DeepMiR2GO: Inferring Functions of Human MicroRNAs Using a Deep Multi-Label Classification Model
Abstract
1. Introduction
2. Results
2.1. Benchmarks
2.2. Parameter Selection
2.3. Incorporating Disease Similarity Network
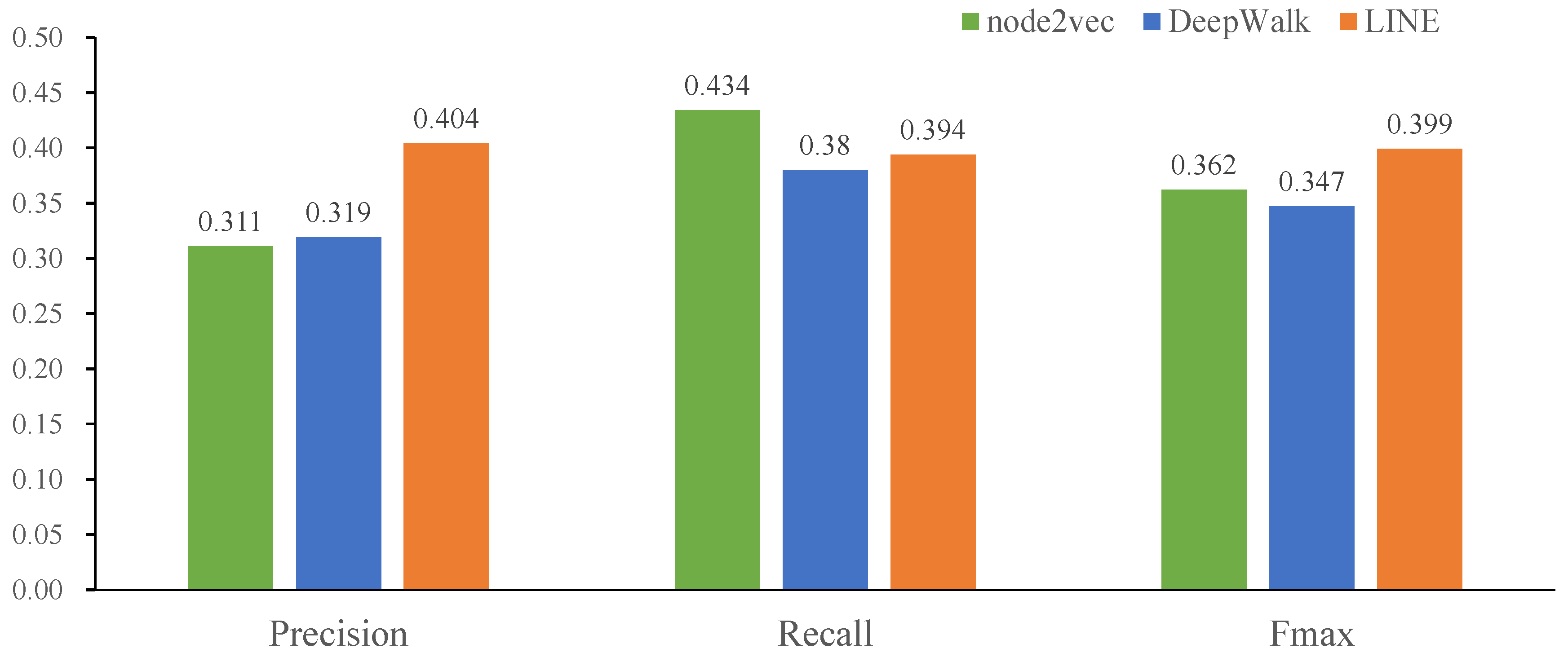
2.4. Comparison of Three Classic Network Representation Algorithms
2.5. The Effect of Hierarchical Multiple Classification Model
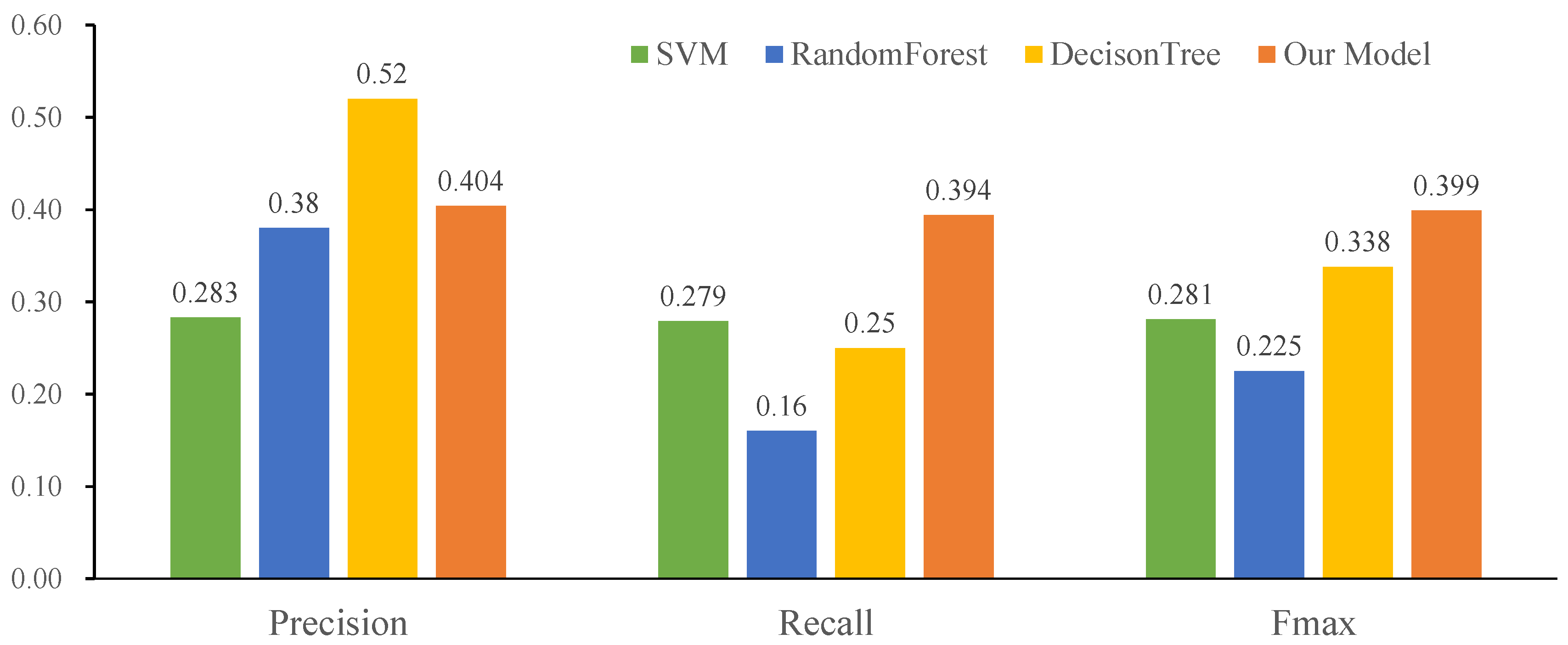
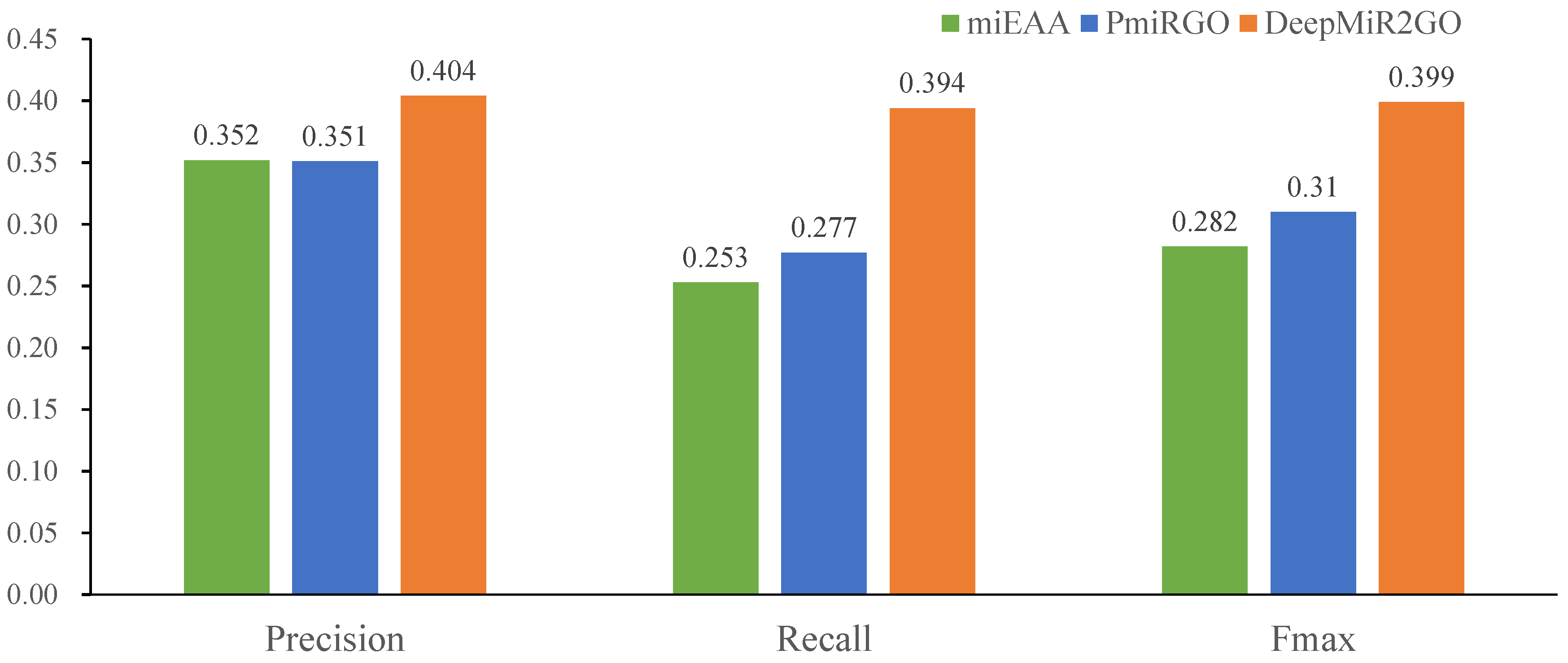
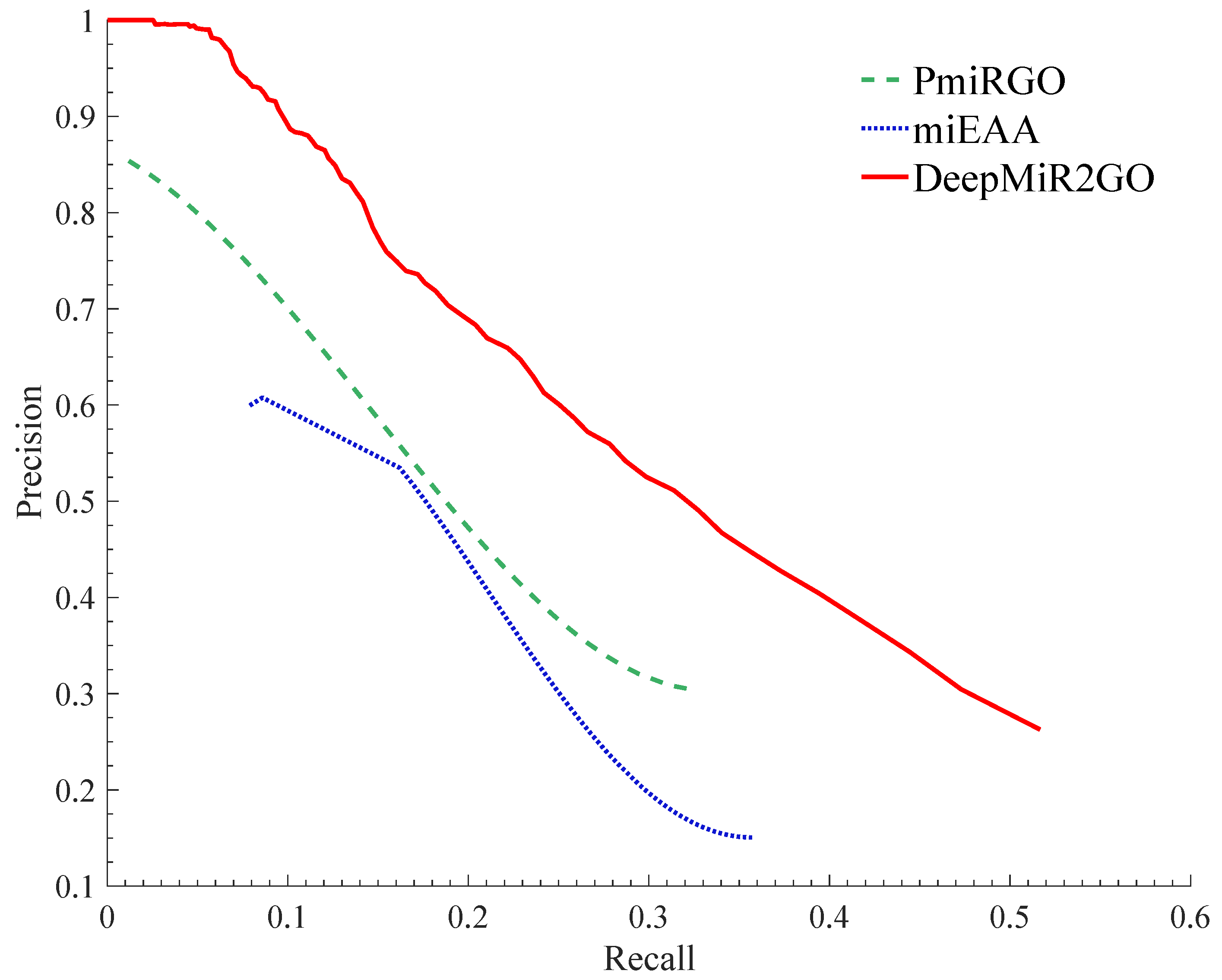
2.6. Performance
2.7. Case Study
3. Discussion
4. Materials and Methods
4.1. MiRNA Co-Expression Similarity Network
4.2. Protein-Protein Interaction Network
4.3. Disease Similarity Network
4.4. miRNA-Target Interaction Network
4.5. miRNA-Disease Association Network
4.6. Gene-Disease Association Network
4.7. Methods
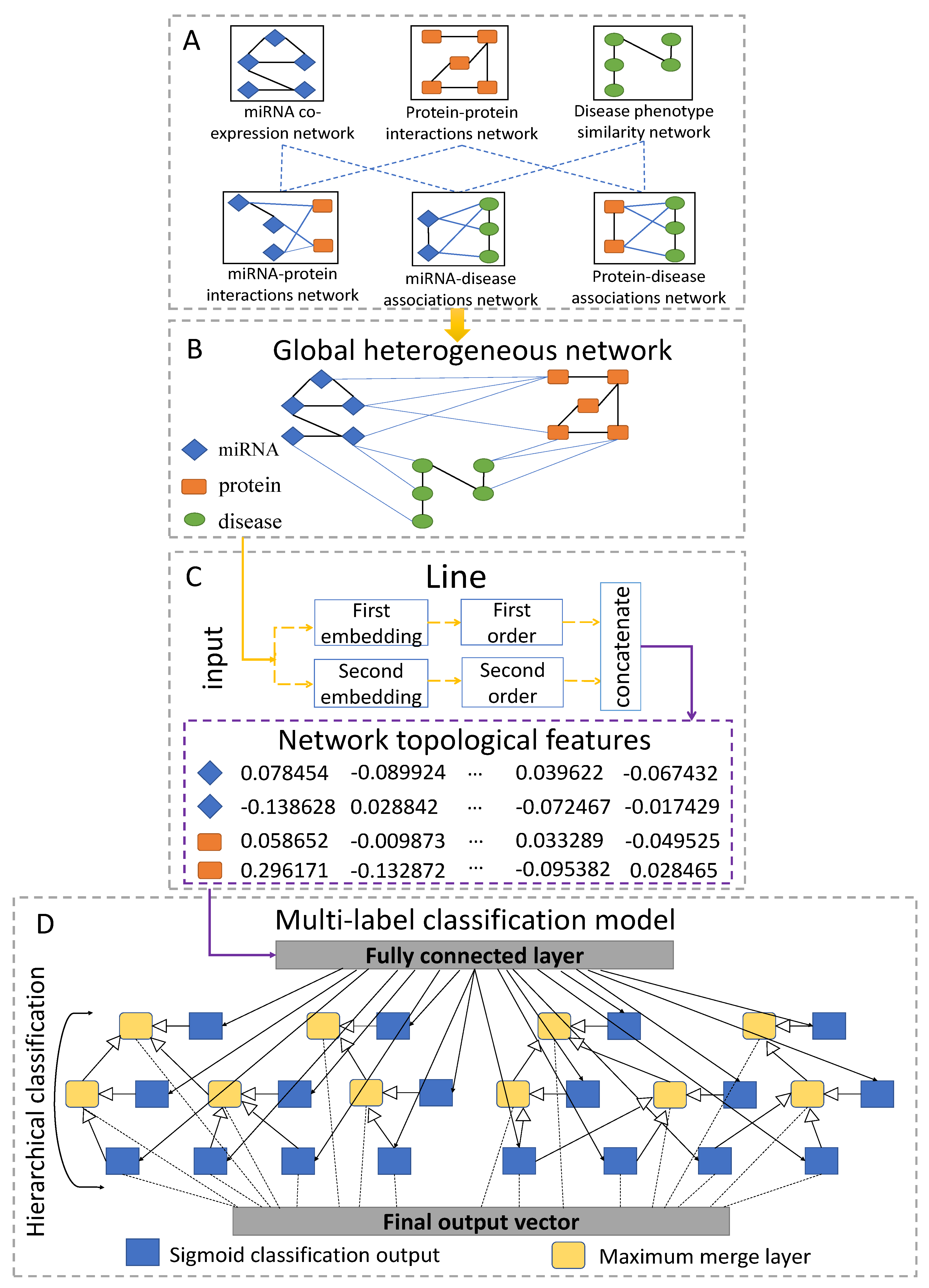
4.7.1. Constructing Global Heterogeneous Network
4.7.2. Learning Topological Features
4.7.3. Training the Hierarchical Multi-Label Classification Network
4.7.4. Evaluation Measures
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PPI | Protein Protein Interaction |
| aa | Amino Acids |
| IEA | Inferred from Electronic Annotation |
| GO | Gene Ontology |
| BP | Biological Process |
| MF | Molecular Function |
| CC | Cellular Component |
References
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, D.P. Micrornas: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V.R. The C. elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 rna regulates developmental timing in caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Griffithsjones, S.; Saini, H.K.; Dongen, S.V.; Enright, A.J. mirbase: Tools for microrna genomics. Nucleic Acids Res. 2007, 36, 154–158. [Google Scholar] [CrossRef]
- Ruby, J.G.; Stark, A.; Johnston, W.K.; Kellis, M.; Bartel, D.P.; Lai, E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of drosophila micrornas. Genome Res. 2007, 17, 1850–1864. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.L.; Sewer, A.; Iovino, N.; Aravin, A.A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microrna expression atlas based on small rna library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Miska, E.A. How micrornas control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005, 15, 563–568. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. Microrna signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Zeng, X.; Liu, L.; Lu, L.; Zou, Q. Prediction of potential disease-associated microRNAs using structural perturbation method. Bioinformatics 2018, 34, 2425–2432. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, W.; Zou, B.; Wang, J.; Deng, Y.; Deng, L. DrugCombDB: A comprehensive database of drug combinations toward the discovery of combinatorial therapy. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wan, S.; Yang, Z.; Teschendorff, A.E.; Zou, Q. Tumor Origin Detection with Tissue-Specific miRNA and DNA methylation Markers. Bioinformatics 2018, 34, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarezsaavedra, E.; Lamb, J.; Peck, D.; Sweetcordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. Microrna expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Garzon, J.I.; Deng, L.; Murray, D.; Shapira, S.; Petrey, D.; Honig, B. A computational interactome and functional annotation for the human proteome. eLife 2016, 5, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.D. Capture hybridization analysis of rna targets (chart). Curr. Protoc. Mol. Biol. 2013, 101. [Google Scholar] [CrossRef]
- Yu, G.; Fu, G.; Wang, J.; Zhao, Y. Newgoa: Predicting new go annotations of proteins by bi-random walks on a hybrid graph. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 15, 1390–1402. [Google Scholar] [CrossRef]
- Costanzo, M.; Vandersluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D.; et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 2016, 353, 1420. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. Micrornas: Small rnas with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Zou, Q.; Li, J.; Song, L.; Zeng, X.; Wang, G. Similarity computation strategies in the microRNA-disease network: A Survey. Briefings Funct. Genom. 2016, 15, 55–64. [Google Scholar] [CrossRef]
- Dam, S.V.; Vosa, U.; Graaf, A.V.D.; Franke, L.; Magalhaes, J.P.D. Gene co-expression analysis for functional classification and gene-disease predictions. Briefings Bioinform. 2017, 19, 575–592. [Google Scholar]
- Pandey, S.P.; Krishnamachari, A. Computational analysis of plant rna pol-ii promoters. BioSystems 2006, 83, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Huang, Y.; Qu, Y.; Jiang, Y.; Zou, Q. Computational analysis of mirna target identification. Curr. Bioinform. 2012, 7, 512–525. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.; Jonesrhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microrna targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Maragkakis, M.; Reczko, M.; Simossis, V.A.; Alexiou, P.; Papadopoulos, G.L.; Dalamagas, T.; Giannopoulos, G.; Goumas, G.I.; Koukis, E.; Kourtis, K.; et al. Diana-microt web server: Elucidating microrna functions through target prediction. Nucleic Acids Res. 2009, 37, 273–276. [Google Scholar] [CrossRef]
- Ulitsky, I.; Laurent, L.C.; Shamir, R. Towards computational prediction of microrna function and activity. Nucleic Acids Res. 2010, 38, e160. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Khaleeq, Q.T.; Meese, E.; Keller, A. mieaa: Microrna enrichment analysis and annotation. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; Macmenamin, P.; Piedade, I.D.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microrna target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Friedman, R.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mrnas are conserved targets of micrornas. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef]
- Deng, L.; Wang, J.; Zhang, J. Predicting gene ontology function of human micrornas by integrating multiple networks. Front. Genet. 2019, 10, 3. [Google Scholar] [CrossRef]
- Tang, J.; Qu, M.; Wang, M.; Zhang, M.; Yan, J.; Mei, Q. Line: Large-scale information network embedding. In Proceedings of the 24th International Conference on World Wide Web, Florence, Italy, 18–22 May 2015; pp. 1067–1077. [Google Scholar]
- Kulmanov, M.; Khan, M.A.; Hoehndorf, R. Deepgo: Predicting protein functions from sequence and interactions using a deep ontology-aware classifier. Bioinformatics 2018, 34, 660–668. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.L.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. the gene ontology consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Carbon, S.; Chan, J.; Kishore, R.; Lee, R.; Muller, H.M.; Raciti, D.; Auken, K.V.; Sternberg, P.W. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar]
- Basith, S.; Manavalan, B.; Shin, T.H.; Lee, G. SDM6A: A Web-Based Integrative Machine-Learning Framework for Predicting 6mA Sites in the Rice Genome. Mol. Ther. Nucleic Acids 2019, 18, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, B.; Lee, J. SVMQA: Support–vector-machine-based protein single-model quality assessment. Bioinformatics 2017, 33, 2496–2503. [Google Scholar] [CrossRef]
- Basith, S.; Manavalan, B.; Shin, T.H.; Lee, G. iGHBP: Computational identification of growth hormone binding proteins from sequences using extremely randomised tree. Comput. Struct. Biotechnol. J. 2018, 16, 412–420. [Google Scholar] [CrossRef]
- Mork, S.; Pletscherfrankild, S.; Caro, A.P.; Gorodkin, J.; Jensen, L.J. Protein-driven inference of mirna-disease associations. Bioinformatics 2014, 30, 392–397. [Google Scholar] [CrossRef]
- Zeng, X.; Zhang, X.; Zou, Q. Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Briefings Bioinform. 2016, 17, 193–203. [Google Scholar] [CrossRef]
- Sen, P.; Namata, G.; Bilgic, M.; Getoor, L.; Gallagher, B.; Eliassirad, T. Collective classification in network data. Ai Magazine 2008, 29, 93–106. [Google Scholar] [CrossRef]
- Tu, C.; Liu, Z.; Sun, M. Inferring correspondences from multiple sources for microblog user tags. Chin. Natl. Conf. Soc. Media Process. 2014, 489, 1–12. [Google Scholar]
- Lu, L.; Zhou, T. Link prediction in complex networks: A survey. Phys. -Stat. Mech. Its Appl. 2011, 390, 1150–1170. [Google Scholar] [CrossRef]
- Yang, C.; Liu, Z.; Zhao, D.; Sun, M.; Chang, E.Y. Network representation learning with rich text information. In Proceedings of the Twenty-Fourth International Joint Conference on Artificial Intelligence (IJCAI 2015), Buenos Aires, Argentina, 25–31 July 2015; pp. 2111–2117. [Google Scholar]
- Perozzi, B.; Alrfou, R.; Skiena, S. Deepwalk: Online learning of social representations. Knowl. Discov. Data Min. 2014, 701–710. [Google Scholar]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. Knowl. Discov. Data Min. 2016, 855–864. [Google Scholar]
- Hearst, M.A.; Dumais, S.T.; Osman, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Liu, Y.; Chang, W.; Han, Y.; Zou, Q.; Guo, M.; Wenbin, L.I. In silico detection of novel micrornas genes in soybean genome. Agric. Sci. China 2011, 10, 1336–1345. [Google Scholar] [CrossRef]
- Zhu, X.; Feng, C.; Lai, H.; Chen, W.; Lin, H. Predicting protein structural classes for low-similarity sequences by evaluating different features. Knowl.-Based Syst. 2019, 163, 787–793. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2011, 45, 5–32. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, Z.; Li, S.; Tan, J.; Chen, W.; Lin, H. Evaluation of different computational methods on 5-methylcytosine sites identification. Briefings Bioinform. 2019. [Google Scholar] [CrossRef]
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.C. mACPpred: A Support Vector Machine-Based Meta-Predictor for Identification of Anticancer Peptides. Int. J. Mol. Sci. 2019, 20, 1964. [Google Scholar] [CrossRef]
- Wei, L.; Su, R.; Luan, S.; Liao, Z.; Manavalan, B.; Zou, Q.; Shi, X. Iterative feature representations improve N4-methylcytosine site prediction. Bioinformatics 2019. [Google Scholar] [CrossRef]
- Manavalan, B.; Subramaniyam, S.; Shin, T.H.; Kim, M.O.; Lee, G. Machine-Learning-Based Prediction of Cell-Penetrating Peptides and Their Uptake Efficiency with Improved Accuracy. J. Proteome Res. 2018, 17, 2715–2726. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, B.; Shin, T.H.; Kim, M.O.; Lee, G. AIPpred: Sequence-Based Prediction of Anti-inflammatory Peptides Using Random Forest. Front. Pharmacol. 2018, 8, 276. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Keller, A.; Kuentzer, J.; Kneissl, B.; Comtesse, N.; Elnakady, Y.A.; Muller, R.; Meese, E.; Lenhof, H. Genetrail–advanced gene set enrichment analysis. Nucleic Acids Res. 2007, 35, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Zhang, Z.; Huang, Z.; Chen, W.; Huang, G.; Meng, F.; Kang, Y. Microrna-92a-3p regulates the expression of cartilage-specific genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthr. Cartil. 2017, 25, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, M.; Salehi, R. Blockage of mir-92a-3p with locked nucleic acid induces apoptosis and prevents cell proliferation in human acute megakaryoblastic leukemia. Cancer Gene Ther. 2016, 23, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shang, S.; Wang, J.; Zhang, T.; Nie, F.; Song, X.; Zhao, H.; Zhu, C.; Zhang, R.; Hao, D. Identification of mir-22-3p, mir-92a-3p, and mir-137 in peripheral blood as biomarker for schizophrenia. Psychiatry-Res.-Neuroimaging 2018, 265, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Casadei, L.; Calore, F.; Creighton, C.J.; Guescini, M.; Batte, K.; Iwenofu, O.H.; Zewdu, A.; Braggio, D.; Bill, K.L.J.; Fadda, P.; et al. Exosome-derived mir-25-3p and mir-92a-3p stimulate liposarcoma progression. Cancer Res. 2017, 77, 3846–3856. [Google Scholar] [CrossRef]
- Deng, L.; Li, W.; Zhang, J. LDAH2V: Exploring meta-paths across multiple networks for lncRNA-disease association prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019. [Google Scholar] [CrossRef]
- Panwar, B.; Omenn, G.S.; Guan, Y. Mirmine: A database of human mirna expression profiles. Bioinformatics 2017, 33, 1554–1560. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Z.; Chen, Z.; Deng, L. Integrating multiple heterogeneous networks for novel lncrna-disease association inference. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 396–406. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huertacepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.; et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Driel, M.A.V.; Bruggeman, J.; Vriend, G.; Brunner, H.G.; Leunissen, J.A.M. A text-mining analysis of the human phenome. Eur. J. Hum. Genet. 2006, 14, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.; Lin, F.; Wu, W.; Liang, C.; Huang, W.C.; Chan, W.; Tsai, W.; Chen, G.; Lee, C.; Chiu, C.; et al. mirtarbase: A database curates experimentally validated microrna-target interactions. Nucleic Acids Res. 2011, 39, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Jiangcheng, S.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. Hmdd v3.0: A database for experimentally supported human microrna-disease associations. Nucleic Acids Res. 2018, 47, 10. [Google Scholar] [CrossRef]
- Pinero, J.; Bravo, A.; Queraltrosinach, N.; Gutierrezsacristan, A.; Deupons, J.; Centeno, E.; Garciagarcia, J.; Sanz, F.; Furlong, L.I. Disgenet: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Fan, C.; Tang, Y.; Deng, L. Katzlgo: Large-scale prediction of lncrna functions by using the katz measure based on multiple networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 16, 407–416. [Google Scholar] [CrossRef]
- Huntley, R.P.; Dimmer, E.; Barrell, D.; Binns, D.; Apweiler, R. The gene ontology annotation (goa) database. Nat. Preced. 2009. [Google Scholar] [CrossRef]
- Deng, L.; Wu, H.; Liu, C.; Zhan, W.; Zhang, J. Probing the functions of long non-coding rnas by exploiting the topology of global association and interaction network. Comput. Biol. Chem. 2018, 74, 360–367. [Google Scholar] [CrossRef]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. Coursera Neural Networks Mach. Learn. 2012, 4, 26–31. [Google Scholar]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence And Statistics, Sardinia, Italy, 13–15 May 2010; pp. 249–256. [Google Scholar]
- Yang, H.; Yang, W.; Dao, F.; Lv, H.; Ding, H.; Chen, W.; Lin, H. A comparison and assessment of computational method for identifying recombination hotspots in Saccharomyces cerevisiae. Briefings Bioinform. 2019. [Google Scholar] [CrossRef]
- Yang, W.; Zhu, X.; Huang, J.; Ding, H.; Lin, H. A brief survey of machine learning methods in protein sub-Golgi localization. Curr. Bioinform. 2019, 14, 234–240. [Google Scholar] [CrossRef]
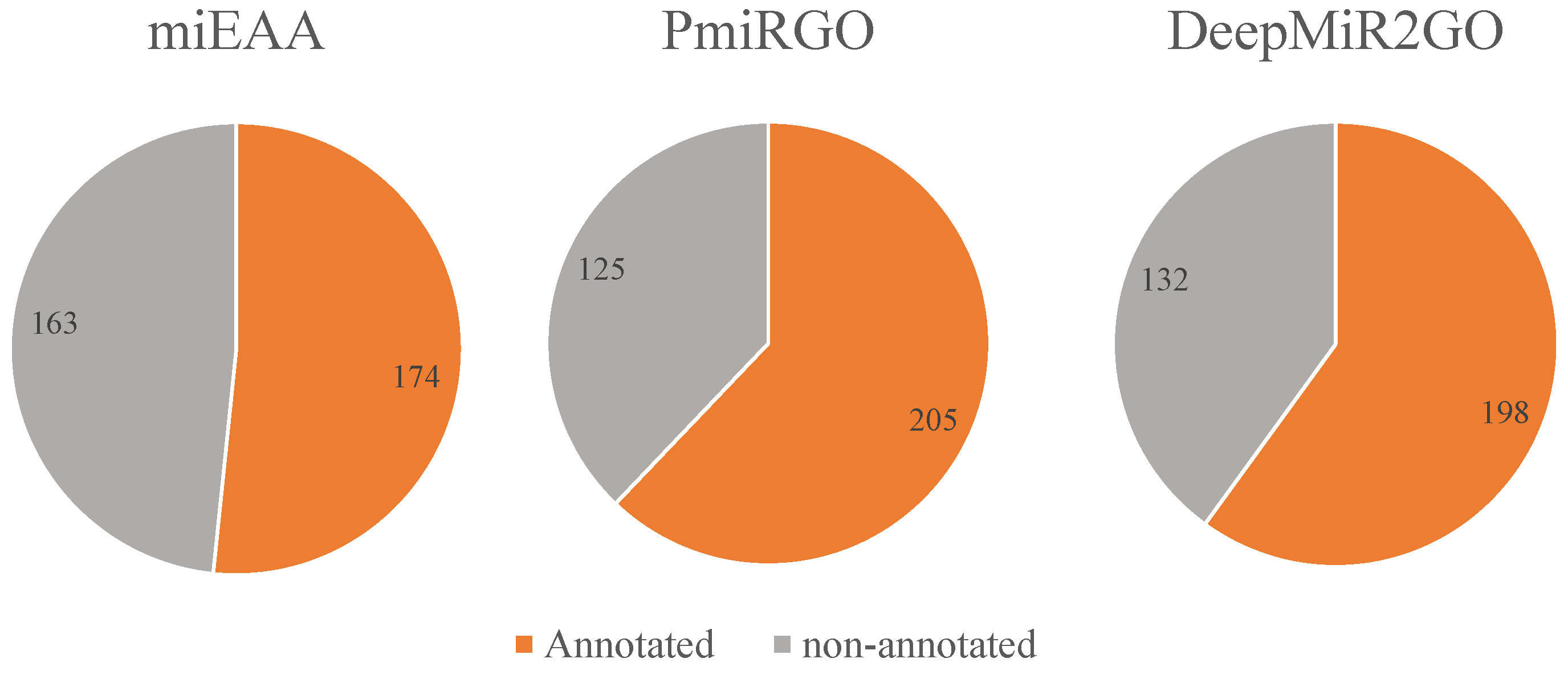
| Biological Processes | |||
| Dimensionality | AvePre | AveRec | |
| 64 | 0.399 | 0.404 | 0.394 |
| 128 | 0.389 | 0.403 | 0.376 |
| 256 | 0.393 | 0.383 | 0.403 |
| 512 | 0.374 | 0.345 | 0.409 |
| Molecular Functions | |||
| Dimensionality | AvePre | AveRec | |
| 64 | 0.510 | 0.500 | 0.520 |
| 128 | 0.399 | 0.376 | 0.424 |
| 256 | 0.504 | 0.510 | 0.498 |
| 512 | 0.411 | 0.470 | 0.364 |
| Biological Processes | |||
| Dimensionality | AvePre | AveRec | |
| With | 0.399 | 0.404 | 0.394 |
| Without | 0.360 | 0.372 | 0.349 |
| Molecular Functions | |||
| Dimensionality | AvePre | AveRec | |
| With | 0.510 | 0.500 | 0.520 |
| Without | 0.279 | 0.238 | 0.339 |
| ID | GO Terms | GO Names |
|---|---|---|
| 1 | GO:0044260 | cellular macromolecule metabolic process |
| 2 | GO:0060255 | regulation of macromolecule metabolic process |
| 3 | GO:0031323 | regulation of cellular metabolic process |
| 4 | GO:0080090 | regulation of primary metabolic process |
| 5 | GO:0043170 | macromolecule metabolic process |
| 6 | GO:0050794 | regulation of cellular process |
| 7 | GO:0050789 | regulation of biological process |
| 8 | GO:0065007 | biological regulation |
| 9 | GO:0044763 | cellular process |
| 10 | GO:0071704 | organic substance metabolic process |
| 11 | GO:0010468 | regulation of gene expression |
| 12 | GO:0044237 | cellular metabolic process |
| 13 | GO:0009987 | cellular process |
| 14 | GO:0044238 | primary metabolic process |
| 15 | GO:0019222 | regulation of metabolic process |
| 16 | GO:0044699 | biological process |
| 17 | GO:0008152 | metabolic process |
| ID | GO Terms | Depth | GO Names |
|---|---|---|---|
| 1 | GO:0045892 | 12 | negative regulation of transcription |
| 2 | GO:0010629 | 7 | negative regulation of gene expression |
| 3 | GO:0045944 | 13 | positive regulation of transcription by RNA polymerase II |
| 4 | GO:1903507 | 11 | negative regulation of nucleic acid-templated transcription |
| 5 | GO:1902679 | 10 | negative regulation of RNA biosynthetic process |
| 6 | GO:0008285 | 6 | negative regulation of cell population proliferation |
| 7 | GO:1903508 | 11 | positive regulation of nucleic acid-templated transcription |
| 8 | GO:0000122 | 13 | negative regulation of transcription by RNA polymerase II |
| 9 | GO:2000113 | 8 | negative regulation of cellular |
| 10 | GO:0051253 | 9 | negative regulation of RNA metabolic process |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Zhang, J.; Cai, Y.; Deng, L. DeepMiR2GO: Inferring Functions of Human MicroRNAs Using a Deep Multi-Label Classification Model. Int. J. Mol. Sci. 2019, 20, 6046. https://doi.org/10.3390/ijms20236046
Wang J, Zhang J, Cai Y, Deng L. DeepMiR2GO: Inferring Functions of Human MicroRNAs Using a Deep Multi-Label Classification Model. International Journal of Molecular Sciences. 2019; 20(23):6046. https://doi.org/10.3390/ijms20236046
Chicago/Turabian StyleWang, Jiacheng, Jingpu Zhang, Yideng Cai, and Lei Deng. 2019. "DeepMiR2GO: Inferring Functions of Human MicroRNAs Using a Deep Multi-Label Classification Model" International Journal of Molecular Sciences 20, no. 23: 6046. https://doi.org/10.3390/ijms20236046
APA StyleWang, J., Zhang, J., Cai, Y., & Deng, L. (2019). DeepMiR2GO: Inferring Functions of Human MicroRNAs Using a Deep Multi-Label Classification Model. International Journal of Molecular Sciences, 20(23), 6046. https://doi.org/10.3390/ijms20236046






