Abstract
The JASMONATE ZIM DOMAIN (JAZ) proteins act as negative regulators in the jasmonic acid (JA) signaling pathways of plants, and these proteins have been reported to play key roles in plant secondary metabolism mediated by JA. In this study, we firstly isolated one JAZ from P. cablin, PatJAZ6, which was characterized and revealed based on multiple alignments and a phylogenic tree analysis. The result of subcellular localization indicated that the PatJAZ6 protein was located in the nucleus of plant protoplasts. The expression level of PatJAZ6 was significantly induced by the methyl jasmonate (MeJA). Furthermore, by means of yeast two-hybrid screening, we identified two transcription factors that interact with the PatJAZ6, the PatMYC2b1 and PatMYC2b2. Virus-induced gene silencing (VIGS) of PatJAZ6 caused a decrease in expression abundance, resulting in a significant increase in the accumulation of patchouli alcohol. Moreover, we overexpressed PatJAZ6 in P. cablin, which down-regulated the patchoulol synthase expression, and then suppressed the biosynthesis of patchouli alcohol. The results demonstrate that PatJAZ6 probably acts as a repressor in the regulation of patchouli alcohol biosynthesis, contributed to a model proposed for the potential JA signaling pathway in P. cablin.
1. Introduction
Pogostemon cablin (P. cablin), the medicinal part of which is dry whole grass, is a kind of Labiatae plant that has long been considered an important Chinese herbal medicine in Lingnan [1]. The main medicine produced from P. cablin is patchouli alcohol, which has been reported to have antibacterial [2], anti-inflammatory [3] and vasodilatory [4] properties, among others. On the one hand, patchouli alcohol may be mainly used in the perfume and cosmetics industry, and on the other hand, it may be frequently used for medical treatment. Due to the large market demand [5], an increasing number of scholars have begun to study P. cablin, especially the molecular synthesis mechanism of patchouli alcohol. Many investigations have focused on pharmacological effects in P. cablin; however, current knowledge on patchouli alcohol biosynthetic pathways is limited. In cultivation and production, the quality of patchouli is often related to planting light conditions, ambient temperature and different abiotic and biotic stresses. Previous studies in our research group found that these factors affected the expression of the patchouli alcohol synthase gene and caused significant differences in patchouli alcohol accumulation [6]. Subsequently, various exogenous hormone treatment experiments showed that patchouli alcohol synthesis was specifically induced by Methyl jasmonate (MeJA). A large number of studies have shown that different environmental signals stimulate plant synthesis of jasmonic acid [7], which affects the synthesis and accumulation of important secondary metabolites and their chemical reactions through JA signaling. Therefore, we predict that the biosynthesis process of patchouli alcohol may be highly dependent on the JA signaling. The pattern of JA signaling regulating the synthesis of secondary metabolites in medicinal plants has progressed in Catharanthus roseus, the main component of which is vinblastine [8]. In addition, this signaling also exists in tobacco (nicotine) [9].
JA is one of the most important signaling molecules in plants, governing responses to abiotic and biotic stresses, as well as in secondary metabolite biosynthesis, signal transduction, stress response [10], growth and development [11]. The JASMONATE ZIM DOMAIN (JAZ) proteins are not only an important component of the JA signaling pathway but also a node that links different signaling pathways in plants [12]. The N-terminus of the JAZ contains a weakly conserved N-terminal (NT) domain. The ZIM domain contains a conserved TIF[F/Y]XG (TIFY) motif, and the Jas domain at the C-terminus is highly conserved and can interact with many proteins [13], such as various transcription factors (TFs). In addition, there are nuclear localization signals in the Jas domain, which causes JAZ proteins to have nuclear localization properties [14]. Thirteen JAZ proteins have been found in Arabidopsis thaliana, playing roles in growth, defense, and reproductive output [15], these proteins are regarded as repressors in the JA signaling pathway, and they interact with the MYC2 transcription factor and repress its function. MYC2 is the initial transcription factor of the JA response gene [16], but the JA induction of plants is controlled by CORONATINE INSENSITIVE 1 (COI1). JAZ family proteins have been identified as COI1 targets and repressors of MYC2 [17]. To date, JAZ proteins have been proven to repress the activity of TFs, and function as repressors in the JA pathway to mediate many developmental processes, including secondary metabolite synthesis.
As a pivotal regulator in the plant JA signaling pathway, JAZ proteins should play important regulatory roles in the biosynthesis of patchouli alcohol in P. cablin. However, little is known about the unambiguous roles of JAZ proteins in patchouli alcohol biosynthesis in P. cablin. In our previous work, we obtained 82,335 raw data of transcriptome using next-generation sequencing (NGS) technology from leaves of P. cablin treated with MeJA [18], 12 unigenes were recognized and identified as JAZ family genes, based on the expression levels of 12 genes under the induction of 300 µM MeJA and our previous screening experiment, we ultimately chose PatJAZ6 (Unigene48011) as the research object, which may participate in the regulation of biosynthesis of patchouli alcohol.
In this study, the PatJAZ6 gene from P. cablin was cloned and characterized, and the relative expression pattern analysis was performed. Then, the subcellular localization study of PatJAZ6 was performed. To further research on the interaction between PatJAZ6 and PatMYC2b1/PatMYC2b2, which was cloned and identified by our laboratory [18], we performed a Yeast two hybrid (Y2H) assay and Firefly Luciferase Complementation Imaging Assay (LCI) verification. Furthermore, to illuminate the roles of PatJAZ6 in patchouli alcohol biosynthesis, gene silencing induced by viruses and overexpression of PatJAZ6 in plants was examined and analyzed. We ultimately elucidated a molecular conduction model between PatJAZ6 and PatMYC2b1/PatMYC2b2 in patchouli alcohol biosynthesis. The present study is the first to analyze the function of the PatJAZ6 gene in P. cablin, which may be instructive for the study of JA signaling molecular mechanisms and secondary metabolites in P. cablin.
2. Results
2.1. Bioinformatics Analysis of PatJAZ6 from P. cablin
To determine the basic bioinformatics of PatJAZ6, several software and websites were used in this work. Analysis of the sequence revealed that PatJAZ6 consists of 1375 bp with a 972-bp open reading frame (ORF). Amino acid alignment confirmed that PatJAZ6 is a member of the JAZ family. JAZ proteins contain three conserved domains, NT, ZIM, and Jas (Figure 1A).Multiple alignments of PatJAZ6 with 12 JAZ proteins from Arabidopsis thaliana showed that PatJAZ6 contained two conserved domains: the ZIM domain (TIFY motif), which is located near the N-terminus, and the Jas domain, which is near the C-terminal (Figure 1B). To further identify the characteristics of PatJAZ6, a phylogenetic tree was constructed using MEGA 7 software. The result illustrated that PatJAZ6 was highly homologous to AtJAZ9 (Figure 1C).
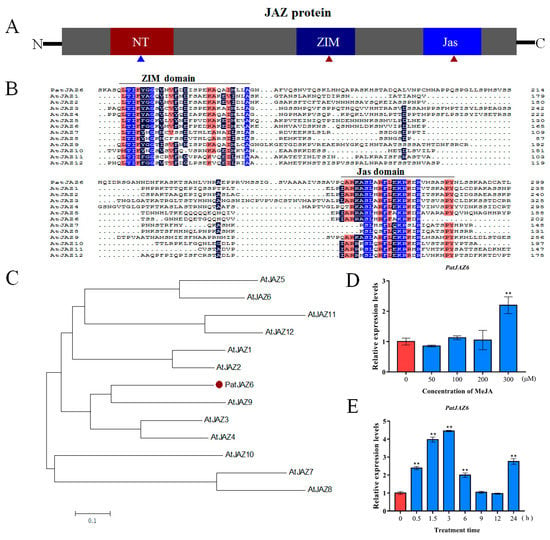
Figure 1.
Bioinformatics analysis and expression profiles of PatJAZ6. (A) A schematic diagram of JAZ (Jasmonate ZIM domain, JAZ) protein. Red triangles represent conserved domains, blue triangle represents a weakly conserved NT domain. (B) Multiple alignments of PatJAZ6 with 12 JAZ proteins from Arabidopsis thaliana. AtJAZ1 (NP_564075.1), AtJAZ2 (NP_565096.1), AtJAZ3 (NP_974330.1), AtJAZ4 (NP_175283.2), AtJAZ5 (NP_001320905.1),AtJAZ6 (NP_001321693.1), AtJAZ7(NP_181007.1), AtJAZ8(NP_564349.1), AtJAZ9(AAL32593.1), AtJAZ10(NP_568287.1), AtJAZ11(NP_001190007.1), AtJAZ12 (NP_197590.1). (C) Phylogenic tree of PatJAZ6 with 12 JAZ proteins from A. thaliana was built using MEGA 7. The red solid dot represents PatJAZ6. (D) The relative expression of PatJAZ6 was calculated after treatment with different concentrations of MeJA for 8 h, and the results were calculated according to the expression of PatJAZ6 at 0 µM MeJA. (E) The relative expression of PatJAZ6 was analyzed at different time points under 300 µM MeJA treatment. The results were calculated based on the expression of PatJAZ6 at 0 h. (One-way ANOVA test; ** p < 0.01).
2.2. Expression Profiles of PatJAZ6 under MeJA Treatments
MeJA plays an important role in regulating secondary metabolite synthesis in a variety of plants. To detect whether PatJAZ6 responds to MeJA, the relative expression of PatJAZ6 was analyzed by qRT-PCR after treatment with different concentrations of exogenous MeJA for 8 h (Figure 1D). This analysis showed that 300 µM MeJA effectively induced the expression of PatJAZ6 in P. cablin; therefore, 300 µM MeJA was selected as the lowest effective concentration to detect the expression of PatJAZ6. Different time points (0, 0.5, 1.5, 3, 6, 9, 12, and 24 h) were set after MeJA treatment to detect PatJAZ6 expression. The results showed that MeJA effectively induced the expression of PatJAZ6 in P. cablin (Figure 1E). Within 3 h of MeJA treatment, the expression levels of PatJAZ6 increased rapidly and reached a maximum at 3 h, subsequently decreased at 6 h, and returned to the initial levels at 9 h and 12 h. After 24 h, the expression of PatJAZ6 began to increase again, which is possible that expression of PatJAZ6 is driven by circadian rhythms.
2.3. Subcellular Localization of PatJAZ6
To determine the subcellular localization of PatJAZ6, the ORF of PatJAZ6 without a termination codon was inserted into the N-terminus of the Green Fluorescent Protein (GFP) tag in vector PAN580. The recombinant plasmid was transformed into A. thaliana protoplasts by the polyethylene glycol (PEG-mediated method [19]. Subcellular localization results showed that PatJAZ6 was localized in the nucleus (Figure 2). We can infer that PatJAZ6 is highly likely to play functional roles in the nucleus, such as regulating transcription factors in the JA signaling pathway.
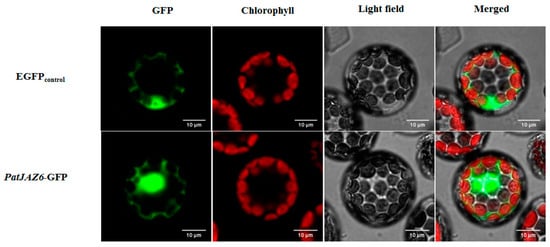
Figure 2.
Subcellular localization of PatJAZ6 in Arabidopsis protoplasts. The open reading frame (ORF) without a termination codon was inserted into the vector named PAN580, Enhanced Green Fluorescent Protein (EGFP) was used as a control.
2.4. PatJAZ6 Protein Interacts with PatMYC2b1 and PatMYC2b2
Based on the reported research that JAZ proteins interact with some TFs [20], such as MYC2, which plays a regulated role in the JA signaling pathway, the ORFs of PatJAZ6 and TFs (PatMYC2b1/PatMYC2b2) used in this study were cloned by the gene primers (Table S1) and fused to digested vectors PGBKT7 and pGADT7, respectively, in our research group. To explore whether PatJAZ6 can interact with PatMYC2b1 and PatMYC2b2, a Y2H screen was chosen to confirm the possible interaction relationship between them. Our screen results showed that pGBKT7-PatJAZ6 + pGADT7-PatMYC2b1 and pGBKT7-PatJAZ6+pGADT7-PatMYC2b2 could grow normally on three types of screening plates and turned blue on SD/-Trp/-Leu/-His/-Ade/ plates containing X-α-Gal, which is consistent with the positive control, indicating that there is an interaction between PatJAZ6 and PatMYC2b1 /PatMYC2b2 in yeast systems, which implied that a relationship between PatJAZ6 and PatMYC2b1 /PatMYC2b2 may have existed in plants to regulate the biosynthesis of secondary metabolites (Figure 3A).
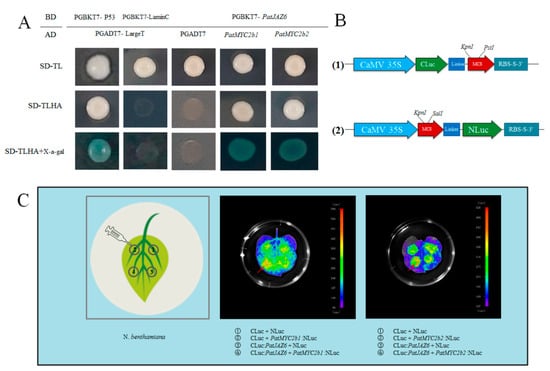
Figure 3.
Protein interaction verification of PatJAZ6. (A) Yeast two-hybrid assay. Plasmids PGADT7-LargeT and pGBKT7-P53 were cotransformed into AH109 as the positive control. Plasmids PGADT7-LargeT and PGBKT7-LaminC were cotransformed into AH109 as the negative control. PGBKT7-PatJAZ6 and pGADT7-TFs were cotransformed into Saccharomyces cerevisiae AH109 competent cells. The blue colonies represent the positive results. (B) (1) The complete ORF of PatJAZ6 was inserted into the vector named PCAMBIA1300CLuc with the restriction enzyme sites KpnI and PstI. (2) The complete ORFs of PatMYC2b1 and PatMYC2b2 were inserted into the vector named PCAMBIA1300NLuc with the restriction enzyme sites KpnI and SalI, respectively. (C) Firefly Luciferase Complementation Imaging Assay. LCI images of N. benthamiana leaves coinfiltrated with the Agrobacterial GV3101-Psoup-p19 strains containing PatMYC2b1/PatMYC2b2:NLuc and CLuc:PatJAZ6. Arrow positions indicate where the signal is strongest.
To verify whether this interaction exists in plants, further confirming the reliability of the Y2H results, a LCI was performed in Nicotiana benthamiana leaves by injecting A. tumefaciens GV3101-Psoup-p19 cultures containing recombinant constructs (Figure 3C). Injection positions ①, ② and ③, which represent different plasmid combinations, were set as negative controls. Injection positions ④ in LCI images showed large red areas, whereas ①, ② and ③ had almost no red areas, indicating that the signal at position ④ was significantly stronger than the control, revealing that PatJAZ6 and PatMYC2b1 /PatMYC2b2 have a strong interaction in N. benthamiana, implying a relationship between them most likely existed in P. cablin.
2.5. Effect on Patchouli Alcohol Biosynthesis by the Virus Induced PatJAZ6 Silencing
To investigate the roles of the PatJAZ6 protein in the JA signaling pathway affecting the synthesis of patchouli alcohol, Virus-induced gene silencing (VIGS) was selected to silence PatJAZ6 to explore the effects of PatJAZ6 on related genes and patchouli alcohol. The buffer containing a 1:1 ratio of PTRV1 and PTRV2 was set as a control. Leaf tissues were collected after PatJAZ6 was silenced 14 days, which was used for qRT-PCR and Gas Chromatography-Mass Spectrometer (GC-MS) analysis. In the virus-induced PatJAZ6 silencing, the relative expression level of PatJAZ6 was clearly downregulated in comparison to the control (Figure 4B), while the expression of patchoulol synthase (PTS), which is the key enzyme for patchouli synthesis, was upregulated by approximately 80%. Moreover, the relative expression of PatMYC2b1 and PatMYC2b2 interacting with PatJAZ6 were all increased, especially PatMYC2b2. The results of GC-MS showed that the content of patchouli alcohol in the VIGS-JAZ6 group (3.67 mg/g Fresh weight (FW)) was significantly higher than that in CK (2.5 mg/g FW), exhibiting an increase of 32% (Figure 4C). GC-MS chromatograms of samples from the standard of patchouli alcohol (top panel), CK (middle panel) and VIGS-JAZ6 (bottom panel) leaves showing abundance of patchouli alcohol (Figure 4D). The expression tendency of PatJAZ6 was contrary to PatMYC2b1 and PatMYC2b2, and based on the results of Y2H and LCI, we can speculate that PatJAZ6 plays a role as a transcriptional repressor in P. cablin. In addition, when PatJAZ6 was silenced, patchouli alcohol synthesis was increased, which may be the result of a common increase in TFs and PTS gene expression.
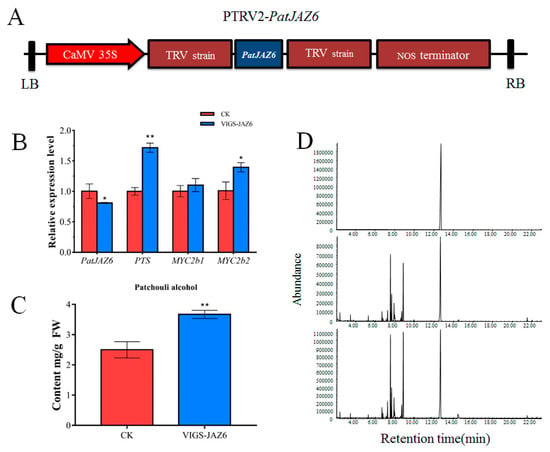
Figure 4.
Analysis of virus-induced PatJAZ6 silencing. (A) The PatJAZ6 gene fragment (less than 500 bp) was cloned into the PTRV2 vector to form the PTRV2-PatJAZ6. (B) The corresponding mRNA expression level of VIGS-JAZ6 analyzed by real-time q-PCR. (C) The content of patchouli alcohol detected in control check (CK) and VIGS-JAZ6 leaves. (D) Gas Chromatography-Mass Spectrometer (GC-MS) chromatograms of samples from the standard of patchouli alcohol (top panel), CK (middle panel) and VIGS-JAZ6 (bottom panel) leaves showing abundance of patchouli alcohol. Asterisks indicate a significant difference from the control. (Student’s t-test; ** p < 0.01, * p < 0.05).
2.6. Effect on Patchouli Alcohol Accumulation by the Overexpression of PatJAZ6
To confirm that PatJAZ6 plays a role as a transcriptional repressor in P. cablin, based on the VIGS-PatJAZ6 experiment, we hypothesized that overexpression of PatJAZ6 may show opposite results to gene silencing of PatJAZ6. The empty PJLTRBO vector was transformed into GV3101-Psoup-p19 as CK. In the PatJAZ6-overexpressing P. cablin leaves, PatJAZ6 expression was upregulated in comparison to the CK, while the transcripts of PTS, PatMYC2b1 and PatMYC2b2 were all reduced at different levels. Among these genes, PatMYC2b2 showed the most significant decrease, exhibiting a nearly 70% reduction (Figure 5B). The content of patchouli alcohol in PJLTRBO-PatJAZ6 was determined by GC-MS, consistent with the gene expression profile of PTS, PatMYC2b1 and PatMYC2b2. The accumulation of patchouli alcohol in PatJAZ6 overexpressing appeared to decrease. Overexpression of PatJAZ6 produced lower levels of patchouli alcohol (5.04 mg/g FW) compared with the control (6.76 mg/g FW) (Figure 5C). The results described above confirmed our conjecture, indicating that PatJAZ6 may repress the biosynthesis of patchouli alcohol.
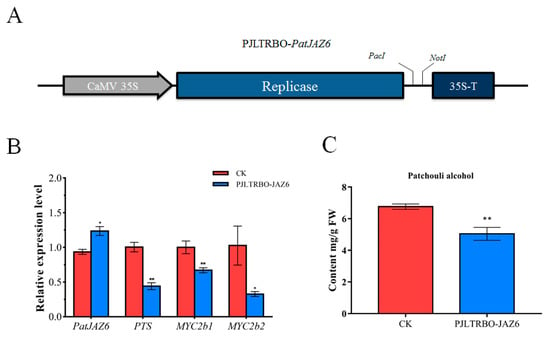
Figure 5.
Overexpression analysis of PatJAZ6. (A) The PatJAZ6 gene fragment was cloned into the PJLTRBO vector to form the PJLTRBO-PatJAZ6 construct with the restriction enzyme sites PacI and NotI. (B) The corresponding mRNA expression level of PJLTRBO-PatJAZ6 analyzed by real-time q-PCR. (C) The content of patchouli alcohol detected in CK and PJLTRBO-PatJAZ6 leaves. Asterisks indicate a significant difference from the control (Student’s t-test; ** p < 0.01, * p < 0.05).
3. Discussion
Patchouli alcohol, a natural tricyclic sesquiterpene compound that is a bioactive ingredient in P. cablin [21], is used worldwide in decorative cosmetics, toilet soaps, perfume industries [22] and medical treatments [23]. With the increasing market demand in the world, a metabolic engineering approach has been considered an effective approach to increase useful metabolites in medical plants, and several strategies have been reported to promote the industrialization process of patchouli alcohol production using Saccharomyces cerevisiae [24], and gene isolation and cloning in MVA and MEP pathways participating in patchouli alcohol biosynthesis have been reported [25]. In addition, full-length transcriptome data reported provide a valuable genetic resource in P. cablin [18]. Despite in-depth research on genes related to patchouli alcohol synthesis, little is known concerning the regulation of patchouli alcohol biosynthesis.
MeJA is an important plant endogenous hormone that is widely present in plants and regulates metabolic and developmental processes in plants [26]. Exogenous application of MeJA can stimulate the expression of defense genes and induce chemical defense in plants, including stimulating the synthesis of a series of secondary metabolites [27]. A growing number of reports indicate that the synthesis of many secondary metabolites in medicinal plants is increased under MeJA induction [28]. Our previous experiments showed that MeJA treatment on P. cablin leaves can significantly increase the accumulation of patchouli alcohol (Figure S1), but the specific molecular mechanisms involved have not been elucidated. We hypothesize that this effect may be related to JA signaling in plants and that the key factors of JA signaling are JAZ proteins. In our current research, the PatJAZ6 gene was cloned from P. cablin and functionally identified as a repressor involved in patchouli alcohol biosynthesis. Bioinformatics analysis revealed that the PatJAZ6 gene showed high homology with 12 JAZs from A. thaliana Arabidopsis and contained highly conserved ZIM and Jas domains, indicating that PatJAZ6 may have similar effects to previously reported JAZ proteins. The expression profiles under MeJA revealed that 300 µM MeJA was the lowest effective concentration to detect the expression of PatJAZ6, this concentration is higher in comparison with other JAZs, such as NtJAZ in tobacco [29] and SmJAZ in Salvia miltiorrhiza [30], that response to 100 µM MeJA. Subcellular localization results showed that PatJAZ6 was localized in the nucleus (Figure 2). This result is consistent with the characteristics of the Jas motif with nuclear localization. Previous studies on the subcellular localization of other JAZs also support this result [31].
JAZ proteins belong to ZIM-domain proteins, are located near the C-terminus, and have a highly conserved Jas motif of 26 amino acids. Studies have now determined that Jas motifs are involved in protein-protein interactions with MYC and COI1 [32]. In our present study, it was found that the PatJAZ6 protein can interact with PatMYC2b1 and PatMYC2b2 through a yeast two-hybrid (Y2H) screening method, which was consistent with previous reports that JAZ proteins interact with the MYC2 transcription factor [33]. Furthermore, an LCI assay was performed in N. benthamiana leaves, which further confirmed the interaction between PatJAZ6 and PatMYC2b1/PatMYC2b2 separately. The above data suggest that PatMYC2b1 and PatMYC2b2 transcription factors in P. cablin may be targets of the PatJAZ6 protein in P. cablin and play key regulatory roles in the accumulation of patchouli alcohol. Of course, there are other transcription factors involved in this synthesis process, which warrants further investigation.
VIGS is widely used to downregulate target genes in a majority of plants [34]. Tobacco rattle virus (TRV) is one of the most widely used viruses in VIGS technology [35]. The VIGS system constructed by this virus is rapidly applied to the model plant N. benthamiana [36] and such crops as pepper [37], cotton [38], and tomato [39]. For the silencing effect, some studies have shown that the silencing effect of the target gene fragment between 300 and 500 bp is the best [40]. Not long ago, the efficient VIGS system in P. cablin was established by our own laboratory (Figure S2A). According to other research reports, gene silencing in wild tobacco revealed that NaJAZi functions as a flower-specific jasmonate repressor that regulates JAs, TPIs, (E)-α-bergamotene and a defensin. Flowers silenced in NaJAZi are more resistant to tobacco budworm attack [41]. In addition, there are reports that knockdown of AsJAZ1 expression through RNA interference led to decreased number of nodules, abnormal development of bacteroids, accumulation of poly-x-hydroxybutyrate (PHB) and loss of nitrogenase activity in legumes–rhizobia symbiosis [42]. However, in our experiments, gene silencing of PatJAZ6 in P. cablin leaves did not exhibit a distinct phenotype, except for slight curling of the leaves, but an increase in the expression of PTS, PatMYC2b1 and PatMYC2b2 was observed, resulting in a significant increase in the accumulation of patchouli alcohol in the VIGS-JAZ6 group (3.67 mg/g FW) compared with the control in CK (2.5 mg/g FW).
Research on JAZs in cash crops, including Oryza sativa [43], Glycine soja [44] and Gossypium hirsutum [45], has progressed rapidly in the past several years. Overexpression of JAZs in these crops produces different phenotypes. For example, overexpression of GsJAZ2 in soybean can increase the sensitivity of plants to salinity; overexpression of GhJAZ2 in cotton impairs the sensitivity to JA, decreases the expression level of JA-response genes (GhPDF1.2 and GhVSP) and enhances the susceptibility to V. dahliae and insect herbivory. However, we did not observe a significant phenotype in the P. cablin plants that overexpressed PatJAZ6, but we observed a decrease in the expression of PTS, PatMYC2b1 and PatMYC2b2, resulting in a lower level of patchouli alcohol (5.04 mg/g FW) in PJLTRBO-JAZ6 compared with the control (6.76 mg/g FW). JAZ proteins have different functions, which may be due to the different roles of transcription factors interacting with JAZ proteins. Since it has been reported that JAZ protein may be involved in the development of glandular trichomes which, in turn, affects the synthesis of secondary metabolites in Artemisia annua [46]; therefore, whether the gene silencing or overexpression of PatJAZ6 also affects the development of glandular trichomes in P. cablin requires further experimental confirmation.
In S. miltiorrhiza hairy roots, SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones [30]. According to the results of our study, it is reasonable to speculate that PatJAZ6 may act as a repressor in patchouli alcohol biosynthesis. Based on the above experimental results, we first present a model for the JA signaling pathway in P. cablin (Figure 6).
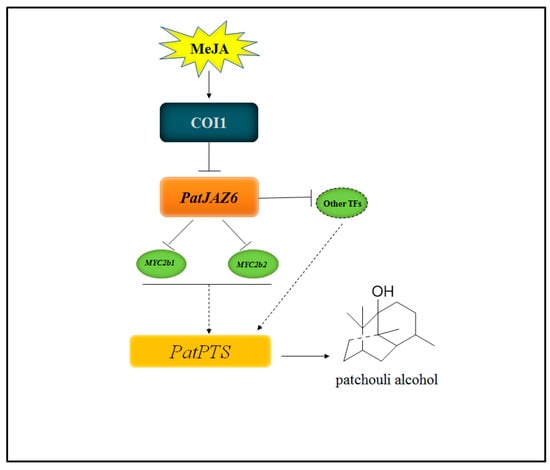
Figure 6.
Model describing the function of PatJAZ6 in JA-induced biosynthesis of patchouli alcohol. PatJAZ6 acts as a repressor regulating JA-induced biosynthesis of patchouli alcohol in Pogostemon cablin.
The model clearly illustrates the connection of how PatJAZ6 acts as a transcription factor suppressor regulating JA-induced biosynthesis of patchouli alcohol in P. cablin. However, further research is needed to determine whether PatJAZ6 interacts with other transcription factors and whether other JAZ proteins are involved in this signaling pathway. We also need to further explore the COI-JAZ-TFs model and how these proteins interact to regulate the synthesis of patchouli alcohol in P. cablin. Taken together, the results of this study help to elucidate the molecular regulation of JA signal-induced patchouli alcohol biosynthesis. Our work indicated that PatJAZ6 acts as a repressor in the regulation of patchouli alcohol biosynthesis. The discovery of the PatJAZ6 function points out a direction for the JA signaling pathway molecular mechanism and patchouli alcohol production in P. cablin.
4. Materials and Methods
4.1. Experimental Materials and Total RNA Extraction
The P. cablin plants were gathered from Yangjiang city, Guangdong Province, China. The cutting propagation method was used to obtain more seedlings that were used for the analysis of the expression patterns of PatJAZ6 and content of patchouli alcohol in leaves. The seeds of N. benthamiana were kept in our laboratory and grown in flower pots in a growth chamber. Well-growing plant materials, which were cultured under a constant environment at 25 °C with a 16/8 h photoperiod treatment, were selected for our experiments. The vector plasmids, E. coli competent cells DH5α and A. tumefaciens competent cells used in this study were all kept in our own laboratory. Total RNA was extracted from P. cablin leaves with the GeneMark Plant Total RNA Purification Kit (GeneMarkBio, Taichung, Taiwan), and then a spectrophotometer (IMPLEN GNBH, Germany) was used to determine the RNA concentration and purity. cDNA synthesis was performed via oligo dT and stored at −20 °C for subsequent experiments.
4.2. MeJA Treatments
MeJA was purchased from Sigma-Aldrich, St. Louis, Missouri, The United States of America, USA, dissolved in ethanol, formulated into 50 mM mother liquor for later use. To screen for the best response concentration, P. cablin plants were sprayed with MeJA solution at 0, 50, 100, 200 and 300 µM concentrations containing 0.1% Tween-80, and leaf samples were collected at 8 h after MeJA treatments.
To investigate the effect of MeJA on PatJAZ6 expression, P. cablin plants were sprayed in the morning with MeJA solution at a 300 µM concentration. Leaf samples were collected at time intervals of 0, 0.5, 1.5, 3, 6, 9, 12, and 24 h after MeJA treatment. All leaf samples were frozen with liquid nitrogen and stored in a −80 °C refrigerator for subsequent RNA extraction.
4.3. Bioinformatics Analysis of PatJAZ6
Bioinformatics analysis of PatJAZ6 was performed using several bioinformatics software and websites. The ORF of PatJAZ6 was determined using ORF finder (http://www.bioinformatics.org/sms2/orf_find.html), and 12 JAZ proteins from A. thaliana were searched from NCBI (https://www.ncbi.nlm.nih.gov/). MEGA v.7 software was used to construct the phylogenetic tree, and DNAMAN software was used to perform multiple sequence alignment.
4.4. Expression Patterns of PatJAZ6 by qRT-PCR
The expression patterns of PatJAZ6 and related genes under different treatments were quantified by qRT-PCR. Plant tissues were gathered after various treatments, and total RNA was isolated. cDNA synthesis was performed using HiScript II QRT SuperMix for qPCR (Vazyme R222-01, Nanjing, China), and the CFX96TM Real-Time System was selected to carry out qRT-PCR analysis with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711-02/03). qRT-PCR conditions were as follows: 95 °C for 3 min for one cycle, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. The relative expression levels of PatJAZ6 and related genes were calculated based on the 2−∆∆Ct method.
4.5. Subcellular Localization of PatJAZ6
The ORF of PatJAZ6 without a termination codon was fused to the N-terminus of the vector pAN580-GFP tag with the restriction enzyme sites SpeI and BamHI. The recombinant plasmids PAN580-PatJAZ6 were transformed into Arabidopsis protoplasts. The empty vector pAN580 was used as a negative control. The result of subcellular localization of PatJAZ6 was observed with ZEISS LSM 800 with Airyscan (ZEISS, Jena city, Germany).
4.6. Yeast Two-Hybrid Assays
The yeast two-hybrid (Y2H) screen was chosen to confirm possible TFs interacting with PatJAZ6. The ORF of PatJAZ6 was cloned into the pGBKT7 vector to generate pGBKT7-PatJAZ6. Both the ORFs of PatMYC2b1 and PatMYC2b2 were inserted into the PGADT7 vector to form pGADT7-PatMYC2b1 and pGADT7-PatMYC2b2, respectively. Plasmids PGADT7-LargeT and pGBKT7-P53 were cotransformed into S. cerevisiae AH109 competent cells as the positive control, while PGADT7-LargeT and PGBKT7-LaminC were cotransformed into AH109 as the negative control. Recombinant plasmids (pGBKT7-PatJAZ6+pGADT7-PatMYC2b1, pGBKT7-PatJAZ6+ pGADT7-PatMYC2b2 were cotransformed separately into AH109 cells and cultured on SD/-Trp/-Leu medium. Then, transformants were vaccinated on SD/-Trp/-Leu/-His/-Ade and SD/-Trp/-Leu/-His/-Ade/X-α-Gal to observe the interaction situation of PatJAZ6 with PatMYC2b1 and PatMYC2b2. All medium plates were incubated in an incubator at 29 °C for 3 days.
4.7. Firefly Luciferase Complementation Imaging Assay
To further understand the interaction between PatJAZ6 and PatMYC2b1/PatMYC2b2 in living plants, Firefly Luciferase Complementation Imaging Assay (LCI) was performed in N. benthamiana leaves. The complete ORF of PatJAZ6 was inserted into the vector PCAMBIA1300CLuc with the restriction enzyme sites KpnI and PstI, while ORFs of PatMYC2b1 and PatMYC2b2 were inserted into the vector PCAMBIA1300NLuc with the restriction enzyme sites KpnI and SalI, respectively (Figure 3B). The A. tumefaciens strain GV3101-Psoup-p19 was transformed with recombinant constructs by the freeze–thaw method. The cultured bacteria solution was mixed at a ratio of 1:1, centrifuged and resuspended in buffer, adjusted optical density (OD) to 0.8–1.0, then placed at room temperature for 2–4 h, injected the back of N. benthamiana leaves with a needle-free syringe (Figure 3C), incubated for 12 h at 23 °C in the dark, and moisturized for 2–4 days at room temperature. The leaves were placed in MS solid medium with the leaves facing up, sprayed with 100 mM d-luciferin potassium salt, and kept in the dark for 6 min. A Berthold Technologies (LB983 NC100,Germany) was used to capture the images with an exposure time of 2 min.
4.8. Virus-Induced PatJAZ6 Silencing
The pTRV1 and pTRV2 vectors were kept in our laboratory and used in this study. A 418-bp fragment (Figure S2B) from the PatJAZ6 ORF was cloned into the EcoRI and BamHI sites of the pTRV2 vector (Figure 4A). The resulting pTRV2-PatJAZ6 constructs, PTRV1 and PTRV2, were transformed into A. tumefaciens GV3101. The mixture of A. tumefaciens cultures containing a 1:1 ratio of PTRV1 and PTRV2 or pTRV2-PatJAZ6 was harvested by centrifugation and resuspended in infiltration buffer to obtain an OD of 1.0 and then incubated at room temperature for 2–4 h. Six-leaf-staged P. cablin plants were selected to infect with 1 mL needleless syringe on the abaxial side of leaves, and two to three leaves per plant needed infiltration. When PatJAZ6 was silenced for 14 days, leaf tissues were collected and frozen for later use.
4.9. Overexpression Analysis
The complete ORF fragment of PatJAZ6 was cloned into the PJLTRBO vector to form the PJLTRBO-PatJAZ6 construct with the restriction enzyme sites PacI and NotI (Figure 5A). The recombinant plasmids PJLTRBO-PatJAZ6 were transformed into GV3101-Psoup-p19, and the empty PJLTRBO vector was transformed into the same strain as the control. A. tumefaciens cultures were harvested and resuspended in infiltration buffer. For P. cablin leaf infiltration, the same injection method described in 4.8 was adopted. Samples were collected 4 days after plants were injected and frozen for later use.
4.10. Patchouli Alcohol Extraction and GC-MS Analysis
200 mg leaf tissues were ground frizzed in liquid nitrogen, 1.5 mL hexane was added into centrifuge tubes, ultrasonic for 30 min with 60 Hz and then heated under a 56 °C water bath for 1 h. After centrifugation, the supernatant was taken and passed through a 0.22-µm organic membrane as the test solution for GC-MS analysis using Agilent 7890B Gas Chromatograph with 5977A inert Mass Selective Detector (Agilent, California, USA). The gas chromatograph was equipped with an HP-5MS capillary column (30 m × 250 mm × 0.25 mm). The instrument was set to an initial temperature of 50 °C and maintained for 0 min. Then, the oven temperature was increased to 130 °C at a rate of 20 °C/min and then increased to 150 °C at a rate of 2 °C/min. The temperature was maintained at 150 °C for 5 min and later increased to 230 °C at a rate of 20 °C/min. The injection volume was 1 µL, and the injection port temperature was 230 °C. In addition, patchouli alcohol standards were purchased from NanTong FeiYu, China. All reagents used were analytical grade.
4.11. Agrobacterium Culture and Buffer Formulation
A. tumefaciens cultures were grown in the shaker with shaking (200 rpm/min) at 28 °C for 20–24 h. The infiltration buffer formulation as follows: 1 M 2-(4-morpholino)-ethane sulfonic acid; 1 M MgCl2 and 200 mM acetosyringone, dissolved in dimethyl sulfoxide.
4.12. Statistical Analysis
Statistical significance was determined by student’s t-test and one-way ANOVA.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/23/6038/s1. Figure S1. MeJA treatment on P. cablin leaves can significantly increase the accumulation of patchouli alcohol. Figure S2. (A) Virus induced PatPDS silencing in P. cablin. (B) Electropherogram of cloning a 418 bp fragment of PatJAZ6 into pTRV2 vector. Table S1: List of primers used in this study.
Author Contributions
L.C. and X.W. designed the research. Y.T. and X.C. performed the experiments and collected the data. X.Z., L.Z., Y.L., X.W. and J.L. analyzed the data and wrote the manuscript. R.Z., H.Z. and L.C. edited the manuscript and provided guidance during experimentation.
Funding
This research was supported by grants from the National Natural Science Foundation of China (81803657).
Acknowledgments
We appreciate Rui He (Guangzhou University of Chinese Medicine) for helping improve our manuscript writing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, M.; Zhang, J.; Lai, Y.; Wang, S.; Li, P.; Xiao, J.; Fu, C.; Hu, H.; Wang, Y. Analysis of Pogostemon cablin from pharmaceutical research to market performances. Expert Opin. Investig. Drugs 2013, 22, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.F.; Lian, D.W.; Chen, Y.Q.; Cai, Y.F.; Zheng, Y.F.; Fan, P.L.; Ren, W.K.; Fu, L.J.; Li, Y.C.; Xie, J.H.; et al. In Vitro and In Vivo Antibacterial Activities of Patchouli Alcohol, a Naturally Occurring Tricyclic Sesquiterpene, against Helicobacter pylori Infection. Antimicrob. Agents Chemother. 2017, 61, e00122-17. [Google Scholar] [CrossRef] [PubMed]
- Lian, D.W.; Xu, Y.F.; Ren, W.K.; Fu, L.J.; Chen, F.J.; Tang, L.Y.; Zhuang, H.L.; Cao, H.Y.; Huang, P. Unraveling the Novel Protective Effect of Patchouli Alcohol Against Helicobacter pylori-Induced Gastritis: Insights Into the Molecular Mechanism in vitro and in vivo. Front. Pharm. 2018, 9, 1347. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.Y.; Peng, C.; Xie, X.F.; Xiong, L.; Zhang, S.Y.; Cao, X.Y. Patchouli alcohol isolated from Pogostemon cablin mediates endothelium-independent vasorelaxation by blockade of Ca(2+) channels in rat isolated thoracic aorta. J. Ethnopharmacol. 2018, 220, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.K.; Sinniah, U.R. Patchouli (Pogostemon cablin Benth.): Botany, agrotechnology and biotechnological aspects. Ind. Crop. Prod. 2016, 87, 161–176. [Google Scholar] [CrossRef]
- Li, J.; Chen, X.; Zhong, L.; Wang, X.; Zhou, X.; Tang, Y.; Liu, Y.; Zheng, H.; Zhan, R.; Chen, L. Comparative iTRAQ-based proteomic analysis provides insight into a complex regulatory network of Pogostemon cablin in response to exogenous MeJA and Ethrel. Ind. Crop. Prod. 2019, 140, 111661. [Google Scholar] [CrossRef]
- Rahnamaie-Tajadod, R.; Goh, H.H.; Mohd Noor, N. Methyl jasmonate-induced compositional changes of volatile organic compounds in Polygonum minus leaves. J. Plant Physiol. 2019, 240, 152994. [Google Scholar] [CrossRef]
- Van Moerkercke, A.; Steensma, P.; Gariboldi, I.; Espoz, J.; Purnama, P.C.; Schweizer, F.; Miettinen, K.; Vanden Bossche, R.; De Clercq, R.; Memelink, J.; et al. The basic helix-loop-helix transcription factor BIS2 is essential for monoterpenoid indole alkaloid production in the medicinal plant Catharanthus roseus. Plant J. 2016, 88, 3–12. [Google Scholar] [CrossRef]
- Shoji, T.; Hashimoto, T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 2011, 52, 1117–1130. [Google Scholar] [CrossRef]
- Reinbothe, C.; Springer, A.; Samol, I.; Reinbothe, S. Plant oxylipins: Role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009, 276, 4666–4681. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qi, T.; Huang, H.; Ren, Q.; Wu, D.; Chang, C.; Peng, W.; Liu, Y.; Peng, J.; Xie, D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Goossens, A. The JAZ Proteins: A Crucial Interface in the Jasmonate Signaling Cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, W.; Vanholme, B.; Pauwels, L.; Plovie, E.; Inzé, D.; Gheysen, G.; Goossens, A. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 2009, 10, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Yoshida, Y.; Major, I.T.; Wang, K.; Sugimoto, K.; Kapali, G.; Havko, N.E.; Benning, C.; Howe, G.A. JAZ repressors of metabolic defense promote growth and reproductive fitness inArabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E10768–E10777. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Sanchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Wang, X.; Zhong, L.; Tang, Y.; Zhou, X.; Liu, Y.; Zhan, R.; Zheng, H.; Chen, W.; et al. Full-length transcriptome sequencing and methyl jasmonate-induced expression profile analysis of genes related to patchoulol biosynthesis and regulation in Pogostemon cablin. BMC Plant Biol. 2019, 19, 266. [Google Scholar] [CrossRef]
- Cao, Y.; Li, H.; Pham, A.Q.; Stacey, G. An Improved Transient Expression System Using Arabidopsis Protoplasts. Curr. Protoc. Plant Biol. 2016, 1, 285–291. [Google Scholar] [CrossRef]
- Fonseca, S.; Fernandez-Calvo, P.; Fernandez, G.M.; Diez-Diaz, M.; Gimenez-Ibanez, S.; Lopez-Vidriero, I.; Godoy, M.; Fernandez-Barbero, G.; Van Leene, J.; De Jaeger, G.; et al. bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS ONE 2014, 9, e86182. [Google Scholar] [CrossRef]
- Hu, G.; Peng, C.; Xie, X.; Zhang, S.; Cao, X. Availability, Pharmaceutics, Security, Pharmacokinetics, and Pharmacological Activities of Patchouli Alcohol. Evid. Based Complement. Altern. Med. 2017, 2017, 4850612. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.P.; Letizia, C.S.; Api, A.M. Fragrance material review on patchouli alcohol. Food Chem. Toxicol. 2008, 46, S255–S256. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.K.; Sinniah, U.R. A Comprehensive Review on the Phytochemical Constituents and Pharmacological Activities of Pogostemon cablin Benth: An Aromatic Medicinal Plant of Industrial Importance. Molecules 2015, 20, 8521–8547. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Liu, M.; Li, Z.H.; Tao, X.; Wei, D.Z.; Wang, F.Q. Significantly Enhanced Production of Patchoulol in Metabolically Engineered Saccharomyces cerevisiae. J. Agric. Food Chem. 2019, 67, 8590–8598. [Google Scholar] [CrossRef]
- Tang, Y.; Zhong, L.; Wang, X.; Zheng, H.; Chen, L. Molecular identification and expression of sesquiterpene pathway genes responsible for patchoulol biosynthesis and regulation in Pogostemon cablin. Bot. Stud. 2019, 60, 11. [Google Scholar] [CrossRef]
- Cao, J.; Li, M.; Chen, J.; Liu, P.; Li, Z. Effects of MeJA on Arabidopsis metabolome under endogenous JA deficiency. Sci. Rep. 2016, 6, 37674. [Google Scholar] [CrossRef]
- Gundlach, H.; Müller, M.J.; Kutchan, T.M.; Zenk, M.H. Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc. Natl. Acad. Sci. USA 1992, 89, 2389–2393. [Google Scholar] [CrossRef]
- Yu, H.; Guo, W.; Yang, D.; Hou, Z.; Liang, Z. Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza. Int. J. Mol. Sci. 2018, 19, 1593. [Google Scholar] [CrossRef]
- Zhang, H.; Li, W.; Niu, D.; Wang, Z.; Yan, X.; Yang, X.; Yang, Y.; Cui, H. Tobacco transcription repressors NtJAZ: Potential involvement in abiotic stress response and glandular trichome induction. Plant Physiol. Biochem. 2019, 141, 388–397. [Google Scholar] [CrossRef]
- Pei, T.; Ma, P.; Ding, K.; Liu, S.; Jia, Y.; Ru, M.; Dong, J.; Liang, Z. SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots. J. Exp. Bot. 2018, 69, 1663–1678. [Google Scholar] [CrossRef]
- Li, W.; Xia, X.C.; Han, L.H.; Ni, P.; Yan, J.Q.; Tao, M.; Huang, G.Q.; Li, X.B. Genome-wide identification and characterization of JAZ gene family in upland cotton (Gossypium hirsutum). Sci. Rep. 2017, 7, 2788. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Sun, W.; Chen, J.; Tan, H.; Xiao, Y.; Li, Q.; Ji, Q.; Gao, S.; Chen, L.; Chen, S.; et al. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 2016, 6, 22852. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Yellina, A.L.; Orashakova, S.; Becker, A. Virus-induced gene silencing (VIGS) in plants: An overview of target species and the virus-derived vector systems. Methods Mol. Biol. 2013, 975, 1–14. [Google Scholar]
- Ratcliff, F.; Martin-Hernandez, A.M.; Baulcombe, D.C. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001, 25, 237–245. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 2014, 9, 1549–1562. [Google Scholar] [CrossRef]
- Li, C.; Hirano, H.; Kasajima, I.; Yamagishi, N.; Yoshikawa, N. Virus-induced gene silencing in chili pepper by apple latent spherical virus vector. J. Virol. Methods 2019, 273, 113711. [Google Scholar] [CrossRef]
- Li, X.; Liu, N.; Sun, Y.; Wang, P.; Ge, X.; Pei, Y.; Liu, D.; Ma, X.; Li, F.; Hou, Y. The cotton GhWIN2 gene activates the cuticle biosynthesis pathway and influences the salicylic and jasmonic acid biosynthesis pathways. BMC Plant Biol. 2019, 19, 379. [Google Scholar] [CrossRef]
- Liu, Y.L.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Li, R.; Wang, M.; Wang, Y.; Schuman, M.C.; Weinhold, A.; Schafer, M.; Jimenez-Aleman, G.H.; Barthel, A.; Baldwin, I.T. Flower-specific jasmonate signaling regulates constitutive floral defenses in wild tobacco. Proc. Natl. Acad. Sci. USA 2017, 114, E7205–E7214. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, M.; Wang, N.; Li, Y. A JAZ Protein in Astragalus sinicus Interacts with a Leghemoglobin through the TIFY Domain and Is Involved in Nodule Development and Nitrogen Fixation. PLoS ONE 2015, 10, e0139964. [Google Scholar] [CrossRef] [PubMed]
- Hori, Y.; Kurotani, K.; Toda, Y.; Hattori, T.; Takeda, S. Overexpression of the JAZ factors with mutated jas domains causes pleiotropic defects in rice spikelet development. Plant Signal. Behav. 2014, 9, e970414. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Cai, H.; Luo, X.; Bai, X.; Deyholos, M.K.; Chen, Q.; Chen, C.; Ji, W.; Zhu, Y. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhu, L.; Wassan, G.M.; Wang, Y.; Miao, Y.; Shaban, M.; Hu, H.; Sun, H.; Zhang, X. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Mol. Plant Pathol. 2018, 19, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Chen, M.; Shen, Q.; Li, L.; Fu, X.; Pan, Q.; Tang, Y.; Shi, P.; Lv, Z.; Jiang, W.; et al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 2017, 213, 1145–1155. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).