Functional Characteristics of Caffeoyl Shikimate Esterase in Larix Kaempferi and Monolignol Biosynthesis in Gymnosperms
Abstract
:1. Introduction
2. Results
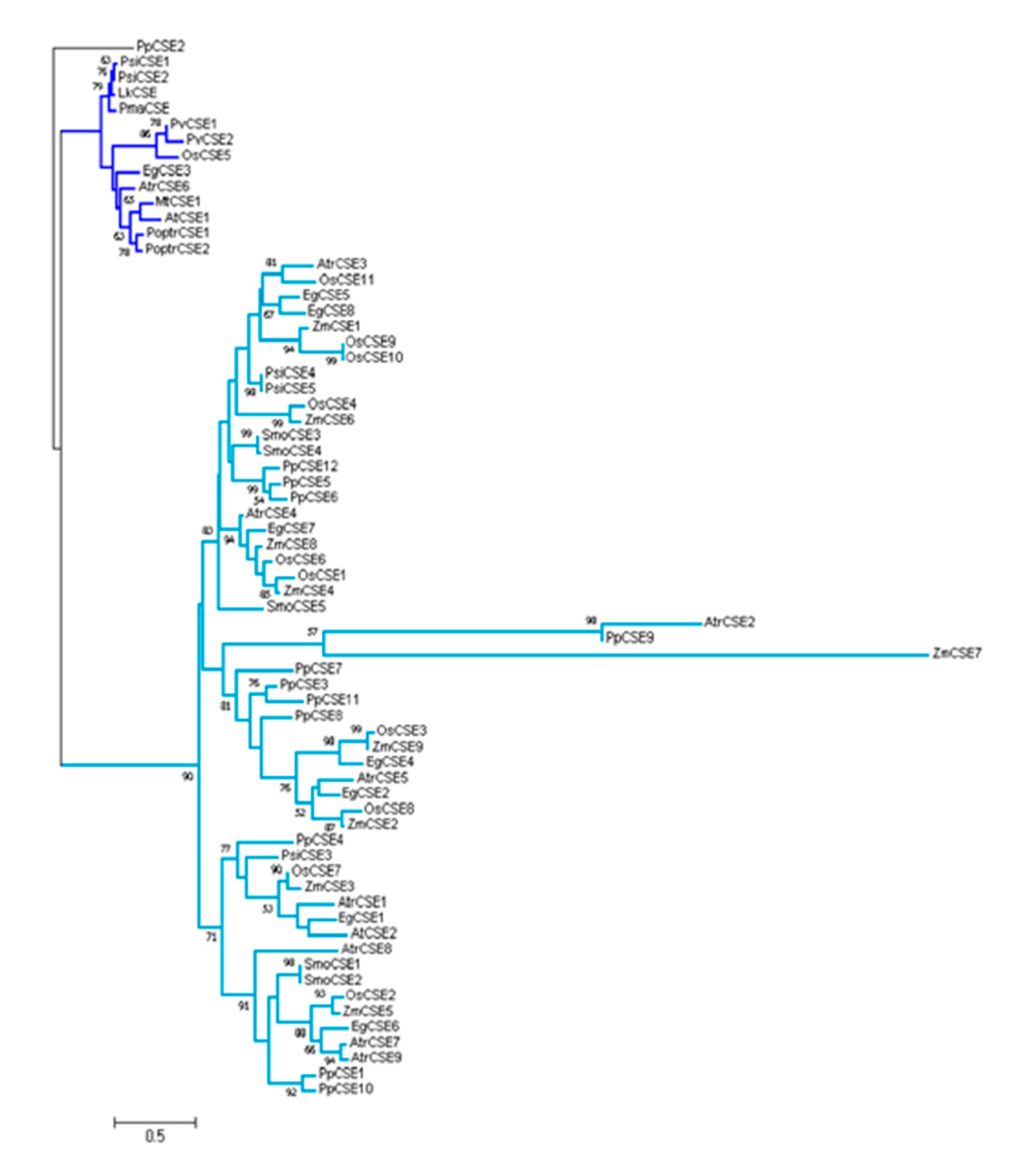
2.1. Identification and Cloning of Caffeoyl Shikimate Esterase (CSE) Ortholog in Larix Kaempferi
2.2. LkCSE —A CSE Ortholog with Conserved Motifs
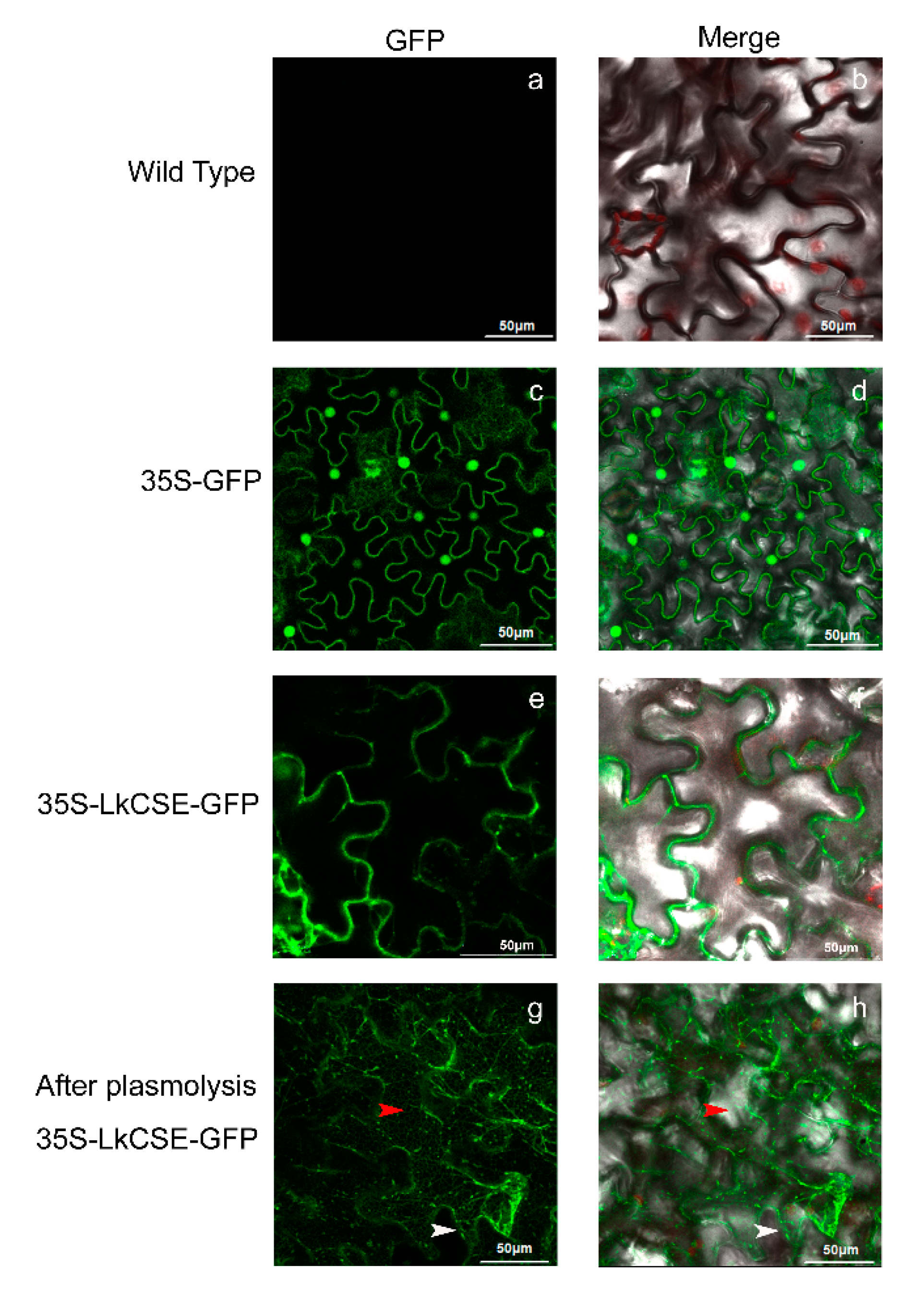
2.3. Subcellular Localization of LkCSE
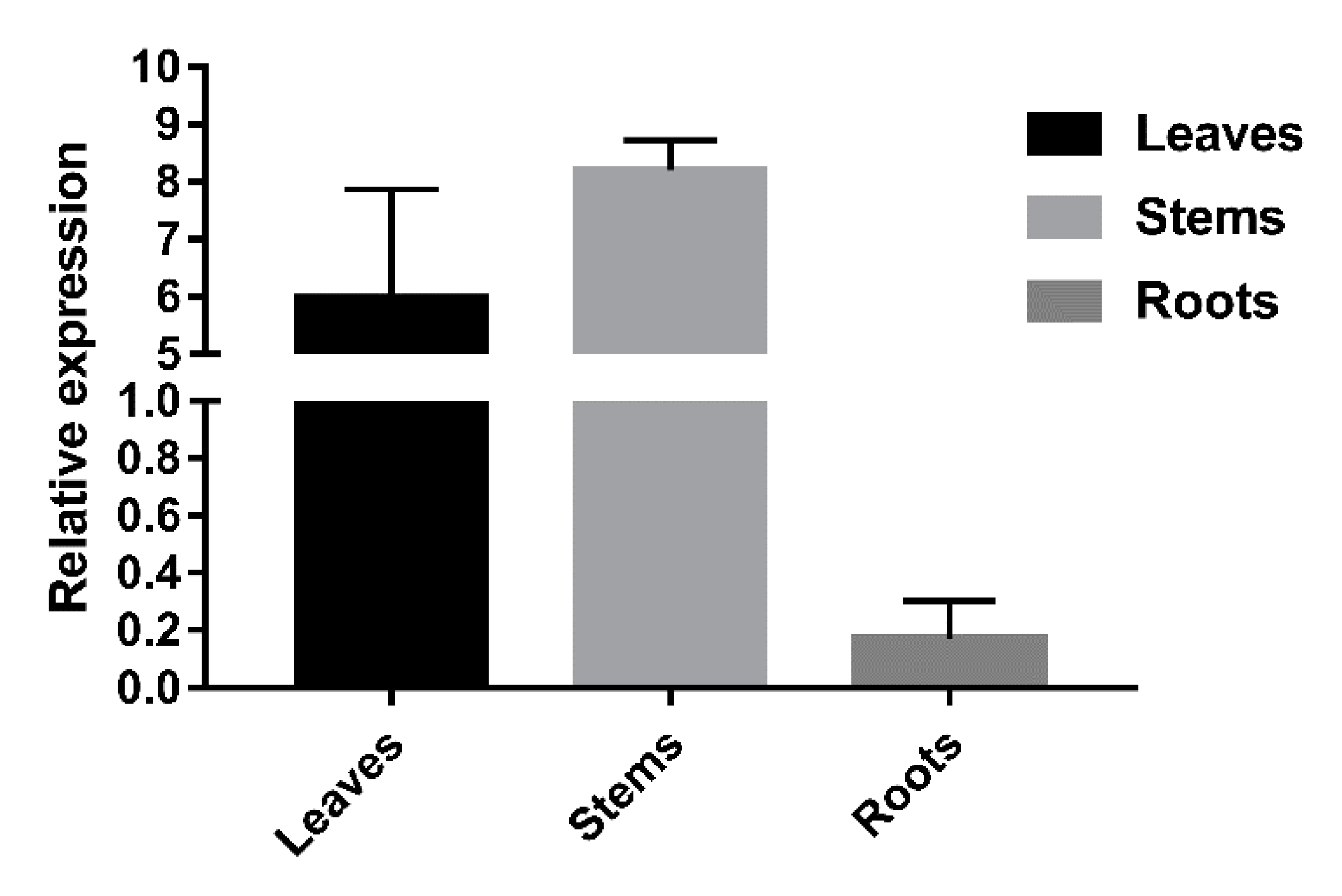
2.4. Tissue Expression Profile of LkCSE Revealed Tissue Specificity
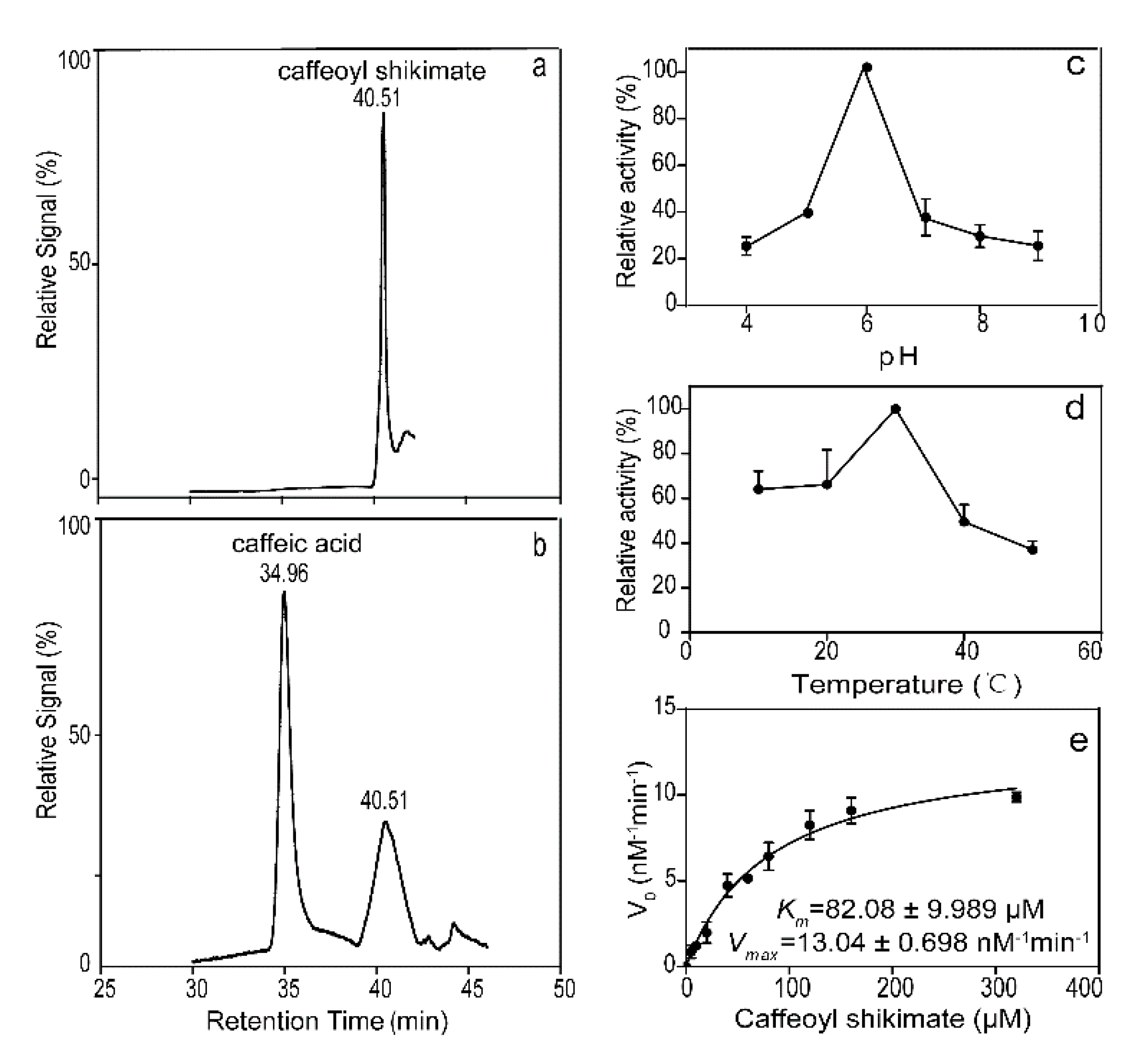
2.5. LkCSE Converts Caffeoyl Shikimate to Caffeate and Shikimate
3. Discussion
3.1. CSE Functions in Gymnosperms
3.2. ER Resident CSE Provides Insight for Monolignol Biosynthesis Flux Regulation
4. Materials and Methods
4.1. Plant Materials
4.2. Transcriptome Based Identification of CSE Genes in L. Kaempferi
4.3. CSE Sequence Alignments and Phylogenetic Analyses
4.4. Subcellular Location of LkCSE
4.5. LkCSE Expression Profile in Different Tissues
4.6. Purification of Recombinant LkCSE
4.7. High-performance Liquid Chromatography–mass Spectrometry Based Enzymatic Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 4CL | 4-coumarate: CoA ligase |
| ACBP2 | Acyl-CoA-binding protein 2 |
| C3’H | P-coumarate 3-hydroxylase |
| C4H | Cinnamate 4-hydroxylase |
| CAD | Cinnamyl alcohol dehydrogenase |
| CCR | Cinnamoyl-CoA reductase |
| EF1A1 | Elongation factor-1 alpha 1 |
| ER | Endoplasmic reticulum |
| F5H | Ferulate 5-hydroxylase |
| GFP | Green fluorescent protein |
| HCT | Hydroxylcinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase |
| LysoPC | Lysophosphatidylcholine |
| LysoPL2 | Lysophospholipase 2 |
| PAL | L-phenylalanine ammonia-lyase |
References
- Bhuiyan, N.H.; Gopalan, S.; Yangdou, W.; John, K. Role of lignification in plant defense. Plant Signal. Behav. 2009, 4, 158–159. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Ligninbiosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; George, B.; John, C.; Eckert, C.A.; Frederick, W.J.; Hallett, J.P.; Liotta, C.L. The path forward for biofuels and biomaterials. Science 2006, 311, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Demedts, B.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin Biosynthesis and Structure. Plant Physiol. 2010, 153, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Poovaiah, C.R.; Madhugiri, N.R.; Soneji, J.R.; Baxter, H.L.; Stewart, C.N. Altered lignin biosynthesis using biotechnology to improve lignocellulosic biofuel feedstocks. Plant Biotechnol. J. 2015, 12, 1163–1173. [Google Scholar] [CrossRef]
- Li, X.; Chapple, C. Understanding Lignification: Challenges Beyond Monolignol Biosynthesis. Plant Physiol. 2010, 154, 449–452. [Google Scholar] [CrossRef]
- Stewart, J.J.; Takuya, A.; Clint, C.; John, R.; Mansfield, S.D. The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar. Plant Physiol. 2009, 150, 621–635. [Google Scholar] [CrossRef]
- Tang, W.; Tang, A.Y. Transgenic woody plants for biofuel. J. For. Res. 2014, 25, 225–236. [Google Scholar] [CrossRef]
- Ruben, V.; Kris, M.; Chiarina, D.; Paula, O.; Grabber, J.H.; John, R.; Wout, B. Metabolic engineering of novel lignin in biomass crops. New Phytol. 2012, 196, 978–1000. [Google Scholar]
- Voelker, S.L.; Barbara, L.; Meinzer, F.C.; Strauss, S.H. Reduced wood stiffness and strength, and altered stem form, in young antisense 4CL transgenic poplars with reduced lignin contents. New Phytol. 2011, 189, 1096–1109. [Google Scholar] [CrossRef]
- Umezawa, T.; Bunzel, M.; Ralph, J. The cinnamate/monolignol pathway. Phytochem. Rev. 2010, 9, 1–17. [Google Scholar] [CrossRef]
- Jing-Ke, W.; Takuya, A.; Bonawitz, N.D.; Xu, L.; John, R.; Clint, C. Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 2010, 22, 1033–1045. [Google Scholar]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation Is a Major Regulator of Phenylpropanoid Availability and Biological Activity in Plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef] [PubMed]
- Saleme, M.L.S.; Cesarino, I.; Vargas, L.; Kim, H.; Vanholme, R.; Goeminne, G.; Van Acker, R.; Fonseca, F.C.A.; Pallidis, A.; Voorend, W.; et al. Silencing CAFFEOYL SHIKIMATE ESTERASE Affects Lignification and Improves Saccharification in Poplar. Plant Physiol. 2017, 175, 1040–1057. [Google Scholar] [CrossRef]
- Rochus, F.; Hemm, M.R.; Denault, J.W.; Ruegger, M.O.; Humphreys, J.M.; Clint, C. Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 2010, 30, 47–59. [Google Scholar]
- Rochus, F.; Humphreys, J.M.; Hemm, M.R.; Denault, J.W.; Ruegger, M.O.; Cusumano, J.C.; Clint, C. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2010, 30, 33–45. [Google Scholar]
- Kim, B.G.; Kim, I.A.; Ahn, J.H. Characterization of hydroxycinnamoyl-coenzyme a Shikimate hydroxycinnamoyltransferase from Populus euramericana. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 817–821. [Google Scholar] [CrossRef]
- Wagner, A.; Ralph, J.; Akiyama, T.; Flint, H.; Phillips, L.; Torr, K.; Nanayakkara, B.; Te Kiri, L. Exploring lignification in conifers by silencing hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata. Proc. Natl. Acad. Sci. USA 2007, 104, 11856–11861. [Google Scholar] [CrossRef]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl Shikimate Esterase (CSE) Is an Enzyme in the Lignin Biosynthetic Pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef]
- Wang, J.P.; Naik, P.P.; Chen, H.-C.; Shi, R.; Lin, C.-Y.; Liu, J.; Shuford, C.M.; Li, Q.; Sun, Y.-H.; Tunlaya-Anukit, S.; et al. Complete Proteomic-Based Enzyme Reaction and Inhibition Kinetics Reveal How Monolignol Biosynthetic Enzyme Families Affect Metabolic Flux and Lignin in Populus trichocarpa. Plant Cell 2014, 26, 894–914. [Google Scholar] [CrossRef]
- Escamilla-Treviño, L.L.; Shen, H.; Hernandez, T.; Yin, Y.; Xu, Y.; Dixon, R.A. Early lignin pathway enzymes and routes to chlorogenic acid in switchgrass (Panicum virgatum L.). Plant Mol. Biol. 2014, 84, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.M.; Escamilla-Trevino, L.; Yarce, J.C.; Kim, H.; Ralph, J.; Chen, F.; Dixon, R.A. An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J. 2016, 86, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Chapple, C. The Genetics of Lignin Biosynthesis: Connecting Genotype to Phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Li, H.-Y.; Xiao, S.; Chye, M.-L. Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J. 2010, 5, 1025–1027. [Google Scholar] [CrossRef]
- Vijayaraj, P.; Jashal, C.B.; Vijayakumar, A.; Rani, S.H.; Venkata Rao, D.K.; Rajasekharan, R. A Bifunctional Enzyme That Has Both Monoacylglycerol Acyltransferase and Acyl Hydrolase Activities. Plant Physiol. 2012, 160, 667–683. [Google Scholar] [CrossRef]
- Chen, H.C.; Li, Q.; Shuford, C.M.; Liu, J.; Muddiman, D.C.; Sederoff, R.R.; Chiang, V.L. Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, 21253–21258. [Google Scholar] [CrossRef]
- Shi, R.; Sun, Y.-H.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V.L. Towards a Systems Approach for Lignin Biosynthesis in Populus trichocarpa: Transcript Abundance and Specificity of the Monolignol Biosynthetic Genes. Plant Cell Physiol. 2010, 51, 144–163. [Google Scholar] [CrossRef]
- Muir, J.W.; Morrison, R.I.; Bown, C.J.; Logan, J. The Mobilization of Iron by Aqueous Extracts of Plants. J. Soil Sci. 1964, 15, 220–225. [Google Scholar] [CrossRef]
- Sarkar, S.K.; Malhotra, S.S. Gas-liquid chromatographic method for separation of organic acids and its application of pine needle extracts. J. Chromatogr. A 1979, 171, 227–232. [Google Scholar] [CrossRef]
- Wang, J.P.; Liu, B.; Sun, Y.; Chiang, V.L.; Sederoff, R.R. Enzyme-Enzyme Interactions in Monolignol Biosynthesis. Front. Plant Sci. 2018, 9, 1942. [Google Scholar] [CrossRef]
- Yan, X.; Liu, J.; Kim, H.; Liu, B.; Huang, X.; Yang, Z.; Lin, Y.-C.J.; Chen, H.; Yang, C.; Wang, J.P.; et al. CAD1 and CCR2 protein complex formation in monolignol biosynthesis in Populus trichocarpa. New Phytol. 2019, 222, 244–260. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Lung, S.-C.; Li, X.; Li, X.D.; Chye, M.-L. Thermodynamic insights into an interaction between ACYL-COA-BINDING PROTEIN2 and LYSOPHOSPHOLIPASE2 in Arabidopsis. J. Biol. Chem. 2019, 294, 6214–6226. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Filipski, A.; Peterson, D.; Stecher, G.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Yamashita, Y.; Fujiki, M.; Todaka, R.; Nishikawa, Y.; Hosoki, A.; Yabe, C.; Nakamura, J.i.; Kawamura, K.; Suwastika, I.N.; et al. Expression of a rice glutaredoxin in aleurone layers of developing and mature seeds: Subcellular localization and possible functions in antioxidant defense. Planta 2015, 242, 1195–1206. [Google Scholar] [CrossRef]
- Ge, L.; Chen, R. Negative gravitropic response of roots directs auxin flow to control root gravitropism. Plant Cell Environ. 2019, 42, 2372–2383. [Google Scholar] [CrossRef]
- Wang, H.; Li, K.; Sun, X.; Xie, Y.; Han, X.; Zhang, S. Isolation and characterization of larch BABY BOOM2 and its regulation of adventitious root development. Gene 2019, 690, 90–98. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Chao, N.; Zhang, M.; Jiang, X.; Gai, Y. Functional Characteristics of Caffeoyl Shikimate Esterase in Larix Kaempferi and Monolignol Biosynthesis in Gymnosperms. Int. J. Mol. Sci. 2019, 20, 6071. https://doi.org/10.3390/ijms20236071
Wang X, Chao N, Zhang M, Jiang X, Gai Y. Functional Characteristics of Caffeoyl Shikimate Esterase in Larix Kaempferi and Monolignol Biosynthesis in Gymnosperms. International Journal of Molecular Sciences. 2019; 20(23):6071. https://doi.org/10.3390/ijms20236071
Chicago/Turabian StyleWang, Xuechun, Nan Chao, Meng Zhang, Xiangning Jiang, and Ying Gai. 2019. "Functional Characteristics of Caffeoyl Shikimate Esterase in Larix Kaempferi and Monolignol Biosynthesis in Gymnosperms" International Journal of Molecular Sciences 20, no. 23: 6071. https://doi.org/10.3390/ijms20236071
APA StyleWang, X., Chao, N., Zhang, M., Jiang, X., & Gai, Y. (2019). Functional Characteristics of Caffeoyl Shikimate Esterase in Larix Kaempferi and Monolignol Biosynthesis in Gymnosperms. International Journal of Molecular Sciences, 20(23), 6071. https://doi.org/10.3390/ijms20236071




