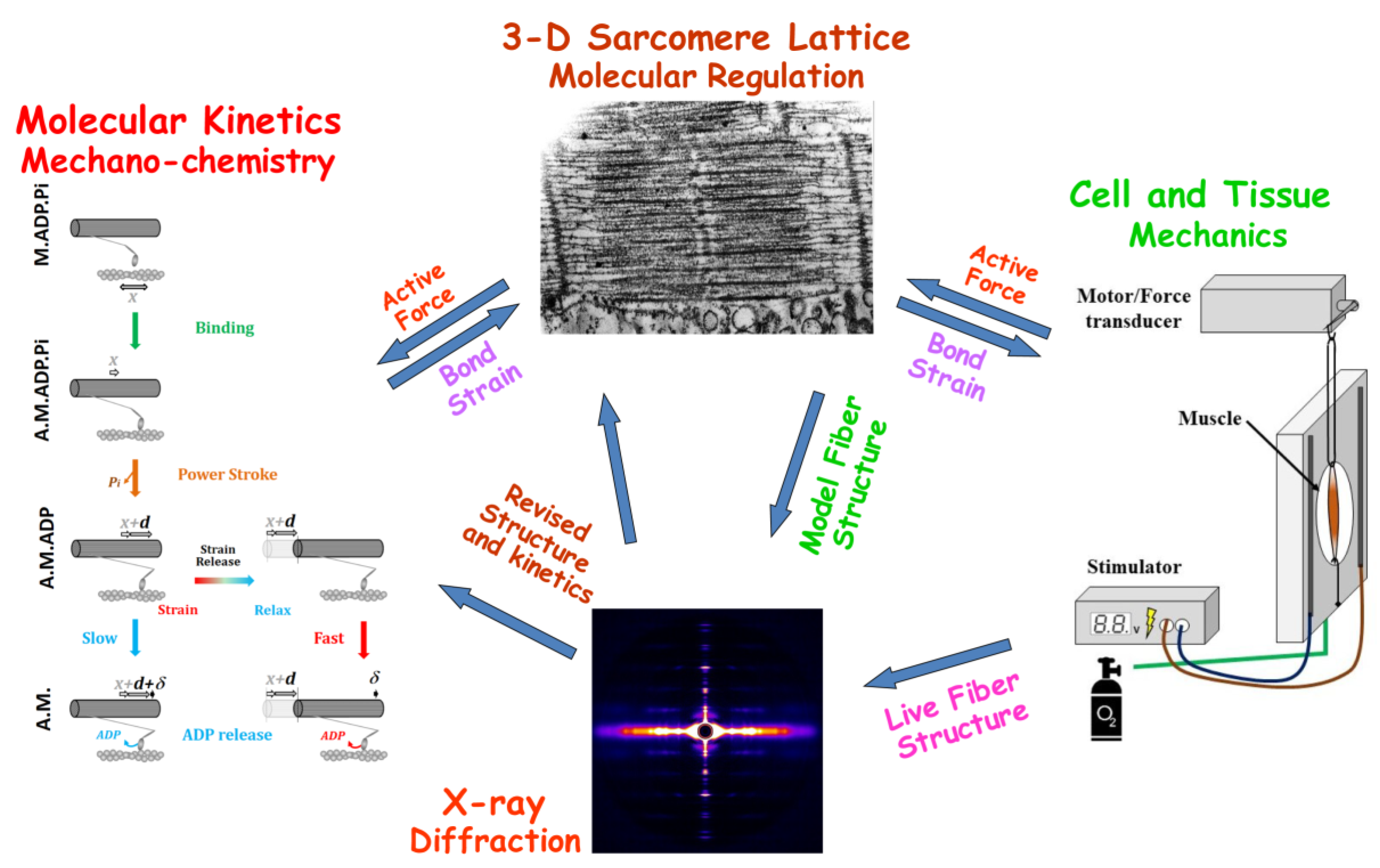
1. Introduction
Mechanical forces acting in living cells are extremely important in understanding mechanotransduction, cell migration and cell physiological function [
1,
2,
3,
4]. Despite their importance, there have been only a few attempts to estimate the magnitude of these forces. For example, Evans et al. measured the magnitude of forces in integrins in adherent cells [
5,
6,
7]. In living cells, the forces in cytoskeletal filaments have been assessed only from observed tension or tractions from mechanical measurements and estimated number of filaments in cross-section [
8,
9,
10,
11]. For assessing forces in cell filaments, probably the best test system is striated muscle because it contains well-defined subcellular structure, with established methodologies for measurements of tension and the deformation of actin and myosin filaments by X-ray diffraction. In contracting muscle, X-ray fiber diffraction patterns contain information concerning the position of all molecules within the three-dimensional sarcomere structures as well as the variable local deformation of filamentous structural elements. The simplest method for interpreting the relationship between deformed sarcomere structure, due to developed tension, and meridional reflections in X-ray diffraction patterns, uses the correlation between an average atomic spacing and X-ray patterns along the meridional axis. Using this approach, Huxley et al. (1994) and Wakabayashi et al. (1994) estimated the elastic characteristics of actin and myosin filaments [
12,
13] by plotting the changes in the axial spacing of the first order actin meridional reflection as a function of developed force. As we show below, however, there is much more information contained in the diffraction patterns if methods can be developed to extract it.
In living cells, fibrous proteins are exposed to mechanical forces interacting with other subcellular structures showing the local strain variations along the fiber length. To account for these variations, a theoretical formalism has been developed for predicting the diffraction patterns from individual helical molecular structures nonuniformly strained along their lengths [
14]. Here, we apply this methodology to predict X-ray patterns from contracting muscle with the goal of more precisely interpreting X-ray diffraction patterns from living muscle fibers. We use the Monte Carlo simulation platform, MUSICO [
8,
15], to generate the geometry of actin filaments deformed by the action of randomly attached myosin heads where, at any instant of time, the distribution of variable strains and intermonomer spacings along myofilaments are available for predicting the X-ray patterns. Adjusting the model parameters in MUSICO to match the X-ray diffraction and mechanical observations of Huxley et al. [
12] provides a window to the multiscale relationship between kinetics of individual molecules and the macroscopic mechanical behavior in contracting muscle. In particular, we show that the newly developed MUSICO X-ray diffraction module can predict several orders of actin meridional diffraction reflections where the first order reflection primarily reports the average strains (or monomer spacings) in the filaments, but the higher order meridional reflections also report on the non-uniformity of the spacings [
14]. Combining the MUSICO simulations and the observed X-ray diffraction patterns provides, for the first time, the opportunity to assess forces acting in living muscle cells, assuming that the elastic characteristics of actin filaments are known.
3. Results
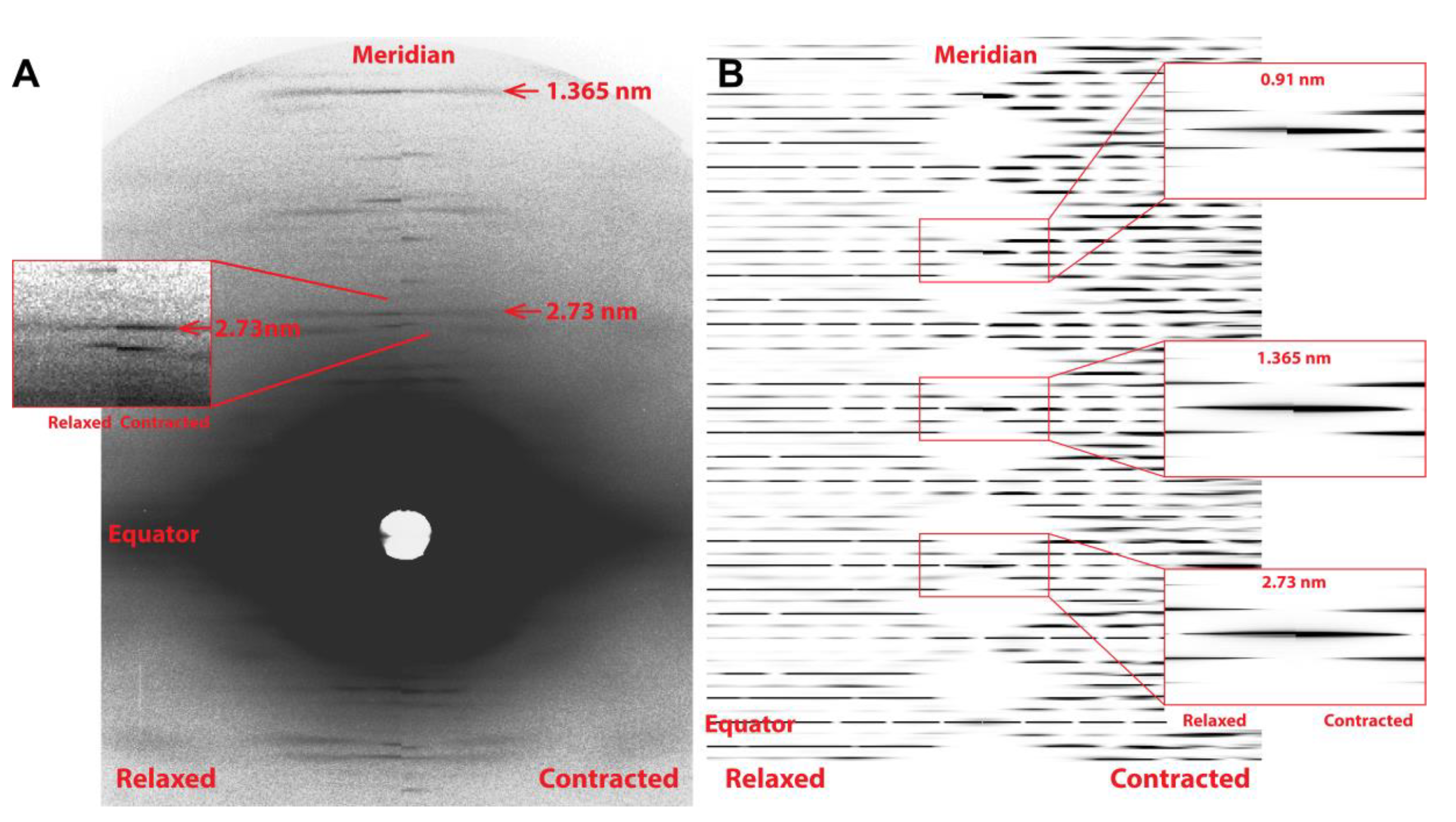
The predicted X-ray pattern from a MUSICO simulation, up to the third actin meridional reflection, is shown in
Figure 3B which closely resembles the observed pattern shown in
Figure 3A. In both panels, the relaxed pattern is on the left and the isometrically contracted pattern is on the right. Contracting muscle has a nonuniform and, on average, longer spacing, which, in the inverse space of the diffraction patterns, is manifested as the meridional reflections move slightly closer toward the equatorial axis. This is clearly seen in the expanded reflection details, on the right of
Figure 3B and to the left of the observed reflections in
Figure 3A. It is also noticeable that the higher order reflections show larger shifts between the relaxed and contracted patterns, and broader meridional reflection profiles. The shifts are the measure of average change in spacing from relaxed to contracted state, and the widths of the higher order reflections are the measure of the degree of nonuniformity of the actin monomer spacings.
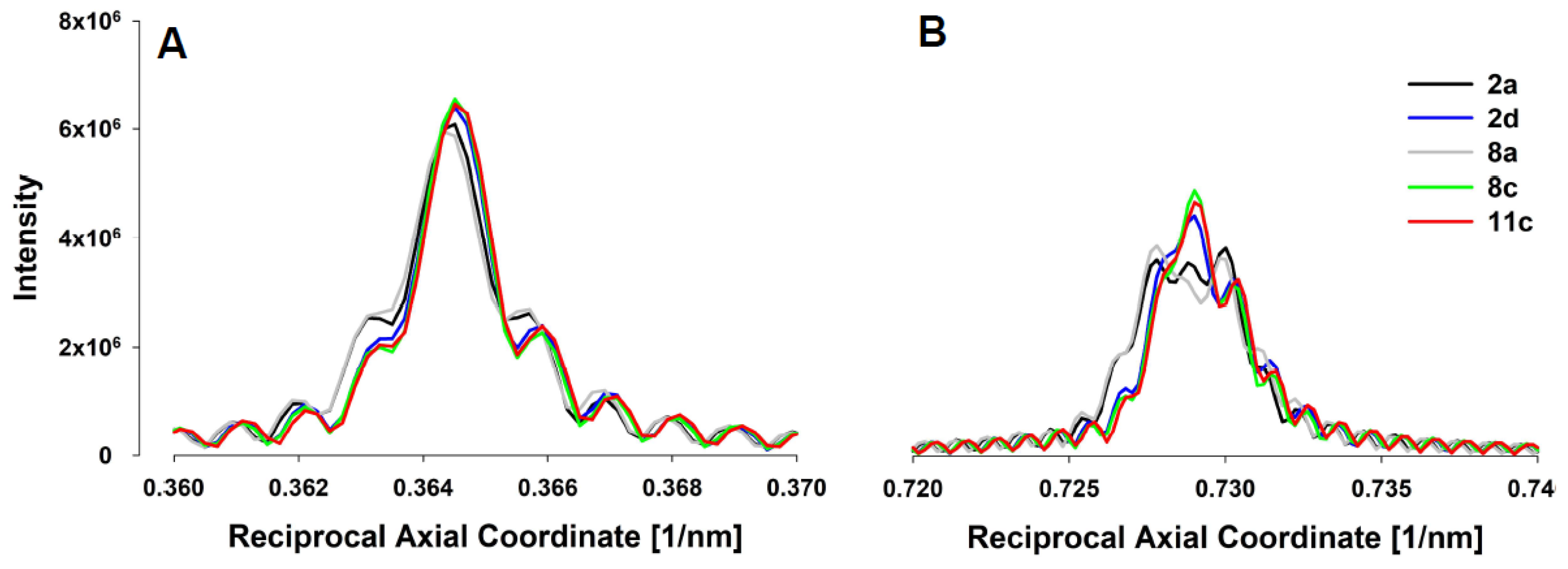
For assessing forces in the thin filaments, we have chosen five experiments from Huxley et al. (1994) [
12], done under isometric conditions where the X-ray patterns (
Figure 3A) and meridional actin reflection profiles, up the second order,
Figure 4, were available. Each experiment displayed slightly different actin reflection profiles with small but important changes in the mean values of actin monomer spacings, likely caused by the variation in levels of activation between the experiments. For each experiment, we titrated the level of activation via the effective myosin binding rate to actin,
, in order to match observed first and second actin meridional profiles.
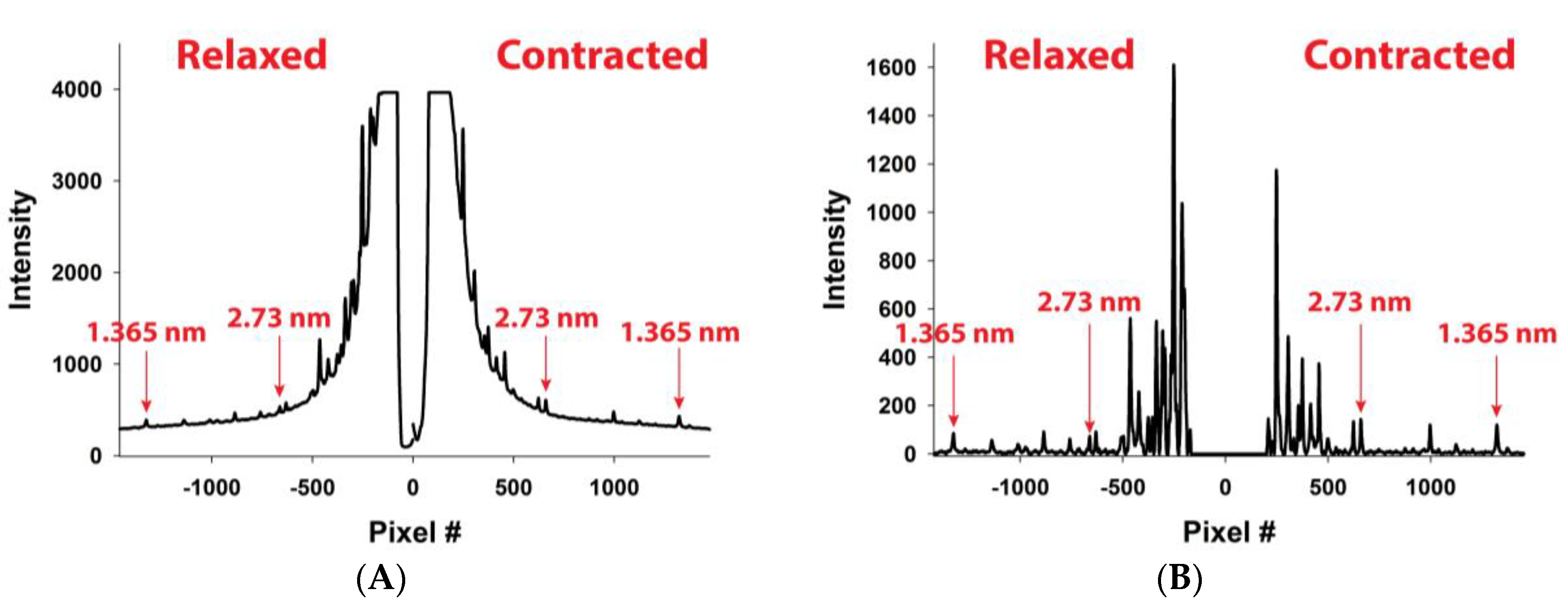
In
Figure 5A, MUSICO predicted profiles for the first and the second reflection are shown, where the solid red lines represent relaxed and the green lines contracted reflection profiles. In order to compare the predicted intensity profiles to the observed, for example, from Experiment No: 2d (
Figure 4B), it is necessary to correct for the axial width of the X-ray beam. We, therefore, convoluted the original predicted profiles (solid lines in
Figure 5A) with the measured axial size of the X-ray beam to obtain the predicted profiles (dashed lines in
Figure 5B) that can be compared to the observed profiles (solid lines with symbols). When we achieved satisfactory fits, the observed X-ray reflections are correlated with MUSICO predicted distribution of spacings, forces in thin filaments and muscle tensions.
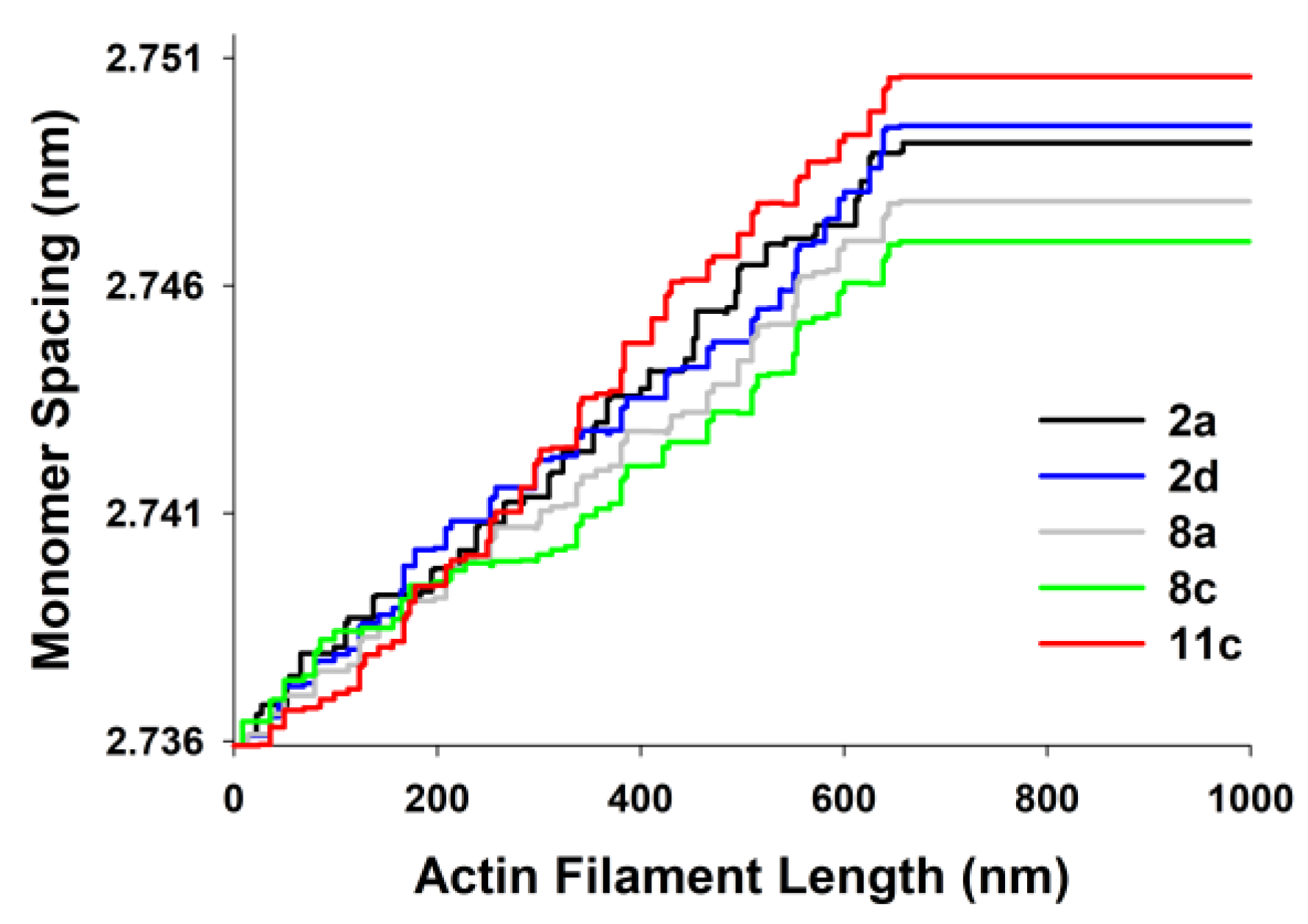
The actin monomer spacings, predicted by MUSICO, as a function of position along the thin filament for the five chosen experiments are shown in
Figure 6. The differences are largest in the part of actin filaments in the I band of the sarcomeres (close to Z-discs) and they reflect the changes in observed tensions, on average, between the experiments.
These differences are also present in the predicted integrated intensity profiles of the first and second order actin meridional reflections along the meridian (
Figure 7A,B, respectively). The quantitative predictions of the average spacing in relaxed and contracted states, average increase in spacing during the contraction, mean strains, mean actin force and its standard deviation in the I band, and muscle tensions are summarized in
Table 1.
From the simultaneous fits of mechanical and X-ray data for each experiment we can derive the relationship between the spacing change and mean forces in the actin filaments in the I band, and, importantly, the deviation of the actin forces from the mean (
Table 1). The predicted tension shows values in the range from 260 to 340 kPa, that is, not only in the range reported by Huxley et al. (1994) [
12], but also in the range between
Rana esculenta fibers of 228 kPa and in
Rana temporaria fibers at 4 °C, that is, about ~342 kPa (Linari et al., 1998 [
9]).
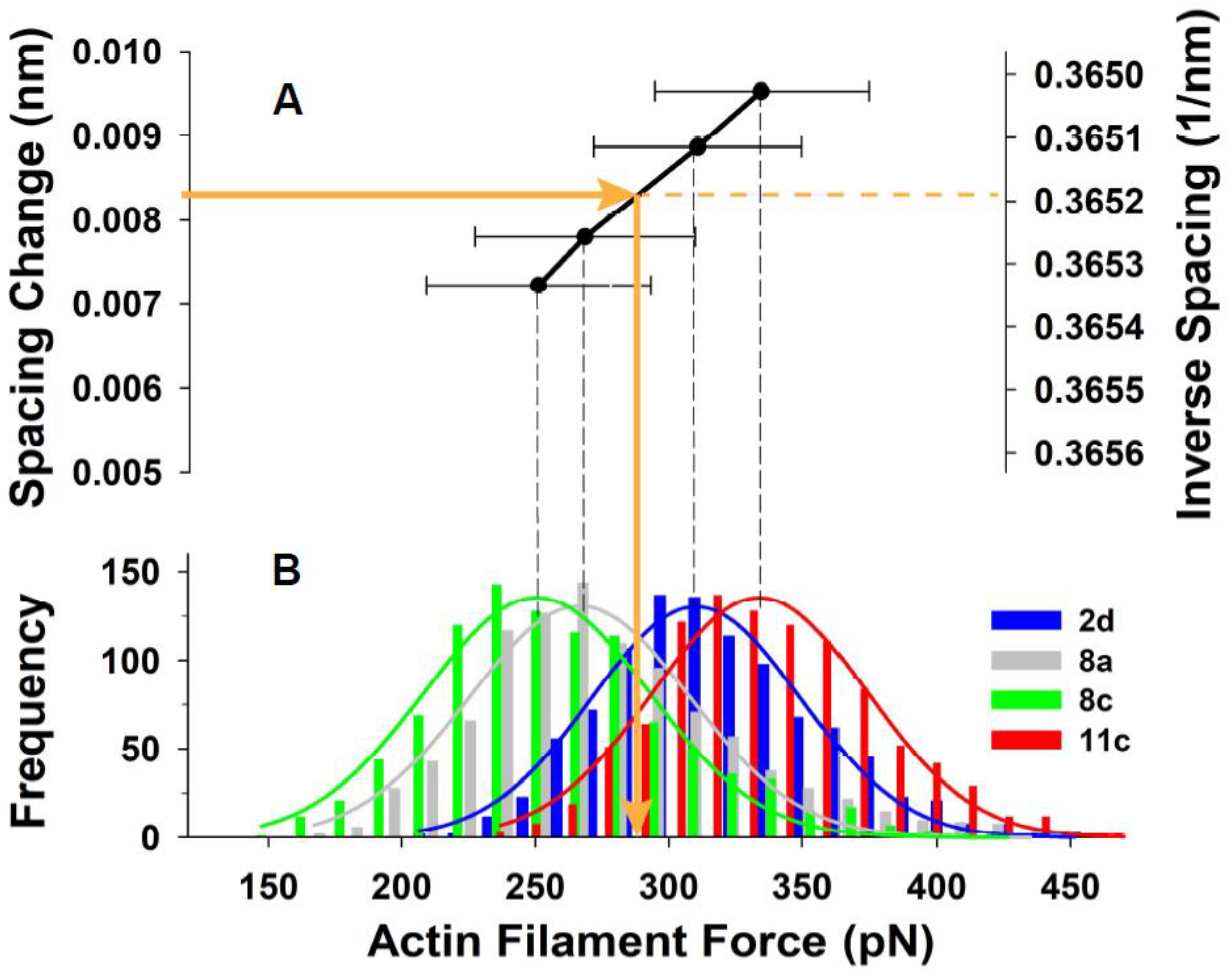
Figure 8A shows the relationship of mean thin filament spacing change (measured from the X-ray patterns) to the mean force (estimated using MUSICO) on the thin filaments for 4 of the 5 individual experiments we analyzed (one experiment was omitted for visual clarity). The horizontal error bars indicate the width of the distributions of forces that gave the mean value in that particular experiment.
Figure 8B shows histograms of the calculated force distributions in the individual thin filaments. These force distributions were derived, as described above, using the widths of the second order actin meridional reflections.
4. Discussion
The addition of the X-ray diffraction module to the MUSICO platform enables simulations that can predict the forces in actin filaments in living muscles. Moreover, when predictions of these simulations are tightly coupled with spacings and peak shapes of the first and higher order X-ray reflections, considered up to second order in this study, they can provide not only the mean forces on actin filaments as assessed earlier [
12,
13], but also the variation of the magnitude of these forces, as shown in
Figure 8B. The variation of forces in myosin and actin filaments are generated by randomly distributed crossbridges bound to actin, and by using the instantaneous mechanical equilibrium, the MUSICO simulations can provide a view of how these forces vary in time along and between the individual filaments. The magnitude of these variations could be one of the key factors in determining mechanisms of mechano-transduction [
23,
24,
25,
26,
27]. This information cannot be estimated by any other experimental or analytical technique.
A unique feature of coupling analysis of X-ray diffraction data to MUSICO simulations is that the measurements at the molecular scale, i.e., actin inter-monomer spacings, and macroscopic measurements of tension in live muscle fibers, taken together with multiscale simulations by MUSICO, provide simultaneous model validation at these two length scales using data from the same experiments. Changes in actin monomer spacings, i.e., the observed inverse spacing, and the width of their distributions, as estimated from the width of the second actin reflection (
Figure 5B, right panel), are tightly coupled with the mean forces and the variation in the thin filament forces (
Figure 8B). The width of the second reflection profile is determined by both the nonuniformity of actin monomer spacings along individual actin filaments and variations between the filaments. The MUSICO simulations can distinguish between these two contributions. These effects could be seen more clearly in higher order actin meridional reflections, as, for example, the third and fourth order reflections observed by Wakabayashi et al. [
13]. To obtain data with the higher order actin meridional reflections would require new experiments designed expressly for this purpose.
A key assumption in the estimation of forces in thin filaments is the known elastic characteristics of thin filaments. From mechanical and X-ray diffraction studies, the estimated stiffness of thin filaments in different muscles and preparations, e.g., frog muscles (
Rana esculenta and
Rana temporaria), skinned rabbit fibers and in the dogfish muscles, ranges from 67 to about 100 pN/nm per 1 μm long filament [
9,
28,
29,
30]. This range of values could be overestimated due to specificities in experimental protocols and complex calculations of the thin filament compliance. The estimated values can depend on species and also differ between fibers from different frog species, for example,
Rana temporaria vs.
Rana esculenta. In our MUSICO simulations, we used an actin filament stiffness of ~65 pN/nm per 1 µm F-actin filament from Kojima et al. [
31], in preference to other measurements since it was the result of a direct measurement rather than inferred indirectly from mechanical or other data. With Kojima’s stiffness estimate, our model predictions matched well the first and second meridional actin reflection axial spacings in the relaxed and contracting state. In addition, our model predicted the observed tension in Huxley’s experiments. If, on the other hand, we use higher estimates for stiffness (~100 pN/nm) [
29,
30], the estimated tension from our measurements will be in the range of 450–500 kPa, which are unrealistically high values. The thin filament stiffness from Kojima et al. [
31] could still be overestimated because the actin filaments in this study were visualized by phalloidin, and the bound phalloidin could increase the stiffness of the actin filaments [
32]. Additionally, recent measurements of Kiss et al. [
33] indicated much lower values of thin filament stiffness in mouse soleus muscle and even smaller stiffness in nebulin deficient muscles. A decrease in thin filament stiffness may change the distribution of positions of bound crossbridges along the filaments, the number of bound crossbridges and the average force per filament. The most significant change will be in magnitudes of the mean force per thin and thick filament, where the mean force per thin filament will decrease approximately linearly with the decrease of thin filament stiffness in order to match X-ray observed mean actin monomer spacings. To establish a firm correlation between thin filament spacings and average force per thin filament, it is necessary to measure, beside the X-ray diffraction higher order actin meridional reflections, the muscle tension and the fraction of myofibrils in a given cross-section of the muscle. Kiss et al. observed the first actin meridional X-ray reflection, tension and the occupancy of soleus muscle cross-section [
33], but the geometry of the soleus muscle is significantly different from frog sartorius muscle, so their estimates may not be appropriate.
From Huxley’s experimental X-ray patterns used here [
12], we estimated a force per actin filament of 297 pN, corresponding to a muscle tension of ~300 kPa. Linearly scaling the estimate for thin filament stiffness from Kiss et al., of 30 pN/nm per 1 μm long thin filament, to Kojima’s stiffness of 65 pN/nm, will bring the estimated tension in Huxley’s experiments on bullfrog sartorius muscle to ~140 kPa. This estimate seems too low, because Huxley et al. reported a range of tensions between 200 and 300 kPa [
12]. From similar measurements, Wakabayashi reported muscle tension of ~280 kPa [
13]. Assuming that the average tension in Huxley’s experiments is in the range from 250 to 280 kPa, the corrected thin filament stiffness for the stiffening by phalloidin could be in the range of 54 to 60 pN/nm per 1 μm long thin filament, respectively. Taking into account these estimated thin filament stiffnesses, the forces and tensions shown in
Figure 8B could be scaled down by 10–15%. If the thin filament stiffness is decreased by 15% (55.25 pN/nm), then the force per actin filament is 251.5 pN and this force can be achieved by reduction in the level of activation, i.e., reduction in apparent
Regardless of the uncertainties discussed above, the mean forces and their variation calculated by simultaneous measurements of X-ray diffraction profiles and muscle tension can be used for the estimation of the number of bound crossbridges, and the mean and variation of the crossbridge forces. This is one of the unresolved fundamental questions in muscle contraction and can be quantitatively assessed by MUSICO simulations employing various crossbridge cycles. For the crossbridge cycle used in MUSICO simulations with parameters appropriate for a muscle tension of 300 kPa, the average number of crossbridges bound is 72.5%, i.e., 108/150; and the number of bound crossbridges may vary between the thick filaments in similar fashion as variation of force shown in
Figure 8B. The predicted number of bound crossbridges per myosin filament is similar to the observed 72% of bound crossbridges or 36% of bound heads in frog (
Rana temporaria) [
34]. In the MUSICO simulations, at any moment of time, we have a mixture of weakly and strongly bound crossbridges. Due to the strain dependence, the state transition rates between crossbridge states, each crossbridge experiences different force even in the same state. From this analysis, the estimated average crossbridge force is ~5.5 pN. A decrease in the stiffness of the thin filaments of about 15% requires a decrease in predicted isometric tension also by about 15%, in order to keep average spacing as observed by X-ray diffraction. In the MUSICO simulations, the decrease in muscle force is achieved by reduction in apparent binding rate,
, i.e., reduced levels of activation; and this decrease is manifested primarily as a decrease in the number of attached crossbridges, to about 61.5% or to 82/150 per half myosin filament with very little change in mean crossbridge force. This estimated crossbridge force is similar to that estimated from simple analysis of mechanical and X-ray observations [
9,
29,
34]. However, crossbridge force estimates from these simple analyses showed a large range of values (from 3.5 to 5.5 pN). This variation in average force could originate in numerous assumptions and simplifications used in their calculation, including rigor vs. fully activated muscle, range of estimates of thick and thin filament stiffnesses, and the range of estimates of crossbridge stiffnesses. Some of thin filament and crossbridge stiffnesses used in previous studies have unreasonable values that can skew the assessed values of the fraction of bound crossbridges and mean crossbridge forces, thus, a comprehensive approach is necessary. The analysis presented here, however, precisely takes into account geometrical factors and requires only a few assumptions regarding the elasticities of the filaments and crossbridges. Importantly, it relates the measured spacings in thin filaments to the forces in actin filaments which vary according to the crossbridge forces. Thus, this approach opens the way toward a more reliable resolution of the enigma of the relative contributions of crossbridge forces and the number of bound crossbridges to the observed tension. The remaining assumptions in the simulations could be removed by constraining MUSICO simulations by multiple experiments on the same muscle type and experimental conditions accompanied with measurements of ATPase and other energetic quantities, along with reliable mechanical measurements of muscle tension accompanying the X-ray diffraction patterns.
The ability to estimate forces in fibrous proteins in living cells could be instrumental in defining the modulation of biochemical processes associated with mechano-transduction. For example, the role of structural proteins in Z-disc and M-band, such as titin, myomesin, obscurin and α-actinin, have been suggested to be involved in active mechano-signaling processes that control gene expression, protein synthesis and protein degradation in the sarcomeres of striated muscle [
23,
24,
25,
26,
27]. The stochastic nature of myosin binding produces a large variation of the forces exerted by individual myosin filaments on the M-band and actin filaments on the Z-disc. Thus, the signaling functions in muscle mechano-transduction might be related rather to differences in forces between neighboring myosin or actin filaments than to the absolute force acting on the M-band or the Z-disc, respectively. The reason is that M-band and Z-disc structural proteins are deformed by local differences in forces, thus the prospective mechano-transduction process may be associated with fluctuations in forces to which these proteins are exposed. Using X-ray diffraction measurements in concert with MUSICO simulations could be used to define the magnitude of these force fluctuations triggering signaling that modulate gene expression and the adaptation of muscle in response to changes in local mechanical strain.
Limitations of the Study
Our goal in this paper was to explain, for the first time, how muscle tension and the values of axial actin spacings are related to the heterogeneity of forces in the thin filaments in tetanically contracting muscle. We demonstrated how the magnitude of the heterogeneity of the forces can be estimated using a Monte Carlo approach and the observed higher order meridional reflections. We believe that this goal has been achieved. There are, however, other structural changes in the thin filaments associated with activation that have been observed in muscles stretched beyond filament overlap [
13,
35], and comparing patterns from resting and actively shortening muscle at velocity close to a maximum [
36]. While all studies indicate that at maximum isometric tension, thin filaments are about 0.3% longer than at rest, these latter studies show that when activated without significant binding of force producing heads, thin filaments are about 0.1% shorter and more twisted than at rest. It is likely that consideration of these activation related events could change the results in
Figure 8. Our current analysis gives numbers consistent with known forces and filament extensibilities, so these changes are likely to be modest. The mechanisms responsible for these structural changes with activation, however, are not known, so we had no basis on which to attempt to model them using MUSICO. A more precise model of structural changes in the thin filaments in contracting muscle that could address all levels of force and length changes would require new experiments expressly designed for this purpose. Our approach is general enough to address any such new data as they become available.
6. Conclusions
The new methodology presented here, combining small angle X-ray diffraction and multi-scale simulation, provides a unique tool for measurement of force acting on a single myosin or actin filament in living muscle fibers. The centroid of the relatively sharp 1st actin meridional peak best describes the average inter-monomer spacing, whereas the much broader width of the 2nd meridional peak reflects both the degree of nonuniformity of spacing along each actin filament and the variation of forces in different actin filaments. MUSICO simulations can separate these two effects on the width and shape of 2nd order actin meridional peak showing that there are large variations in the forces in different actin filaments in a sarcomere during contraction.
The MUSICO simulations generally predicted well the changes in molecular spacings during force development and relaxation in skeletal muscle observed experimentally by H.E. Huxley et al. [
12]. The estimated myofilament forces in living muscle fibers provide the possibility for a more precise assessment of crossbridge forces. Moreover, the effect of local differences in myofilament forces acting on signaling molecules in M-band and Z-discs could be important in activating protein synthesis and degradation, and triggering muscle adaptation in response to changes in mechanical strain.
This approach can be extended to any muscle system, and it could ultimately provide an interpretive framework for studying mechanisms of inherited or acquired muscle diseases in transgenic animal models and, prospectively, humans. By extracting the maximum information from the X-ray patterns, in combination with the physiological data, this approach provides a template to test hypotheses concerning crossbridge and regulatory protein action in working muscle.