Scholar] [CrossRef]
- Aoki, J.I.; Muxel, S.M.; Zampieri, R.A.; Müller, K.E.; Nerland, A.H.; Floeter-Winter, L.M. Differential immune response modulation in early leishmania amazonensis infection of balb/c and c57bl/6 macrophages based on transcriptome profiles. Sci. Rep. 2019. under review. [Google Scholar]
- Dillon, L.A.; Suresh, R.; Okrah, K.; Corrada Bravo, H.; Mosser, D.M.; El-Sayed, N.M. Simultaneous transcriptional profiling of leishmania major and its murine macrophage host cell reveals insights into host-pathogen interactions. BMC Genom. 2015, 16, 1108. [Google Scholar] [CrossRef] [PubMed]
- Camargo, E.P.; Coelho, J.A.; Moraes, G.; Figueiredo, E.N. Trypanosoma spp., leishmania spp. And leptomonas spp.: Enzymes of ornithine-arginine metabolism. Exp. Parasitol. 1978, 46, 141–144. [Google Scholar] [CrossRef]
- Roberts, S.C.; Tancer, M.J.; Polinsky, M.R.; Gibson, K.M.; Heby, O.; Ullman, B. Arginase plays a pivotal role in polyamine precursor metabolism in leishmania. Characterization of gene deletion mutants. J. Biol. Chem. 2004, 279, 23668–23678. [Google Scholar] [CrossRef] [PubMed]
- Muleme, H.M.; Reguera, R.M.; Berard, A.; Azinwi, R.; Jia, P.; Okwor, I.B.; Beverley, S.; Uzonna, J.E. Infection with arginase-deficient leishmania major reveals a parasite number-dependent and cytokine-independent regulation of host cellular arginase activity and disease pathogenesis. J. Immunol. 2009, 183, 8068–8076. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.M.; Mosser, D.M. The role of il-10 in promoting disease progression in leishmaniasis. J. Immunol. 2001, 166, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Corraliza, I.M.; Soler, G.; Eichmann, K.; Modolell, M. Arginase induction by suppressors of nitric oxide synthesis (il-4, il-10 and pge2) in murine bone-marrow-derived macrophages. Biochem. Biophys. Res. Commun. 1995, 206, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Laranjeira-Silva, M.F.; Floeter-Winter, L.M. Arginase in leishmania. In Proteins and Proteomics of Leishmania and Trypanossoma; Andre, L.S., Santos, M.H.B., Claudia, M., d´Avila-Levy, C.M., Kneipp, L.F., Sodre, C.L., Eds.; Springer: Berlin, Germany, 2014; pp. 103–118. [Google Scholar]
- Vieira, L.Q.; Goldschmidt, M.; Nashleanas, M.; Pfeffer, K.; Mak, T.; Scott, P. Mice lacking the tnf receptor p55 fail to resolve lesions caused by infection with leishmania major, but control parasite replication. J. Immunol. 1996, 157, 827–835. [Google Scholar]
- O’Neill, L.A.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef]
- Wu, G.; Morris, S.M., Jr. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336, 1–17. [Google Scholar] [CrossRef]
- Liese, J.; Schleicher, U.; Bogdan, C. Tlr9 signaling is essential for the innate nk cell response in murine cutaneous leishmaniasis. Eur. J. Immunol. 2007, 37, 3424–3434. [Google Scholar] [CrossRef]
- Nathan, C.; Shiloh, M.U. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 2000, 97, 8841–8848. [Google Scholar] [CrossRef] [PubMed]
- Lima-Junior, D.S.; Costa, D.L.; Carregaro, V.; Cunha, L.D.; Silva, A.L.; Mineo, T.W.; Gutierrez, F.R.; Bellio, M.; Bortoluci, K.R.; Flavell, R.A.; et al. Inflammasome-derived il-1beta production induces nitric oxide-mediated resistance to leishmania. Nat. Med. 2013, 19, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Kropf, P.; Freudenberg, M.A.; Modolell, M.; Price, H.P.; Herath, S.; Antoniazi, S.; Galanos, C.; Smith, D.F.; Muller, I. Toll-like receptor 4 contributes to efficient control of infection with the protozoan parasite leishmania major. Infect. Immun. 2004, 72, 1920–1928. [Google Scholar] [CrossRef] [PubMed]
- Gaur, U.; Roberts, S.C.; Dalvi, R.P.; Corraliza, I.; Ullman, B.; Wilson, M.E. An effect of parasite-encoded arginase on the outcome of murine cutaneous leishmaniasis. J. Immunol. 2007, 179, 8446–8453. [Google Scholar] [CrossRef] [PubMed]
- Reczkowski, R.S.; Ash, D.E. Rat liver arginase: Kinetic mechanism, alternate substrates, and inhibitors. Arch. Biochem. Biophys. 1994, 312, 31–37. [Google Scholar] [CrossRef]
- Shen, L.J.; Beloussow, K.; Shen, W.C. Accessibility of endothelial and inducible nitric oxide synthase to the intracellular citrulline-arginine regeneration pathway. Biochem. Pharmacol. 2005, 69, 97–104. [Google Scholar] [CrossRef]
- Diotallevi, M.; Checconi, P.; Palamara, A.T.; Celestino, I.; Coppo, L.; Holmgren, A.; Abbas, K.; Peyrot, F.; Mengozzi, M.; Ghezzi, P. Glutathione fine-tunes the innate immune response toward antiviral pathways in a macrophage cell line independently of its antioxidant properties. Front. Immunol. 2017, 8, 1239. [Google Scholar] [CrossRef]
- Oza, S.L.; Shaw, M.P.; Wyllie, S.; Fairlamb, A.H. Trypanothione biosynthesis in leishmania major. Mol. Biochem. Parasitol. 2005, 139, 107–116. [Google Scholar] [CrossRef]
- Canuto, G.A.; Castilho-Martins, E.A.; Tavares, M.; López-Gonzálvez, A.; Rivas, L.; Barbas, C. Ce-esi-ms metabolic fingerprinting of leishmania resistance to antimony treatment. Electrophoresis 2012, 33, 1901–1910. [Google Scholar] [CrossRef]
- Singh, R.J.; Hogg, N.; Joseph, J.; Kalyanaraman, B. Mechanism of nitric oxide release from s-nitrosothiols. J. Biol. Chem. 1996, 271, 18596–18603. [Google Scholar] [CrossRef]
- Nakano, T.; Goto, S.; Takaoka, Y.; Tseng, H.P.; Fujimura, T.; Kawamoto, S.; Ono, K.; Chen, C.L. A novel moonlight function of glyceraldehyde-3-phosphate dehydrogenase (gapdh) for immunomodulation. Biofactors 2018, 44, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Rojo, D.; Barbas, C.; Lopez-Gonzalvez, A. Metabolomics analysis of leishmania by capillary electrophoresis and mass spectrometry. Methods Mol. Biol. 2019, 1859, 253–260. [Google Scholar] [PubMed]
- Godzien, J.; Alonso-Herranz, V.; Barbas, C.; Armitage, E.G. Controlling the quality of metabolomics data: New strategies to get the best out of the qc sample. SpringerPlus 2015, 11, 518–528. [Google Scholar] [CrossRef]

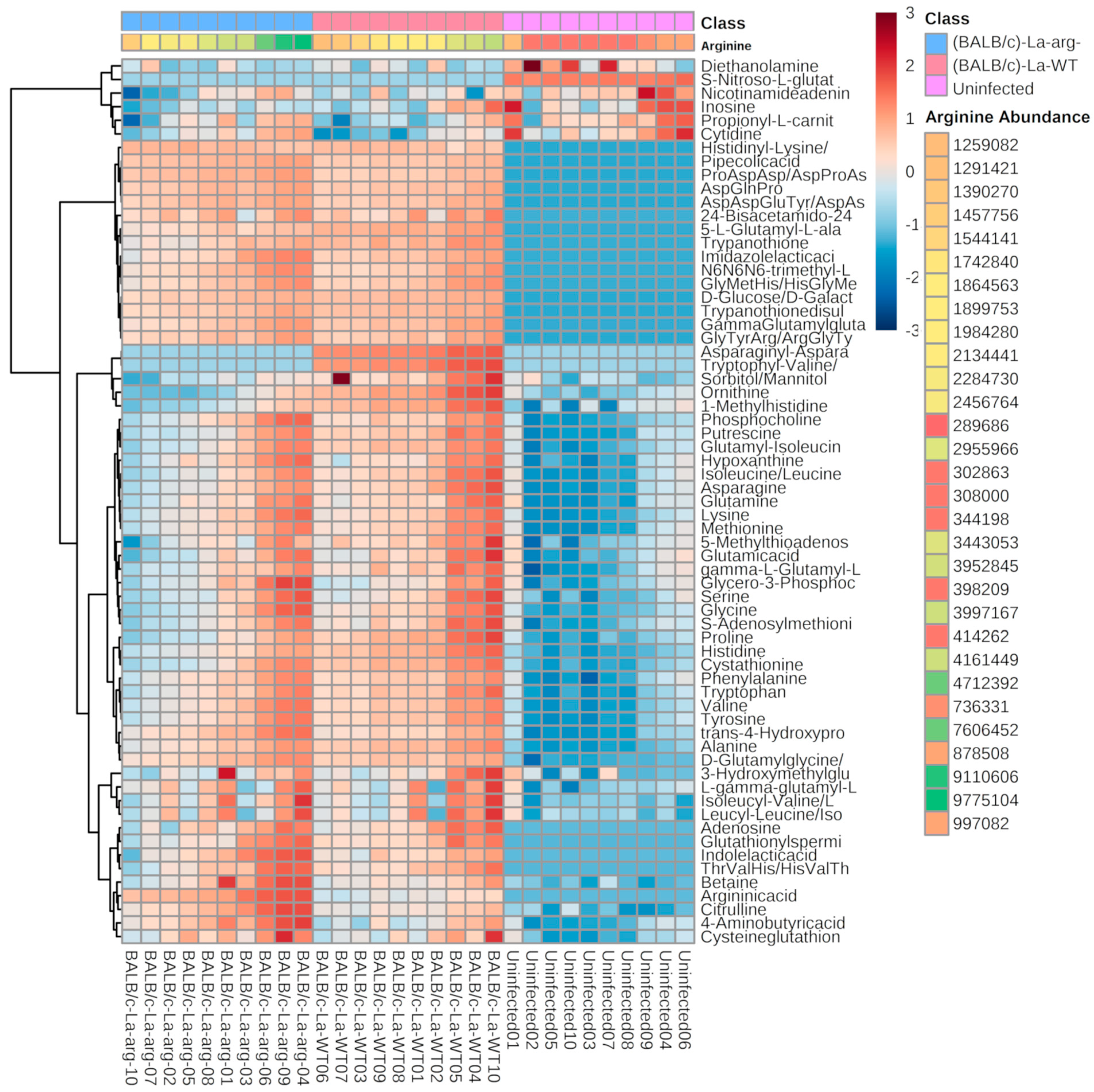
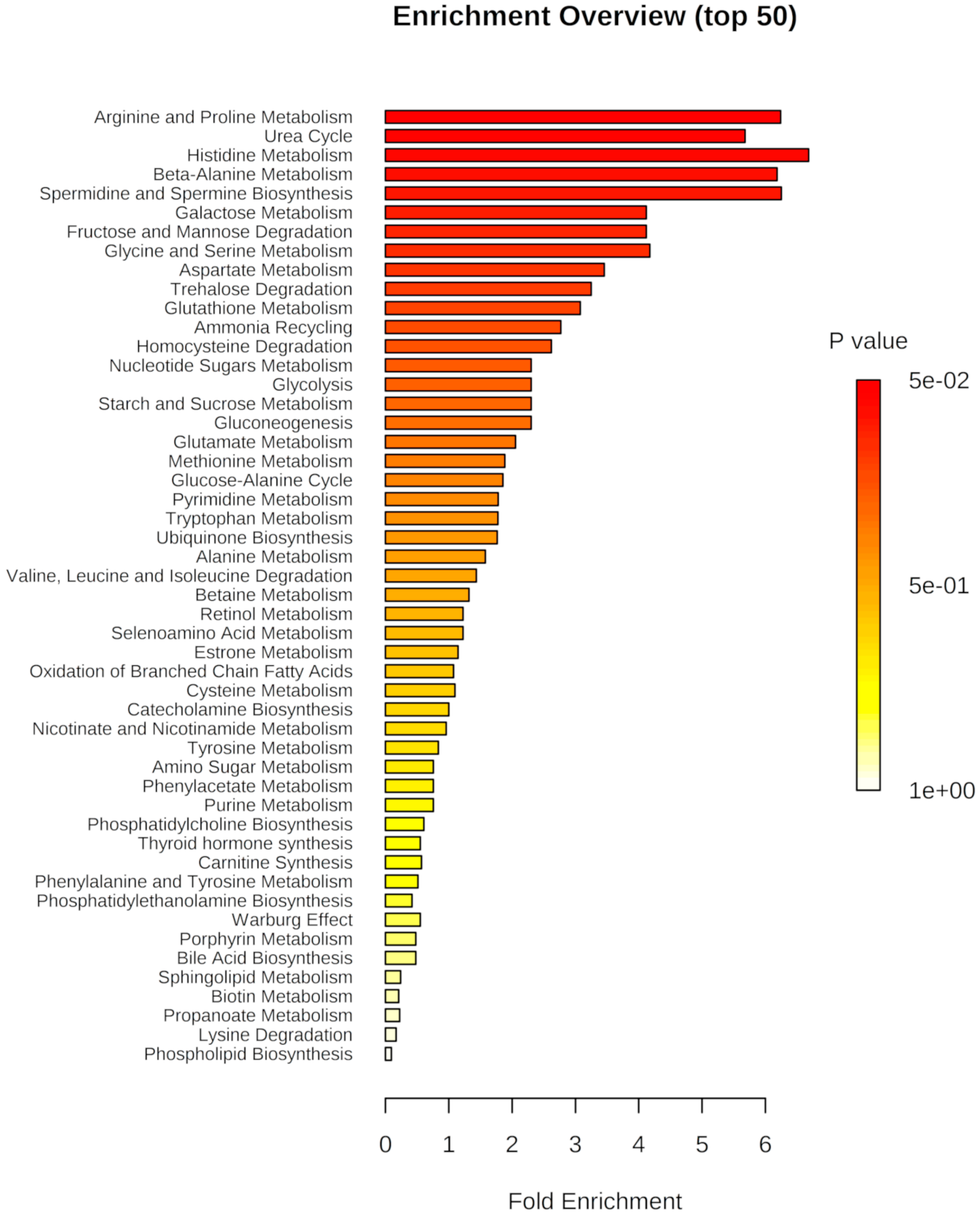
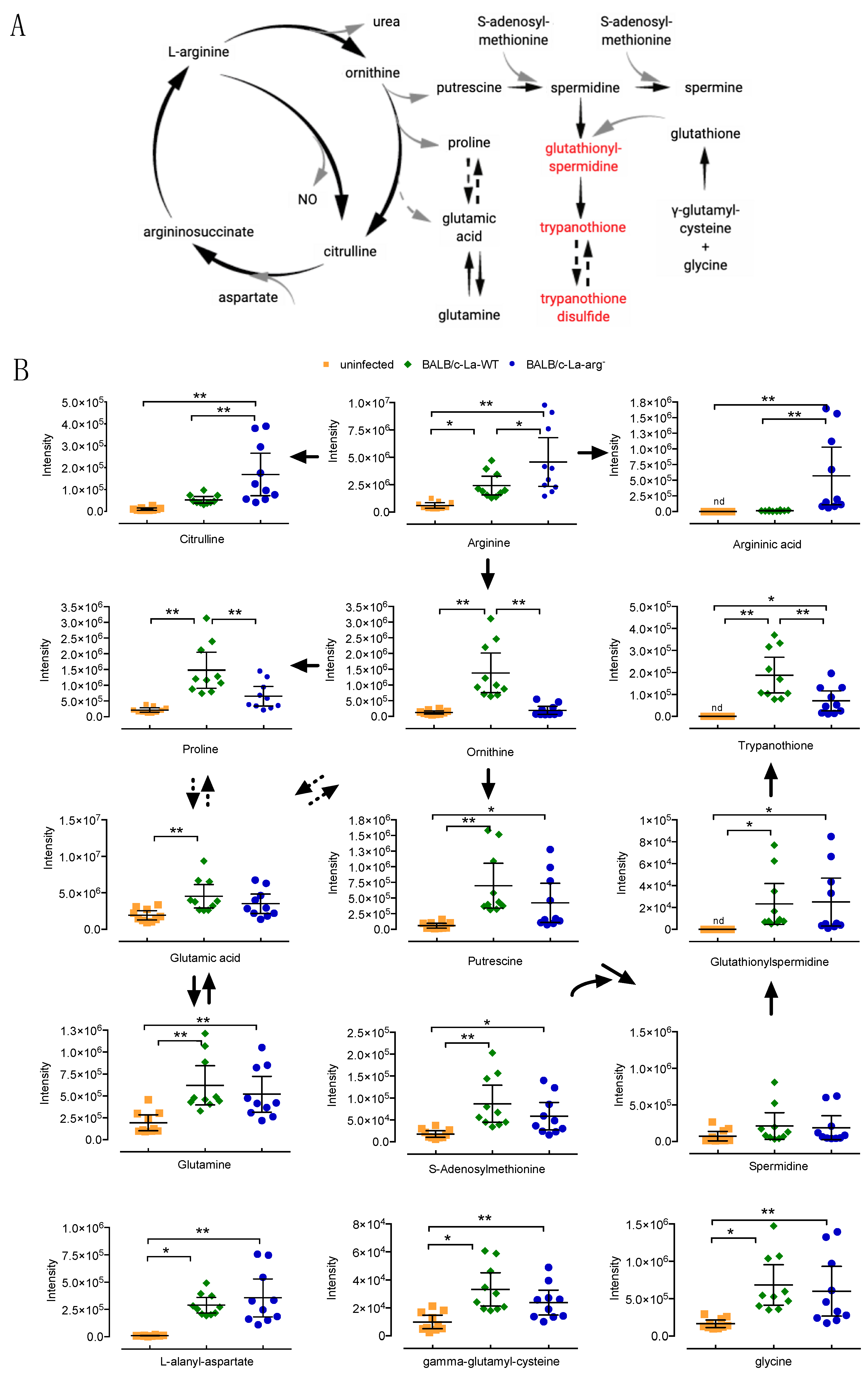
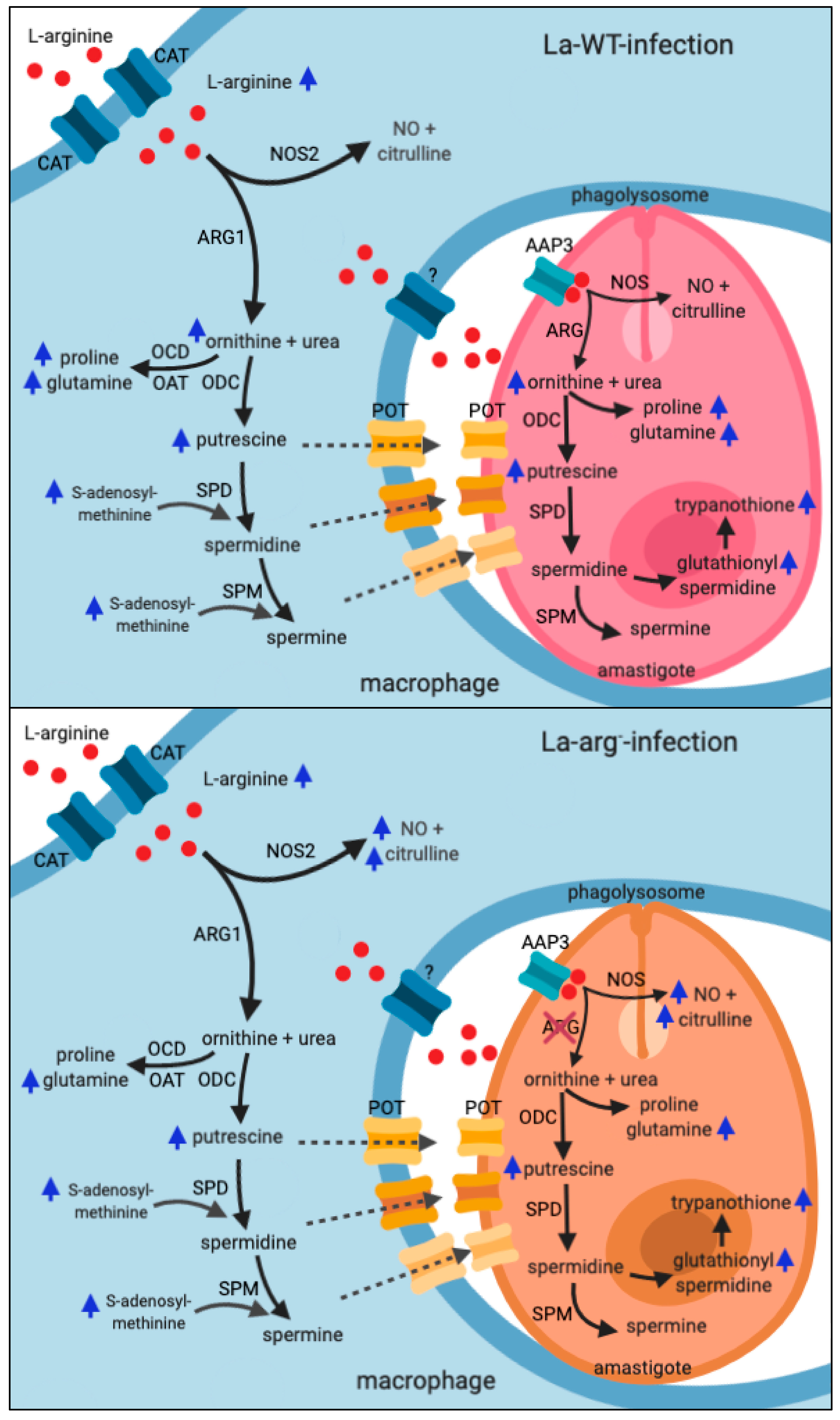
| (BALB/c)-La-WT vs. Uninfected | (BALB/c)-La-arg− vs. Uninfected | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Mass | RMT | %CV of CQ | p ANOVA | p FDR | p Value | % Change | P (Corr) | VIP | p Value | % Change | P (Corr) | VIP |
| Glycine | 75.0328 | 0.71 | 3.27 | 5.5 × 10−3 | 1.5 × 10−2 | 2.6 × 10−3 | 315.1 | 0.72 | <1 | 9.7 × 10−3 | 263.8 | 0.56 | 1.21 |
| Putrescine | 88.1001 | 0.41 | 1.58 | 3.8 × 10−3 | 1.1 × 10−2 | 9.6 × 10−4 | 1093.3 | 0.70 | <1 | 4.4 × 10−2 | 624.8 | 0.52 | 1.11 |
| Alanine | 89.0482 | 0.76 | 3.64 | 2.3 × 10−5 | 2.0 × 10−4 | 7.6 × 10−6 | 1964.2 | 0.85 | 4.44 | 4.4 × 10−4 | 1422.9 | 0.72 | 3.18 |
| 4-Aminobutyric acid | 103.0646 | 0.66 | 3.91 | 7.5 × 10−5 | 4.6 × 10−4 | 7.9 × 10−2 | NS | 0.61 | <1 | 1.9 × 10−5 | 356.8 | 0.73 | <1 |
| Serine | 105.0429 | 0.84 | 2.02 | 8.1 × 10−3 | 2.0 × 10−2 | 3.4 × 10−3 | 173.3 | 0.66 | <1 | 1.6 × 10−2 | 139.0 | 0.56 | 1.13 |
| Diethanolamine | 105.0780 | 0.65 | 7.09 | 1.5 × 10−2 | 3.3 × 10−2 | 1.3 × 10−2 | −72.0 | −0.44 | <1 | 1.0 × 10−2 | −74.2 | 0.46 | <1 |
| Proline | 115.0633 | 0.91 | 1.72 | 4.3 × 10−5 | 3.1 × 10−4 | 1.1 × 10−5 | 602.4 | 0.78 | 1.40 | 7.2 × 10−2 | NS | 0.59 | 1.21 |
| Valine | 117.0790 | 0.84 | 1.37 | 1.2 × 10−3 | 4.6 × 10−3 | 1.1 × 10−3 | 906.4 | 0.81 | 2.29 | 1.3 × 10−3 | 890.3 | 0.62 | 2.62 |
| Betaine | 117.0792 | 0.95 | 6.45 | 1.9 × 10−4 | 1.0 × 10−3 | 2.3 × 10−2 | 131.0 | 0.82 | <1 | 4.1 × 10−5 | 264.6 | 0.74 | <1 |
| Pipecolic acid | 129.0788 | 0.86 | 1.26 | 3.1 × 10−5 | 2.3 × 10−4 | 3.6 × 10−2 | ↑ | 0.87 | 1.50 | 6.9 × 10−6 | ↑ | 0.73 | 3.47 |
| trans-4-Hydroxyproline | 131.0585 | 1.01 | 2.18 | 2.8 × 10−4 | 1.4 × 10−3 | 8.0 × 10−3 | 725.3 | 0.81 | <1 | 6.8 × 10−5 | 1190.2 | 0.69 | 1.53 |
| Isoleucine/Leucine | 131.0944 | 0.87 | 1.74 | 2.1 × 10−3 | 6.6 × 10−3 | 5.0 × 10−4 | 467.8 | 0.70 | 4.42 | 3.8 × 10−2 | 257.9 | 0.56 | 2.63 |
| Asparagine | 132.0529 | 0.88 | 1.87 | 1.2 × 10−3 | 4.6 × 10−3 | 2.9 × 10−4 | 493.3 | 0.68 | <1 | 4.1 × 10−2 | 253.7 | 0.62 | <1 |
| Ornithine | 132.0896 | 0.59 | 4.29 | 8.4 × 10−6 | 1.2 × 10−4 | 9.8 × 10−6 | 1006.6 | 0.74 | 1.42 | 7.7 × 10−1 | NS | 0.28 | <1 |
| Hypoxanthine | 136.0383 | 1.01 | 2.84 | 7.1 × 10−3 | 1.8 × 10−2 | 5.8 × 10−3 | 391.5 | 0.66 | 1.48 | 5.8 × 10−3 | 391.2 | 0.59 | 2.00 |
| Glutamine | 146.0689 | 0.91 | 6.30 | 2.4 × 10−3 | 7.3 × 10−3 | 8.9 × 10−4 | 221.7 | 0.69 | <1 | 8.2 × 10−3 | 169.4 | 0.61 | 1.03 |
| Lysine | 146.1053 | 0.6 | 2.98 | 1.5 × 10−3 | 5.0 × 10−3 | 1.0 × 10−3 | 371.2 | 0.75 | 1.26 | 2.1 × 10−3 | 343.1 | 0.63 | 1.84 |
| Glutamic acid | 147.0530 | 0.92 | 1.55 | 9.5 × 10−3 | 2.3 × 10−2 | 2.6 × 10−3 | 135.5 | 0.65 | 3.21 | 5.4 × 10−2 | NS | 0.50 | 2.33 |
| Methionine | 149.0511 | 0.9 | 2.82 | 1.9 × 10−3 | 6.1 × 10−3 | 6.6 × 10−4 | 459.2 | 0.72 | <1 | 8.1 × 10−3 | 340.8 | 0.61 | 1.16 |
| Histidine | 155.0694 | 0.63 | 2.33 | 1.0 × 10−5 | 1.3 × 10−4 | 2.3 × 10−6 | 1552.6 | 0.80 | 1.74 | 2.5 × 10−2 | 615.4 | 0.68 | 1.45 |
| Imidazolelactic acid | 156.0560 | 0.75 | 9.11 | 1.5 × 10−2 | 3.3 × 10−2 | 8.7 × 10−2 | NS | NS | NS | 4.1 × 10−3 | ↑ | 0.53 | <1 |
| 3-Hydroxymethylglutaric acid/2-Hydroxyadipic acid/2-Ethylmalate | 162.0524 | 1.81 | 6.41 | 3.3 × 10−2 | 6.9 × 10−2 | 1.5 × 10−2 | 72.8 | 0.61 | <1 | 3.8 × 10−2 | 61.4 | 0.60 | <1 |
| Phenylalanine | 165.0803 | 0.93 | 1.84 | 4.1 × 10−4 | 1.7 × 10−3 | 2.9 × 10−4 | 538.5 | 0.82 | 1.05 | 8.2 × 10−4 | 487.2 | 0.65 | 1.71 |
| 1-Methylhistidine | 169.0863 | 0.65 | 4.07 | 1.2 × 10−6 | 2.5 × 10−5 | 1.0 × 10−6 | 549.1 | 0.79 | <1 | 4.7 × 10−1 | NS | 0.34 | <1 |
| Arginine | 174.1116 | 0.62 | 2.43 | 3.9 × 10−4 | 1.7 × 10−3 | 4.5 × 10−2 | 306.5 | 0.74 | 2.04 | 8.8 × 10−5 | 670.4 | 0.68 | 3.62 |
| Argininic acid | 175.0966 | 0.78 | 3.86 | 2.3 × 10−3 | 7.2 × 10−3 | 9.5 × 10−1 | NS | NS | NS | 2.0 × 10−3 | ↑ | 0.55 | 1.38 |
| Citrulline | 175.0975 | 0.93 | 2.33 | 3.9 × 10−4 | 1.7 × 10−3 | 2.5 × 10−1 | NS | 0.83 | <1 | 1.3 × 10−4 | 1497.9 | 0.64 | <1 |
| α-d-Glucose/d-Galactose/d-Fructose/d-(+)-Mannose/myo-Inositol | 180.0608 | 1.82 | 13.22 | 3.5 × 10−7 | 8.3 × 10−6 | 1.1 × 10−7 | ↑ | 0.90 | <1 | 2.6 × 10−5 | ↑ | 0.78 | 1.07 |
| Tyrosine | 181.0738 | 0.95 | 1.81 | 3.1 × 10−4 | 1.5 × 10−3 | 2.2 × 10−4 | 543.7 | 0.84 | <1 | 6.6 × 10−4 | 490.4 | 0.65 | 1.39 |
| Sorbitol/Mannitol | 182.0802 | 1.81 | 7.76 | 1.0 × 10−4 | 5.9 × 10−4 | 8.1 × 10−5 | 82.3 | 0.72 | <1 | 6.8 × 10−1 | NS | 0.39 | <1 |
| Phosphocholine | 183.0660 | 1.69 | 2.86 | 1.4 × 10−3 | 4.9 × 10−3 | 1.6 × 10−3 | 409.2 | 0.80 | 1.36 | 1.3 × 10−3 | 418.7 | 0.62 | 2.06 |
| N6, N6, N6-trimethyl-L-lysine | 188.1511 | 0.62 | 9.39 | 1.3 × 10−3 | 4.6 × 10−3 | 4.2 × 10−3 | ↑ | 0.73 | <1 | 5.3 × 10−4 | ↑ | 0.67 | <1 |
| γ-D-Glutamylglycine/L-beta-aspartyl-L-alanine/Alanyl-Aspartate/Aspartyl-Alanine | 204.0744 | 1.01 | 2.53 | 4.9 × 10−5 | 3.3 × 10−4 | 3.4 × 10−4 | 2788.0 | 0.91 | <1 | 2.3 × 10−5 | 3469.4 | 0.72 | 1.07 |
| Tryptophan | 204.0895 | 0.93 | 2.19 | 1.3 × 10−4 | 6.9 × 10−4 | 4.5 × 10−5 | 431.3 | 0.81 | <1 | 1.2 × 10−3 | 320.6 | 0.68 | <1 |
| Indolelactic acid | 205.0735 | 1.85 | 6.11 | 7.6 × 10−3 | 1.9 × 10−2 | 3.1 × 10−1 | NS | NS | NS | 2.4 × 10−3 | ↑ | 0.54 | <1 |
| Propionyl-L-carnitine | 217.1340 | 0.79 | 21.78 | 1.1 × 10−2 | 2.6 × 10−2 | 3.5 × 10−3 | −46.7 | −0.60 | <1 | 3.9 × 10−2 | −31.7 | −0.42 | <1 |
| 5-L-Glutamyl-L-alanine/Hydroxyprolyl-Serine | 218.0909 | 1.03 | 4.53 | 1.4 × 10−10 | 7.9 × 10−9 | 3.4 × 10−11 | ↑ | 0.93 | <1 | 9.8 × 10−4 | ↑ | 0.88 | <1 |
| Cystathionine | 222.0682 | 0.85 | 1.52 | 1.2 × 10−4 | 6.6 × 10−4 | 2.6 × 10−5 | 1112.1 | 0.84 | <1 | 9.5 × 10−3 | 613.3 | 0.57 | <1 |
| Isoleucyl-Valine/Leucyl-Valine/Valyl-Isoleucine/Valyl-Leucine | 230.1618 | 0.89 | 5.45 | 1.1 × 10−2 | 2.5 × 10−2 | 1.0 × 10−2 | 150.3 | 0.67 | <1 | 7.0 × 10−3 | 159.3 | 0.63 | <1 |
| Cytidine | 243.0889 | 0.82 | 5.47 | 5.8 × 10−3 | 1.5 × 10−2 | 1.6 × 10−3 | −67.6 | −0.59 | <1 | 3.8 × 10−2 | −42.0 | −0.38 | <1 |
| Leucyl-Leucine/Isoleucyl-Leucine/Leucyl-Isoleucine | 244.1798 | 0.9 | 6.33 | 3.0 × 10−2 | 6.3 × 10−2 | 1.4 × 10−2 | 192.3 | 0.61 | <1 | 3.5 × 10−2 | 162.7 | 0.57 | <1 |
| Asparaginyl-Asparagine/N2-Oxalylarginine | 246.0966 | 1.26 | 3.24 | 2.9 × 10−7 | 8.1 × 10−6 | 6.7 × 10−7 | ↑ | 0.79 | <1 | 1.0 × 100 | NS | NS | NS |
| 2,4Bis(acetamido)2,4,6trideoxy-betaL-altropyranose/L-beta-aspartyl-L-leucine/L-gamma-glutamyl-L-valine/Aspartyl-Isoleucine/aspartyl-Leucine/Isoleucyl-Aspartate/LeucylAspartate/Glu-Val | 246.1227 | 0.91 | 10.84 | 2.6 × 10−3 | 7.8 × 10−3 | 1.3 × 10−3 | ↑ | 0.72 | <1 | 4.9 × 10−3 | ↑ | 0.72 | <1 |
| gamma-L-Glutamyl-L-cysteine | 250.0628 | 1.07 | 15.49 | 1.2 × 10−3 | 4.6 × 10−3 | 3.1 × 10−4 | 233.1 | 0.71 | <1 | 2.1 × 10−2 | 138.7 | 0.58 | <1 |
| Glycero-3-Phosphocholine | 257.1028 | 1.78 | 4.73 | 1.5 × 10−2 | 3.3 × 10−2 | 3.6 × 10−2 | 136.4 | 0.61 | <1 | 5.3 × 10−3 | 188.1 | 0.57 | 1.69 |
| L-gamma-glutamyl-L-isoleucine | 260.1390 | 0.91 | 7.49 | 4.7 × 10−2 | 9.1 × 10−2 | 1.9 × 10−2 | 427.3 | 0.57 | <1 | 6.8 × 10−2 | NS | 0.55 | <1 |
| Adenosine | 267.0975 | 0.84 | 5.60 | 3.4 × 10−2 | 7.0 × 10−2 | 1.2 × 10−2 | ↑ | 0.56 | <1 | 7.3 × 10−2 | NS | 0.54 | <1 |
| Inosine | 268.0799 | 1.65 | 2.07 | 2.5 × 10−2 | 5.4 × 10−2 | 7.3 × 10−2 | NS | −0.36 | <1 | 8.0 × 10−3 | −78.4 | −0.53 | <1 |
| Gamma Glutamylglutamic acid | 276.0941 | 1.1 | 7.21 | 9.8 × 10−5 | 5.8 × 10−4 | 6.0 × 10−5 | ↑ | 0.82 | <1 | 3.5 × 10−4 | ↑ | 0.71 | <1 |
| Histidinyl-Lysine/Lysyl-Histidine | 283.1573 | 0.83 | 28.48 | 1.3 × 10−11 | 1.1 × 10−9 | 1.0 × 10−9 | ↑ | 0.90 | <1 | 7.5 × 10−12 | ↑ | 0.95 | <1 |
| 5’-Methylthioadenosine | 297.0882 | 0.86 | 25.00 | 4.1 × 10−3 | 1.2 × 10−2 | 1.0 × 10−3 | 262.7 | 0.67 | <1 | 9.4 × 10−2 | NS | 0.50 | <1 |
| Tryptophyl-Valine/Valyl-Tryptophan | 303.1548 | 0.75 | 6.01 | 1.4 × 10−5 | 1.6 × 10−4 | 2.3 × 10−5 | ↑ | 0.71 | <1 | 1.0 × 100 | NS | NS | NS |
| S-Nitroso-L-glutathione | 336.0745 | 1.12 | ND | 5.0 × 10−18 | 8.3 × 10−16 | 3.0 × 10−17 | ↓ | −0.96 | <1 | 3.0 × 10−17 | ↓ | −0.96 | <1 |
| Gly Met His/His Gly Met/Met His Gly/Met Gly His/Gly His Met/His Met Gly | 343.1361 | 0.84 | 11.05 | 4.7 × 10−3 | 1.3 × 10−2 | 3.4 × 10−3 | ↑ | 0.69 | <1 | 4.9 × 10−3 | ↑ | 0.61 | <1 |
| Pro Asp Asp/Asp Pro Asp/Asp Asp Pro | 345.1186 | 1.8 | 2.76 | 3.0 × 10−6 | 5.0 × 10−5 | 1.6 × 10−2 | ↑ | 0.85 | <1 | 6.4 × 10−7 | ↑ | 0.79 | 1.32 |
| Thr Val His/His Val Thr/Leu Ser His/His Thr Val/Val Thr His/His Ile Ser/Ser His Ile/His Leu Ser/Ile Ser His/Val His Thr/His Ser Leu/His Ser Ile/Ser Ile His/Ser Leu His/Ser His Leu/Ile His Ser/Leu His Ser/Thr His Val | 355.1791 | 0.84 | 23.12 | 3.6 × 10−2 | 7.2 × 10−2 | 9.4 × 10−2 | NS | 0.59 | <1 | 1.1 × 10−2 | ↑ | 0.51 | <1 |
| Asp Gln Pro | 358.1424 | 0.83 | 11.91 | 1.5 × 10−7 | 5.1 × 10−6 | 2.7 × 10−6 | ↑ | 0.88 | <1 | 8.9 × 10−8 | ↑ | 0.86 | <1 |
| Gly Tyr Arg/Arg Gly Tyr/Tyr Gly Arg/Gly Arg Tyr/Arg Tyr Gly/Tyr Arg Gly | 394.1954 | 0.99 | 5.96 | 1.9 x 10−5 | 1.9 × 10−4 | 2.8 × 10−5 | ↑ | 0.84 | <1 | 3.3 × 10−5 | ↑ | 0.80 | <1 |
| S-Adenosylmethionine | 398.1381 | 0.62 | 2.50 | 4.7 × 10−3 | 1.3 × 10−2 | 1.3 × 10−3 | 389.4 | 0.67 | <1 | 4.2 × 10−2 | 230.3 | 0.55 | <1 |
| Cysteineglutathione disulfide | 426.0907 | 0.98 | 27.11 | 2.4 × 10−2 | 5.1 × 10−2 | 5.0 × 10−2 | 253.3 | 0.54 | <1 | 8.3 × 10−3 | 351.3 | 0.60 | <1 |
| Glutathionylspermidine | 434.2371 | 0.64 | 15.79 | 4.1 × 10−2 | 8.2 × 10−2 | 3.4 × 10−2 | ↑ | 0.58 | <1 | 2.3 × 10−2 | ↑ | 0.52 | <1 |
| Asp Asp Glu Tyr/Asp Asp Tyr Glu/Asp Glu Asp Tyr/Asp Glu Tyr Asp/Asp Tyr Asp Glu/Asp Tyr Glu Asp/Glu Asp Asp Tyr/Glu Asp Tyr Asp/Glu Tyr Asp Asp/Tyr Asp Asp Glu/Tyr Asp Glu Asp/Tyr Glu Asp Asp | 540.1670 | 0.83 | 3.91 | 7.7 × 10−6 | 1.2 × 10−4 | 1.3 × 10−4 | ↑ | 0.82 | <1 | 3.0 × 10−6 | ↑ | 0.83 | <1 |
| Nicotinamide adenine dinucleotide (NAD) | 663.1087 | 1.81 | 4.11 | 1.6 × 10−2 | 3.5 × 10−2 | 1.0 × 10−2 | −51.8 | −0.44 | <1 | 1.4 × 10−2 | −49.4 | −0.42 | <1 |
| Trypanothione disulfide | 721.2894 | 0.81 | 13.64 | 2.2 × 10−4 | 1.1 × 10−3 | 6.4 × 10−5 | ↑ | 0.81 | <1 | 3.2 × 10−3 | ↑ | 0.66 | 1.34 |
| Trypanothione | 723.3058 | 0.83 | 26.04 | 2.6 × 10−5 | 2.2 × 10−4 | 6.1 × 10−6 | ↑ | 0.78 | <1 | 4.4 × 10−2 | ↑ | 0.63 | <1 |
| (BALB/c)-La-arg− vs. (BALB/c)-La-WT | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | MASS | RMT | % CV of QC | p ANOVA | p FDR | p Value | % Change | p (Corr) | VIP |
| 4-Aminobutyric acid | 103.0646 | 0.66 | 3.91 | 7.5 × 10−5 | 4.6 × 10−4 | 2.4 × 10−3 | 102.17 | 0.56 | <1 |
| Proline | 115.0633 | 0.91 | 1.72 | 4.3 × 10−5 | 3.1 × 10−4 | 1.6 × 10−3 | −55.90 | −0.59 | 1.74 |
| Betaine | 117.0792 | 0.95 | 6.45 | 1.9×10−4 | 1.0 × 10−3 | 2.0 × 10−2 | 57.84 | 0.50 | <1 |
| Pipecolic acid | 129.0788 | 0.86 | 1.26 | 3.1 × 10−5 | 2.3 × 10−4 | 2.4 × 10−3 | 152.06 | 0.56 | 4.90 |
| Ornithine | 132.0896 | 0.59 | 4.29 | 8.4 × 10−6 | 1.2 × 10−4 | 2.2 × 10−5 | −86.03 | −0.75 | 2.44 |
| Histidine | 155.0694 | 0.63 | 2.33 | 1.0 × 10−5 | 1.3 × 10−4 | 1.2 × 10−3 | −56.71 | −0.59 | 2.03 |
| 1-Methylhistidine | 169.0863 | 0.65 | 4.07 | 1.2 × 10−6 | 2.5 × 10−5 | 7.0 × 10−6 | −74.72 | −0.76 | <1 |
| Arginine | 174.1116 | 0.62 | 2.43 | 3.9 × 10−4 | 1.7 × 10−3 | 1.9 × 10−2 | 89.54 | 0.44 | 5.13 |
| Argininic acid | 175.0966 | 0.78 | 3.86 | 2.3 × 10−3 | 7.2 × 10−3 | 2.4 × 10−3 | 4881.40 | 0.56 | 1.24 |
| Citrulline | 175.0975 | 0.93 | 2.33 | 3.9 × 10−4 | 1.7 × 10−3 | 3.0 × 10−3 | 220.22 | 0.55 | <1 |
| α-D-Glucose/D-Galactose/D-Fructose/D-(+)-Mannose/myo-Inositol | 180.0608 | 1.82 | 13.22 | 3.5 × 10−7 | 8.3 × 10−6 | 4.5 × 10−2 | −29.34 | −0.35 | <1 |
| Sorbitol/Mannitol | 182.0802 | 1.81 | 7.76 | 1.0 × 10−4 | 5.9 × 10−4 | 2.5 × 10−4 | −41.14 | −0.61 | <1 |
| Indolelactic acid | 205.0735 | 1.85 | 6.11 | 7.6 × 10−3 | 1.9 × 10−2 | 2.9 × 10−2 | 221.22 | 0.43 | <1 |
| 5-L-Glutamyl-L-alanine/Hydroxyprolyl-Serine | 218.0909 | 1.03 | 4.53 | 1.4 × 10−10 | 7.9 × 10−9 | 1.6 × 10−7 | −65.39 | −0.79 | <1 |
| Cystathionine | 222.0682 | 0.85 | 1.52 | 1.2 × 10−4 | 6.6 × 10−4 | 3.1 × 10−2 | −41.16 | −0.38 | <1 |
| Asparaginyl-Asparagine/N2-Oxalylarginine | 246.0966 | 1.26 | 3.24 | 2.9 × 10−7 | 8.1 × 10−6 | 6.7 × 10−7 | ↓ | −0.81 | <1 |
| Histidinyl-Lysine/Lysyl-Histidine | 283.1573 | 0.83 | 28.48 | 1.3 × 10−11 | 1.1 × 10−9 | 2.8 × 10−2 | 25.52 | 0.37 | <1 |
| Tryptophyl-Valine/Valyl-Tryptophan | 303.1548 | 0.75 | 6.01 | 1.4 × 10−5 | 1.6 × 10−4 | 2.3 × 10−5 | ↓ | −0.76 | <1 |
| Pro Asp Asp/Asp Pro Asp/Asp Asp Pro | 345.1186 | 1.8 | 2.76 | 3.0 × 10−6 | 5.0 × 10−5 | 5.8 × 10−4 | 152.34 | 0.62 | <1 |
| Trypanothione | 723.3058 | 0.83 | 26.04 | 2.6 × 10−5 | 2.2 × 10−4 | 1.7 × 10−3 | −62.23 | −0.57 | <1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muxel, S.M.; Mamani-Huanca, M.; Aoki, J.I.; Zampieri, R.A.; Floeter-Winter, L.M.; López-Gonzálvez, Á.; Barbas, C. Metabolomic Profile of BALB/c Macrophages Infected with Leishmania amazonensis: Deciphering L-Arginine Metabolism. Int. J. Mol. Sci. 2019, 20, 6248. https://doi.org/10.3390/ijms20246248
Muxel SM, Mamani-Huanca M, Aoki JI, Zampieri RA, Floeter-Winter LM, López-Gonzálvez Á, Barbas C. Metabolomic Profile of BALB/c Macrophages Infected with Leishmania amazonensis: Deciphering L-Arginine Metabolism. International Journal of Molecular Sciences. 2019; 20(24):6248. https://doi.org/10.3390/ijms20246248
Chicago/Turabian StyleMuxel, Sandra Marcia, Maricruz Mamani-Huanca, Juliana Ide Aoki, Ricardo Andrade Zampieri, Lucile Maria Floeter-Winter, Ángeles López-Gonzálvez, and Coral Barbas. 2019. "Metabolomic Profile of BALB/c Macrophages Infected with Leishmania amazonensis: Deciphering L-Arginine Metabolism" International Journal of Molecular Sciences 20, no. 24: 6248. https://doi.org/10.3390/ijms20246248
APA StyleMuxel, S. M., Mamani-Huanca, M., Aoki, J. I., Zampieri, R. A., Floeter-Winter, L. M., López-Gonzálvez, Á., & Barbas, C. (2019). Metabolomic Profile of BALB/c Macrophages Infected with Leishmania amazonensis: Deciphering L-Arginine Metabolism. International Journal of Molecular Sciences, 20(24), 6248. https://doi.org/10.3390/ijms20246248







