Analysis of Orthologous SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1) Promotor Activity in Herbaceous and Woody Angiosperms
Abstract
1. Introduction
2. Results
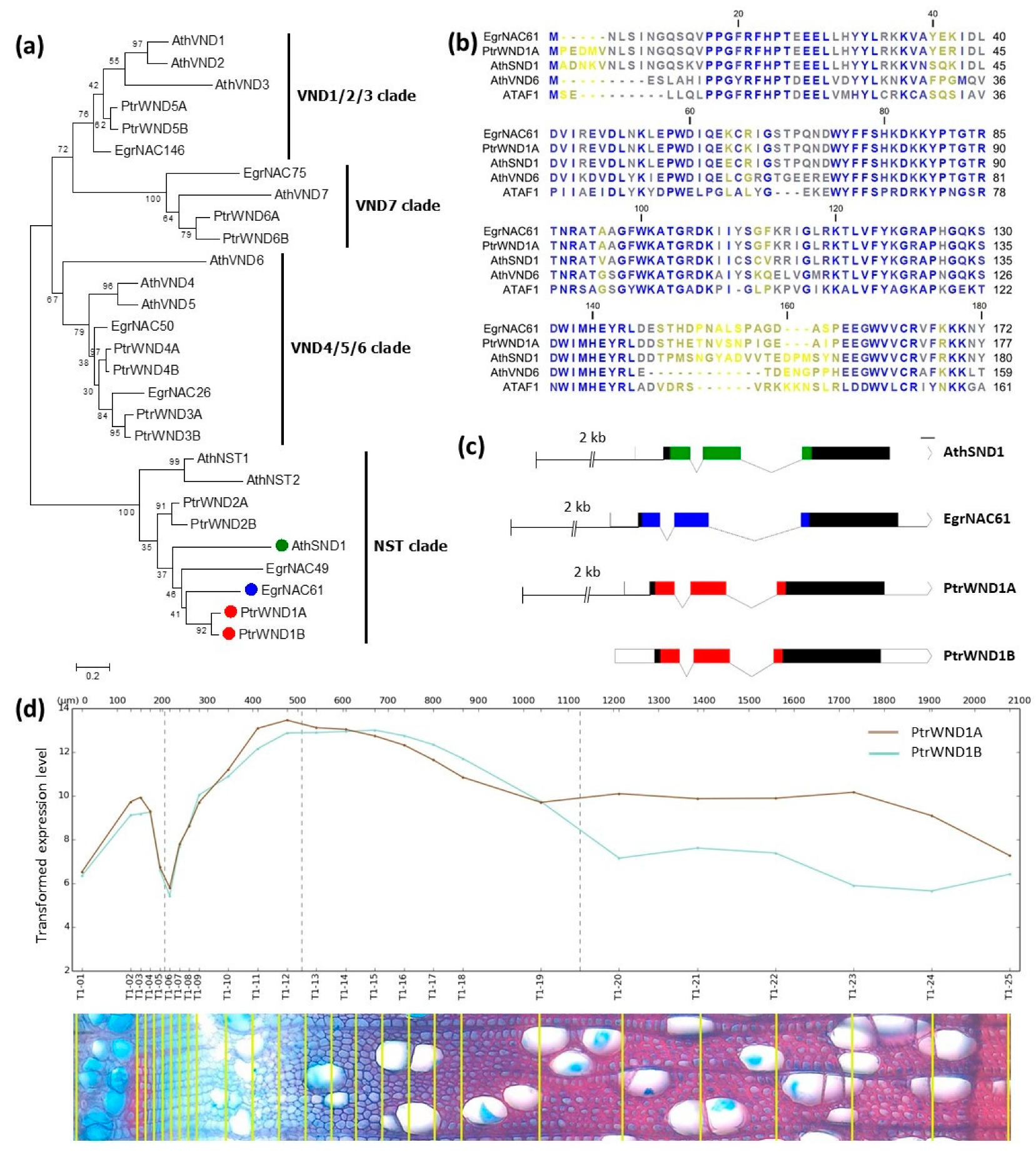
2.1. Phylogenetic Analysis of SWN Proteins from Arabidopsis, Eucalyptus and Populus
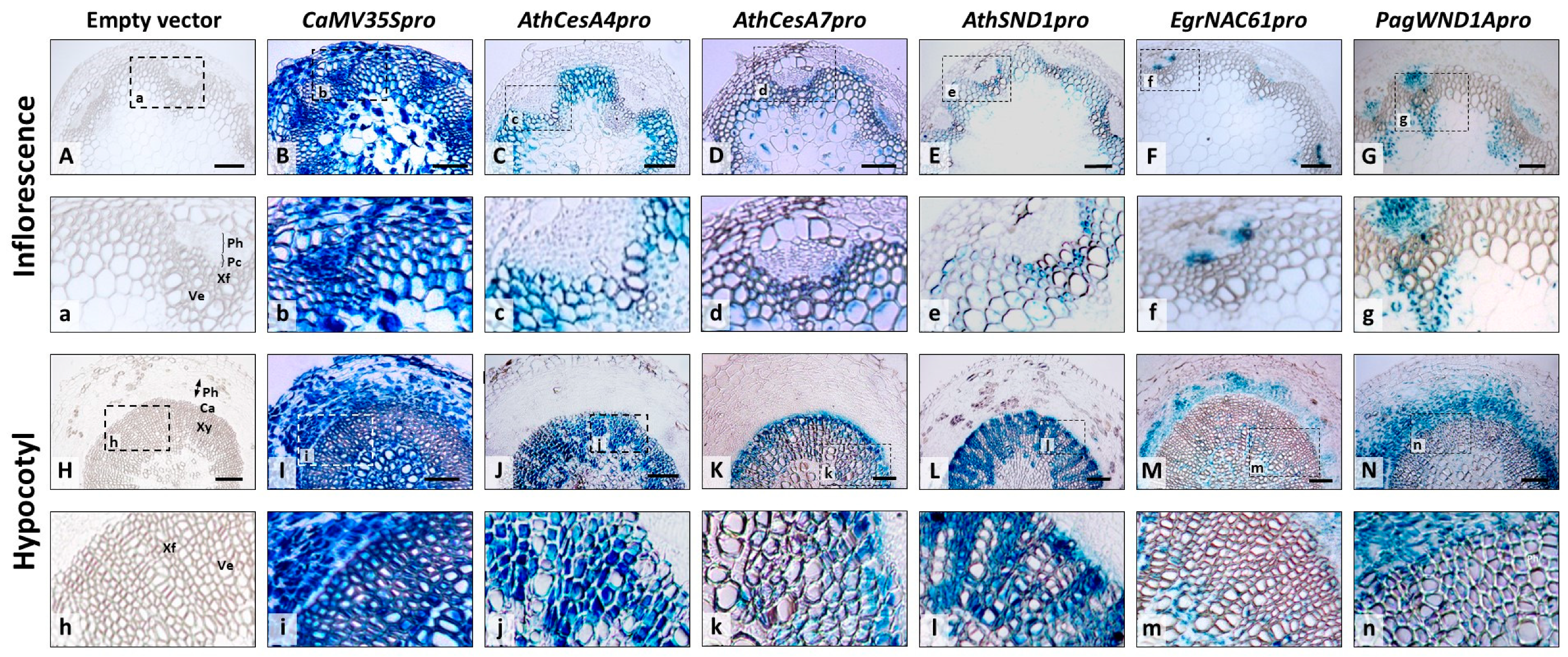
2.2. Tissue-Specific Expression of Orthologous SND1 Promoters in Arabidopsis Inflorescence Stems and Hypocotyls
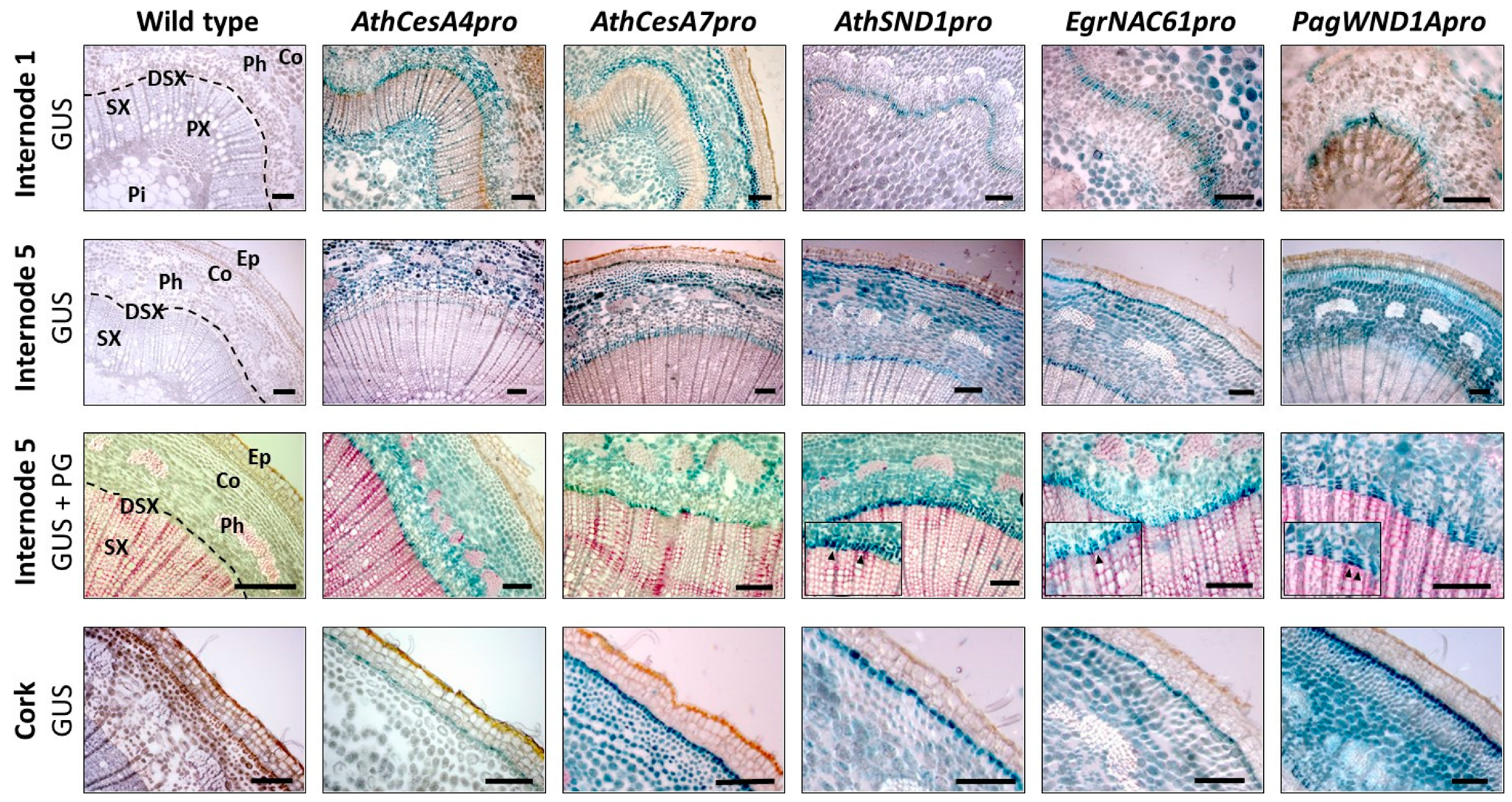
2.3. Analysis of Orthologous SND1 Promoters in Hybrid Poplar Trees
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Analysis
4.2. Reporter Construct Preparation
4.3. Plant Transformation and Growth
4.4. Microscopy and Histochemical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Ath | Arabidopsis thaliana |
| BRN | bearskin |
| Ca | cambium |
| Co | cortex |
| DSX | developing secondary xylem |
| eGFP | enhanced Green Fluorescent Protein |
| Egr | Eucalyptus grandis |
| Ep | epidermis |
| GUS | β-glucuronidase |
| IF | interfascicular fibres |
| Mx | metaxylem |
| NST | NAC secondary wall thickening promoting factor |
| Pag | Populus alba × P. grandidentata |
| Pc | procambium |
| Ph | phloem |
| Pi | pith |
| Ptr | Populus trichocarpa |
| Px | protoxylem |
| PX | primary xylem |
| SCW | secondary cell wall |
| SND | secondary wall-associated NAC domain protein |
| SMB | somrero |
| SWN | Secondary Wall NAC |
| SX | secondary xylem |
| TF | transcription factor |
| UTR | untranslated region |
| Ve | vessels |
| VND | vascular related NAC domain |
| WND | wood associated NAC domain |
| Xf | xylary fibres |
| Xy | xylem |
References
- Carroll, A.; Somerville, C. Cellulosic biofuels. Annu. Rev. Plant Biol. 2009, 60, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Loqué, D.; Scheller, H.V.; Pauly, M. Engineering of plant cell walls for enhanced biofuel production. Curr. Opin. Plant Biol. 2015, 25, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Ragauskas, A.J.; Beckham, G.T.; Biddy, M.J.; Chandra, R.; Chen, F.; Davis, M.F.; Davison, B.H.; Dixon, R.A.; Gilna, P.; Keller, M.; et al. Lignin valorization: Improving lignin processing in the biorefinery. Science 2014, 344, 1246843. [Google Scholar] [CrossRef] [PubMed]
- Thornley, J.H.M.; Cannell, M.G.R. Managing forests for wood yield and carbon storage: A theoretical study. Tree Physiol. 2000, 20, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.-R.; Helariutta, Y.; He, X.-Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The plant vascular system: Evolution, development and functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef]
- Didi, V.; Jackson, P.; Hejatko, J. Hormonal regulation of secondary cell wall formation. J. Exp. Bot. 2015, 66, 5015–5027. [Google Scholar] [CrossRef] [PubMed]
- Hussey, S.G.; Mizrachi, E.; Creux, N.M.; Myburg, A.A. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 2013, 4, 325. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, M.; Smith, R.; Ellis, B. Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 2013, 64, 11–31. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.-H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Ohme-Takagi, M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 2008, 56, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Demura, T.; Ye, Z.H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.-H.; Yang, S.H.; Park, A.H.; Lerouxel, O.; Han, K.-H. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 2007, 50, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yu, H.; Li, L. SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana. PLoS Genet. 2019, 15, e1007928. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Q.; Chen, F.; Wang, M.; Dixon, R.A. NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 2011, 68, 1104–1114. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.H. The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis. Plant Signal. Behav. 2010, 5, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Ye, Z.H. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 2010, 152, 1044–1055. [Google Scholar] [CrossRef]
- Li, Q.; Lin, Y.C.; Sun, Y.H.; Song, J.; Chen, H.; Zhang, X.H.; Sederoff, R.R.; Chiang, V.L. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2012, 109, 14699–14704. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Li, W.; Sun, Y.-H.; Kumari, S.; Wei, H.; Li, Q.; Tunlaya-Anukit, S.; Sederoff, R.R.; Chiang, V.L. SND1 transcription factor—Directed quantitative functional hierarchical genetic regulatory network in wood formation in Populus trichocarpa. Plant Cell 2013, 25, 4324–4341. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, J.; Xu, P.; Zhang, R.; Li, L. Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 2014, 164, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Hou, Y.; Zhao, X.; Lu, W.; Li, Y.; Yang, F.; Tang, S.; Luo, K. Identification and characterization of a wood-associated NAC domain transcription factor PtoVNS11 from Populus tomentosa Carr. Trees 2015, 29, 1091–1101. [Google Scholar] [CrossRef]
- Takata, N.; Awano, T.; Nakata, M.T.; Sano, Y.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol. 2019, 39, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; McCarthy, R.L.; Lee, C.; Ye, Z.-H. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 2011, 157, 1452–1468. [Google Scholar] [CrossRef]
- Hussey, S.G.; Saidi, M.N.; Hefer, C.A.; Myburg, A.A.; Grima-Pettenati, J. Structural, evolutionary and functional analysis of the NAC domain protein family in Eucalyptus. New Phytol. 2015, 206, 1337–1350. [Google Scholar] [CrossRef]
- Laubscher, M.; Brown, K.; Tonfack, L.B.; Myburg, A.A.; Mizrachi, E.; Hussey, S.G. Temporal analysis of Arabidopsis genes activated by Eucalyptus grandis NAC transcription factors associated with xylem fibre and vessel development. Sci. Rep. 2018, 8, 10983. [Google Scholar] [CrossRef]
- Golfier, P.; Volkert, C.; He, F.; Rausch, T.; Wolf, S. Regulation of secondary cell wall biosynthesis by a NAC transcription factor from Miscanthus. Plant Direct 2017, 1, e00024. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; McCarthy, R.L.; Reeves, C.K.; Jones, E.G.; Ye, Z.-H. Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 2011, 52, 1856–1871. [Google Scholar] [CrossRef]
- Pascual, M.B.; Llebrés, M.T.; Craven-Bartle, B.; Cañas, R.A.; Cánovas, F.M.; Ávila, C. PpNAC1, a main regulator of phenylalanine biosynthesis and utilization in maritime pine. Plant Biotechnol. J. 2017, 16. [Google Scholar] [CrossRef]
- Duval, I.; Lachance, D.; Giguère, I.; Bomal, C.; Morency, M.J.; Pelletier, G.; Boyle, B.; MacKay, J.J.; Séguin, A. Large-scale screening of transcription factor-promoter interactions in spruce reveals a transcriptional network involved in vascular development. J. Exp. Bot. 2014, 65, 2319–2333. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Nishikubo, N.; Xu, B.; Yamaguchi, M.; Mitsuda, N.; Goue, N.; Shi, F.; Ohme-Takagi, M.; Demura, T. A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 2011, 67, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.J.; Chen, H.; Li, Q.; Li, W.; Wang, J.P.; Shi, R.; Tunlaya-Anukit, S.; Shuai, P.; Wang, Z.; Ma, H.; et al. Reciprocal cross-regulation of VND and SND multigene TF families for wood formation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2017, 114, E9722–E9729. [Google Scholar] [CrossRef] [PubMed]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, C.; Jin, X.; Xue, W.; Dubreuil, C.; Lezhneva, L.; Fischer, U. The plant hormone auxin directs timing of xylem development by inhibition of secondary cell wall deposition through repression of secondary wall NAC-domain transcription factors. Physiol. Plant. 2018, 165, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Takata, N.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. The Arabidopsis NST3/SND1 promoter is active in secondary woody tissue in poplar. J. Wood Sci. 2017, 63, 396–400. [Google Scholar] [CrossRef]
- Kalluri, U.C.; Yin, H.; Yang, X.; Davison, B.H. Systems and synthetic biology approaches to alter plant cell walls and reduce biomass recalcitrance. Plant Biotechnol. J. 2014, 12, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Moummou, H.; Tonfack, L.B.; Chervin, C.; Benichou, M.; Youmbi, E.; Ginies, C.; Latché, A.; Pech, J.C.; van der Rest, B. Functional characterization of SlscADH1, a fruit-ripening-associated short-chain alcohol dehydrogenase of tomato. J. Plant. Physiol. 2012, 169, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Mishra, L.S.; Kumar, M.; Campbell, L.; Turner, S.R. Wood Formation in Trees Is Increased by Manipulating PXY-Regulated Cell Division. Curr. Biol. 2015, 25, 1050–1055. [Google Scholar] [CrossRef]
- Ratke, C.; Pawar, P.M.-A.; Balasubramanian, V.K.; Naumann, M.; Duncranz, M.L.; Derba-Maceluch, M.; Gorzsás, A.; Endo, S.; Ezcurra, I.; Mellerowicz, E.J. Populus GT43 family members group into distinct sets required for primary and secondary wall xylan biosynthesis and include useful promoters for wood modification. Plant Biotechnol. J. 2015, 13, 26–37. [Google Scholar] [CrossRef]
- Xie, L.; Yang, C.; Wang, X. Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 2011, 62, 4495–4506. [Google Scholar] [CrossRef] [PubMed]
- Hauffe, K.D.; Paszkowski, U.; Schulze-Lefert, P.; Hahlbrock, K.; Dangl, J.L.; Douglas, C.J. A parsley 4CL-1 promoter fragment specifies complex expression patterns in transgenic tobacco. Plant Cell 1991, 3, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Schuetz, M.; Roach, M.; Mansfield, S.D.; Ellis, B.; Samuels, L. Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 2013, 25, 3988–3999. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 2012, 194, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Wang, S.; Liu, Y.; Chen, J.-G.; Douglas, C.J. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 2011, 67, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Creux, N.M.; Ranik, M.; Berger, D.K.; Myburg, A.A. Comparative analysis of orthologous cellulose synthase promoters from Arabidopsis, Populus and Eucalyptus: Evidence of conserved regulatory elements in angiosperms. New Phytol. 2008, 179, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, E.; Porth, I.; Chen, J.G.; Mansfield, S.D.; Douglas, C.J. Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci. Rep. 2014, 4, 5054. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [PubMed]
- Patron, N.J.; Orzaez, D.; Marillonnet, S.; Warzecha, H.; Matthewman, C.; Youles, M.; Raitskin, O.; Leveau, A.; Farré, G.; Rogers, C.; et al. Standards for plant synthetic biology: A common syntax for exchange of DNA parts. New Phytol 2015, 208, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Nevo, E.; Sun, D.; Peng, J. Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution 2012, 66, 1833–1848. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Fawcett, J.A.; Proost, S.; Sterck, L.; Vandepoele, K. The flowering world: A tale of duplications. Trends Plant Sci. 2009, 14, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Proost, S.; Van Bel, M.; Vaneechoutte, D.; Van de Peer, Y.; Inze, D.; Mueller-Roeber, B.; Vandepoele, K. PLAZA 3.0: An access point for plant comparative genomics. Nucleic Acids Res. 2015, 43, D974–D981. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Yuan, Y.; Spiekerman, J.J.; Guley, J.T.; Egbosiuba, J.C.; Ye, Z.H. Functional Characterization of NAC and MYB Transcription Factors Involved in Regulation of Biomass Production in Switchgrass (Panicum virgatum). PLoS ONE 2015, 10, e0134611. [Google Scholar] [CrossRef] [PubMed]
- Voordeckers, K.; Pougach, K.; Verstrepen, K.J. How do regulatory networks evolve and expand throughout evolution? Curr. Opin. Biotechnol. 2015, 34, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Takata, N.; Taniguchi, T. Expression divergence of cellulose synthase (CesA) genes after a recent whole genome duplication event in Populus. Planta 2015, 241, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.D.; Evert, R.F. Seasonal Development of the Secondary Phloem in Populus tremuloides. Bot. Gaz. 1968, 129, 1–8. [Google Scholar] [CrossRef]
- Chaffey, N.; Cholewa, E.; Regan, S.; Sundberg, B. Secondary xylem development in Arabidopsis: A model for wood formation. Physiol. Plant. 2002, 114, 594–600. [Google Scholar] [CrossRef]
- Marques, A.V.; Pereira, H.; Meier, D.; Faix, O. Structural Characterization of Cork Lignin by Thioacidolysis and Permanganate Oxidation. Holzforschung 1999, 53, 167–174. [Google Scholar] [CrossRef]
- Jones, R.; Ougham, H.; Thomas, H.; Waaland, S. The Molecular Life of Plants; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; p. 38. [Google Scholar]
- Rains, M.K.; Gardiyehewa de Silva, N.D.; Molina, I. Reconstructing the suberin pathway in poplar by chemical and transcriptomic analysis of bark tissues. Tree Physiol. 2018, 38, 340–361. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.Q.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Coleman, H.D.; Canam, T.; Kang, K.Y.; Ellis, D.D.; Mansfield, S.D. Over-expression of UDP-glucose pyrophosphorylase in hybrid poplar affects carbon allocation. J. Exp. Bot. 2007, 58, 4257–4268. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Spokevicius, A.V.; van Beveren, K.S.; Bossinger, G. Agrobacterium-mediated transformation of dormant lateral buds in poplar trees reveals developmental patterns in secondary stem tissues. Funct. Plant Biol. 2006, 33, 133–139. [Google Scholar] [CrossRef]
- Pradhan Mitra, P.; Loqué, D. Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J. Vis. Exp. 2014, 87, e51381. [Google Scholar] [CrossRef] [PubMed]
| Constructs | Inflorescence Stem | Hypocotyl | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ep | Co | IF | Pc-Ph | Mx | Px | Pi | Ep | Co | Ca-Ph | Xy | Pi | ||||
| Xf | Ve | Xf | Ve | Xf | Ve | ||||||||||
| AthCesA4pro::eGFP-GUS (n = 4) | - | - | +++ | + | ++ | ++ | ++ | - | + | - | - | - | +++ | ++ | - |
| AthCesA7pro::eGFP-GUS (n = 4) | - | + | +++ | + | +++ | ++ | ++ | - | + | - | - | - | +++ | ++ | - |
| AthSND1pro::eGFP-GUS (n = 6) | - | - | +++ | - | +++ | - | + | - | - | - | - | - | +++ | - | - |
| EgrNAC61pro::eGFP-GUS (n = 5) | - | - | - | +++ | + | - | +++ | - | - | - | + | +++ | +++ | - | +++ |
| PagWND1Apro::eGFP-GUS (n = 4) | - | + | + | +++ | +++ | + | +++ | + | + | - | +++ | +++ | +++ | + | +++ |
| CaMV35S::GUS (n = 3) | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tonfack, L.B.; Hussey, S.G.; Veale, A.; Myburg, A.A.; Mizrachi, E. Analysis of Orthologous SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1) Promotor Activity in Herbaceous and Woody Angiosperms. Int. J. Mol. Sci. 2019, 20, 4623. https://doi.org/10.3390/ijms20184623
Tonfack LB, Hussey SG, Veale A, Myburg AA, Mizrachi E. Analysis of Orthologous SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1) Promotor Activity in Herbaceous and Woody Angiosperms. International Journal of Molecular Sciences. 2019; 20(18):4623. https://doi.org/10.3390/ijms20184623
Chicago/Turabian StyleTonfack, Libert B., Steven G. Hussey, Adri Veale, Alexander A. Myburg, and Eshchar Mizrachi. 2019. "Analysis of Orthologous SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1) Promotor Activity in Herbaceous and Woody Angiosperms" International Journal of Molecular Sciences 20, no. 18: 4623. https://doi.org/10.3390/ijms20184623
APA StyleTonfack, L. B., Hussey, S. G., Veale, A., Myburg, A. A., & Mizrachi, E. (2019). Analysis of Orthologous SECONDARY WALL-ASSOCIATED NAC DOMAIN1 (SND1) Promotor Activity in Herbaceous and Woody Angiosperms. International Journal of Molecular Sciences, 20(18), 4623. https://doi.org/10.3390/ijms20184623





