Endoplasmic Reticulum Stress Cooperates in Silica Nanoparticles-Induced Macrophage Apoptosis via Activation of CHOP-Mediated Apoptotic Signaling Pathway
Abstract
1. Introduction
2. Results
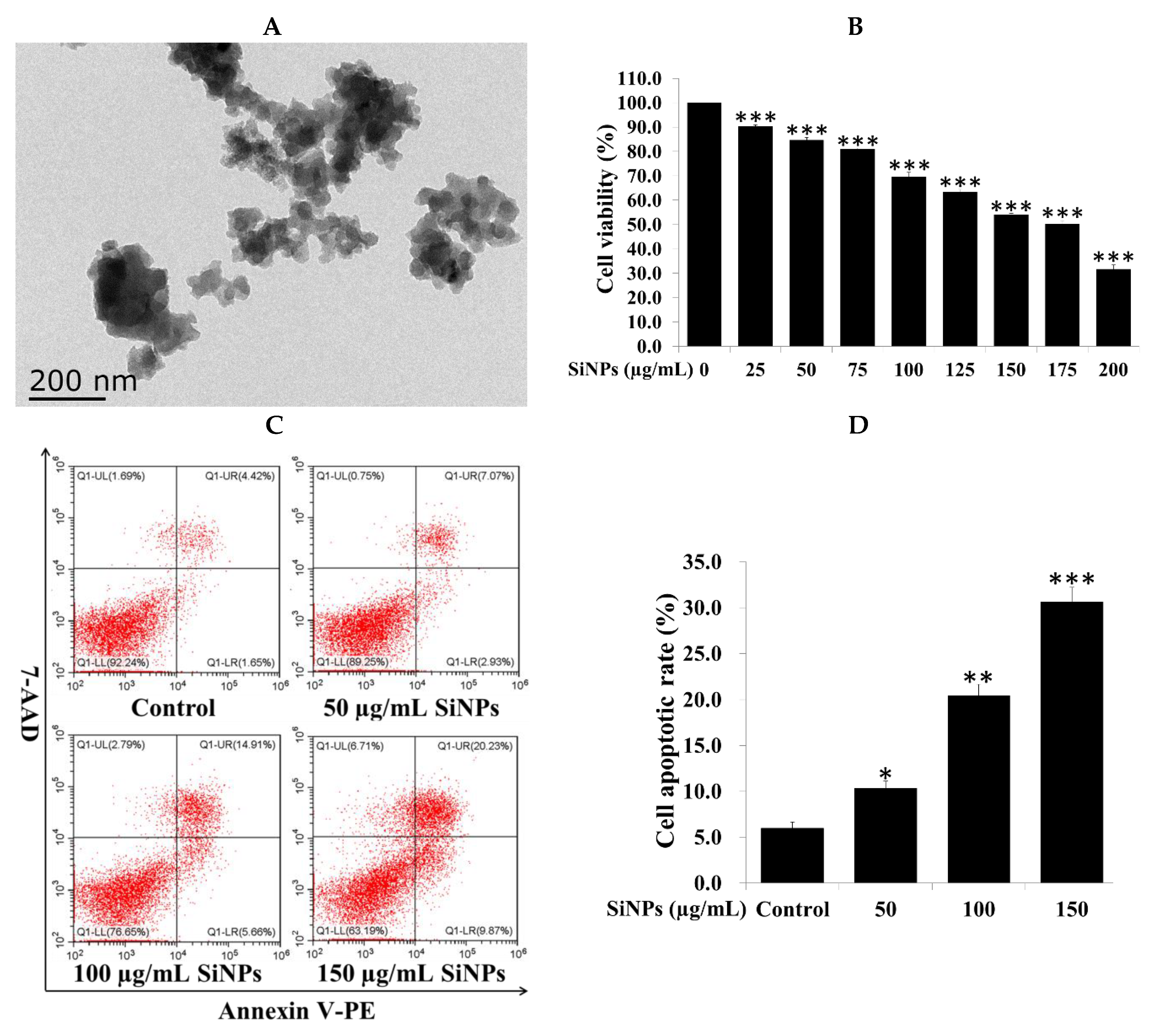
2.1. Silica Nanoparticle (SiNP) Characterization and Cell Viability and Apoptosis in RAW 264.7 Macrophage Cells
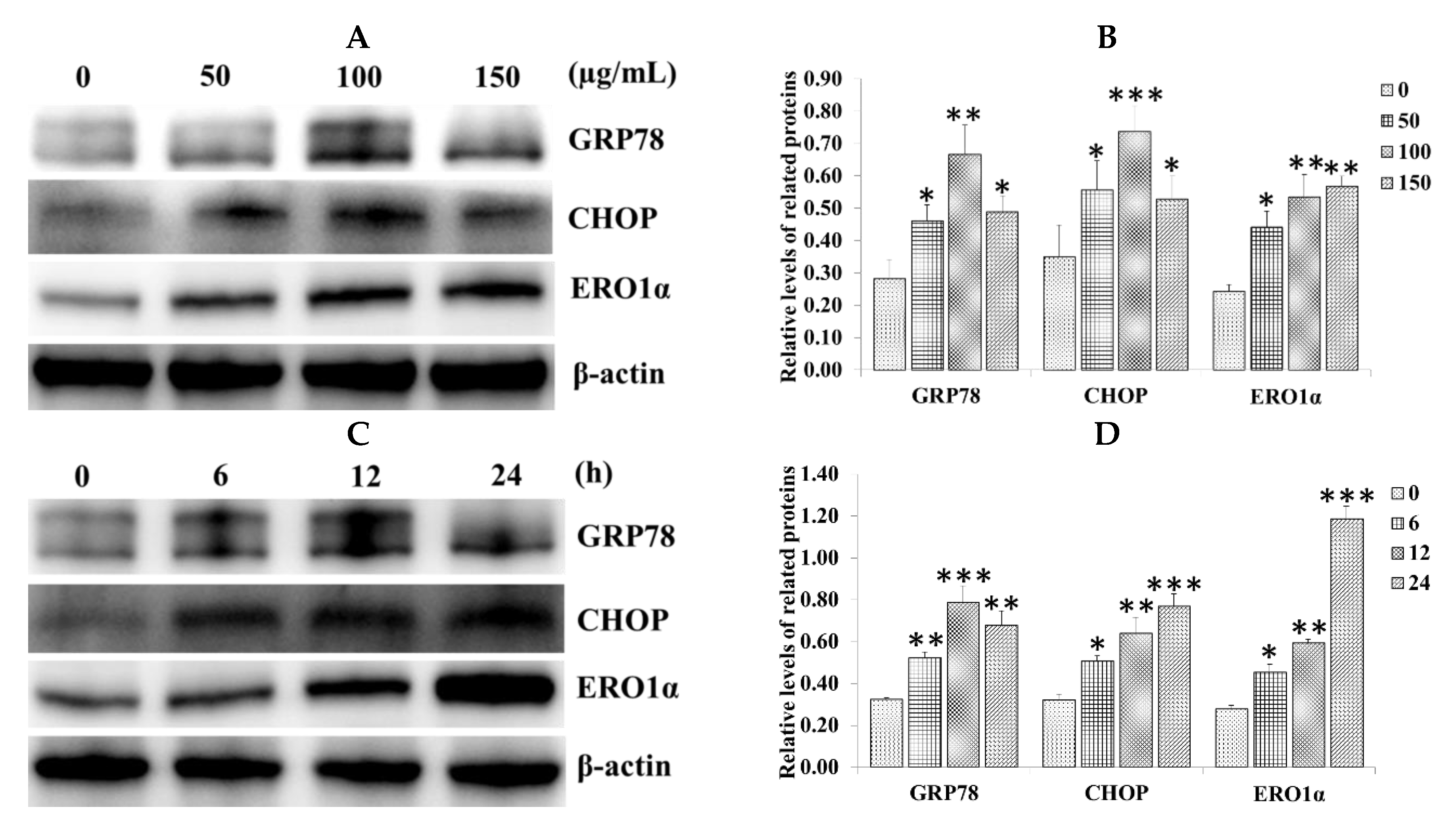
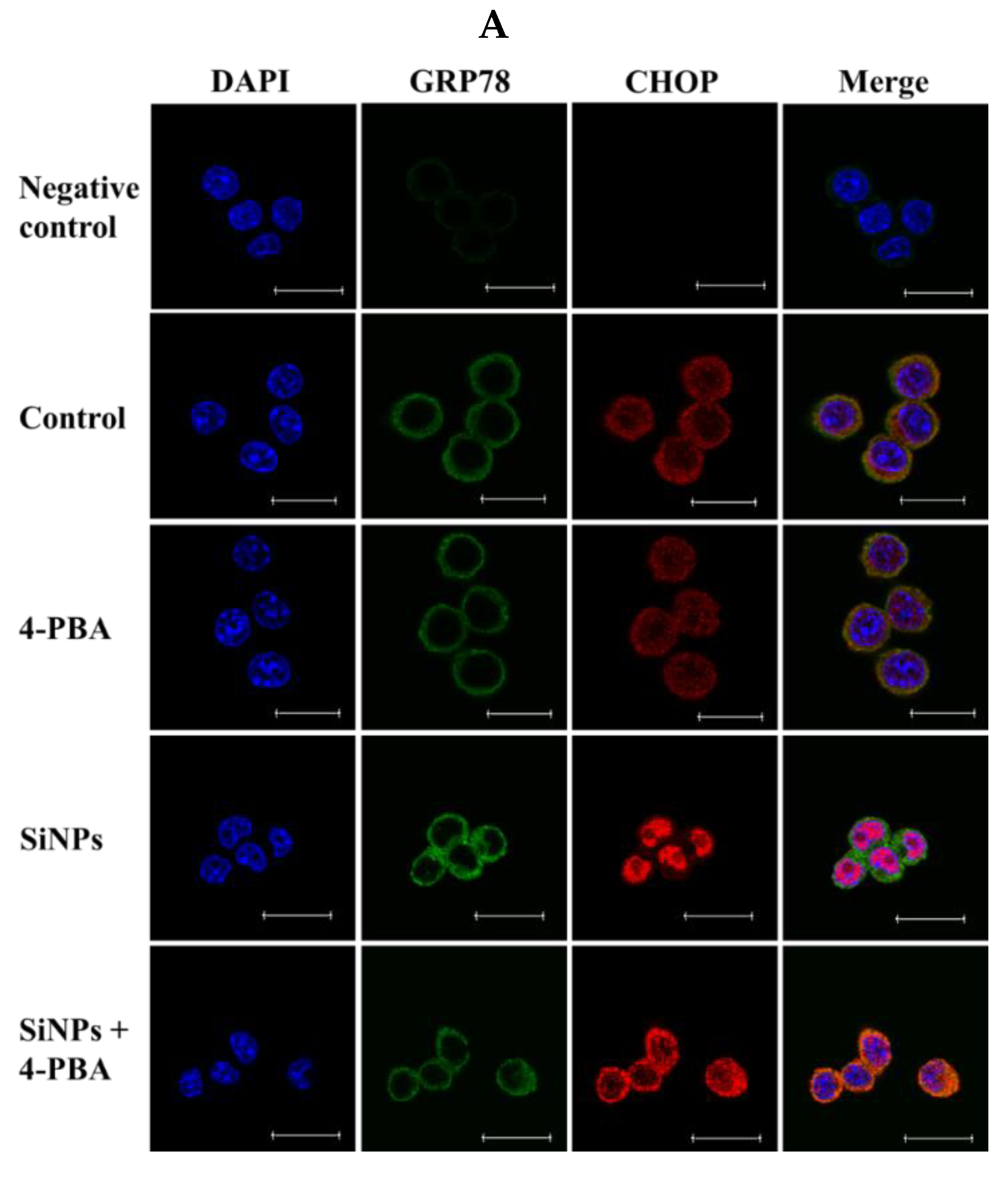
2.2. Effect of SiNPs on the Expression of Endoplasmic Reticulum (ER) Stress-Related Proteins in RAW 264.7 Macrophage Cells
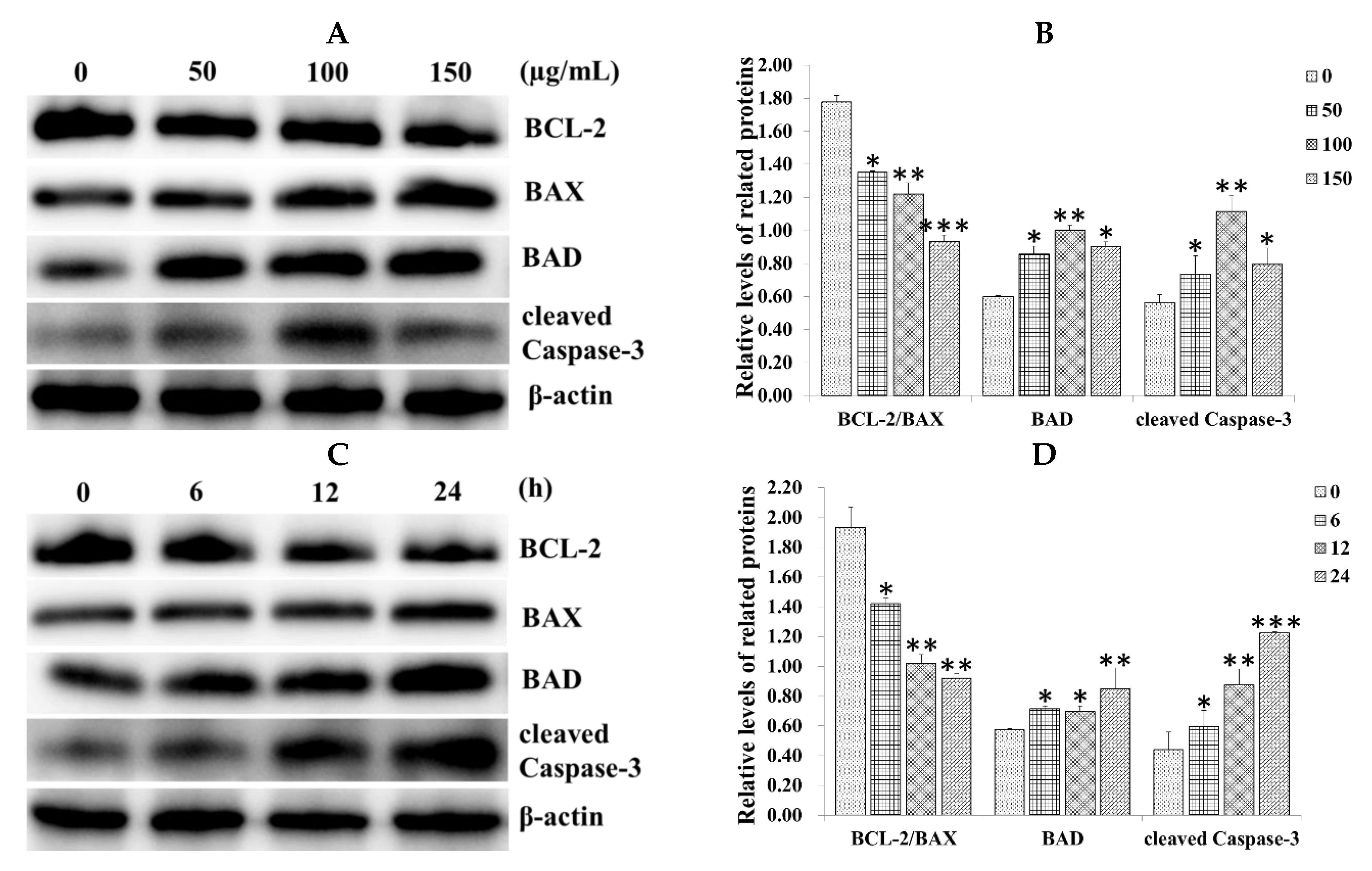
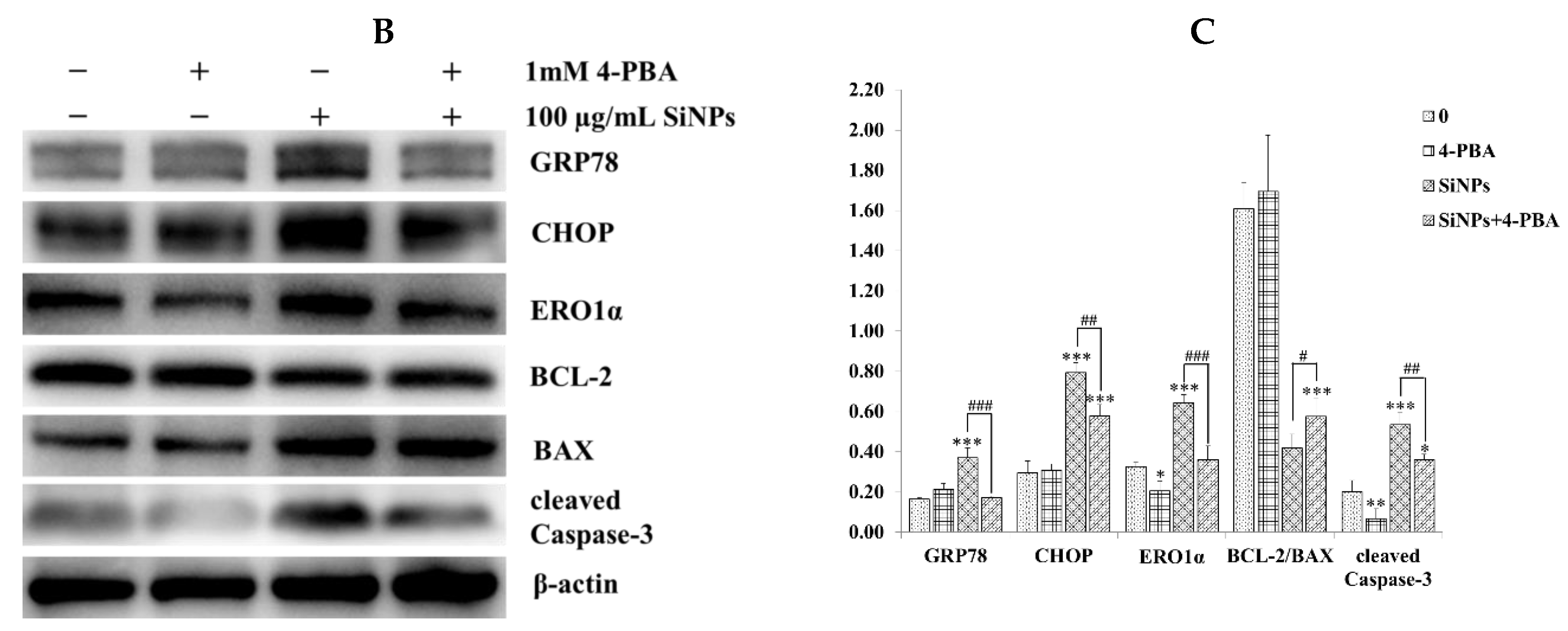
2.3. Effect of SiNPs on the Expression of Apoptosis-Related Proteins in RAW 264.7 Macrophage Cells
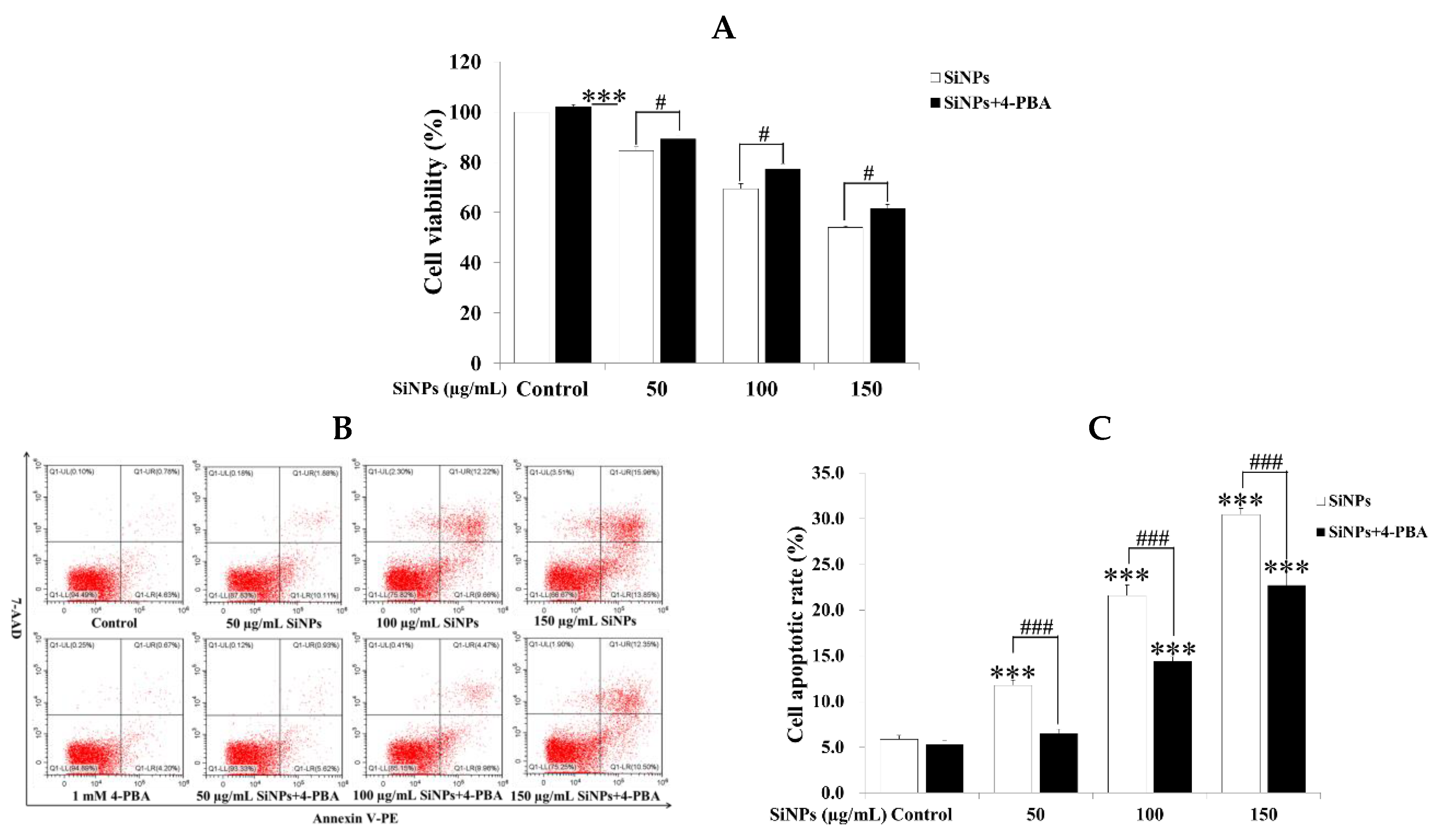
2.4. Effect of 4-PBA on SiNP-Induced Apoptosis in RAW 264.7 Macrophage Cells
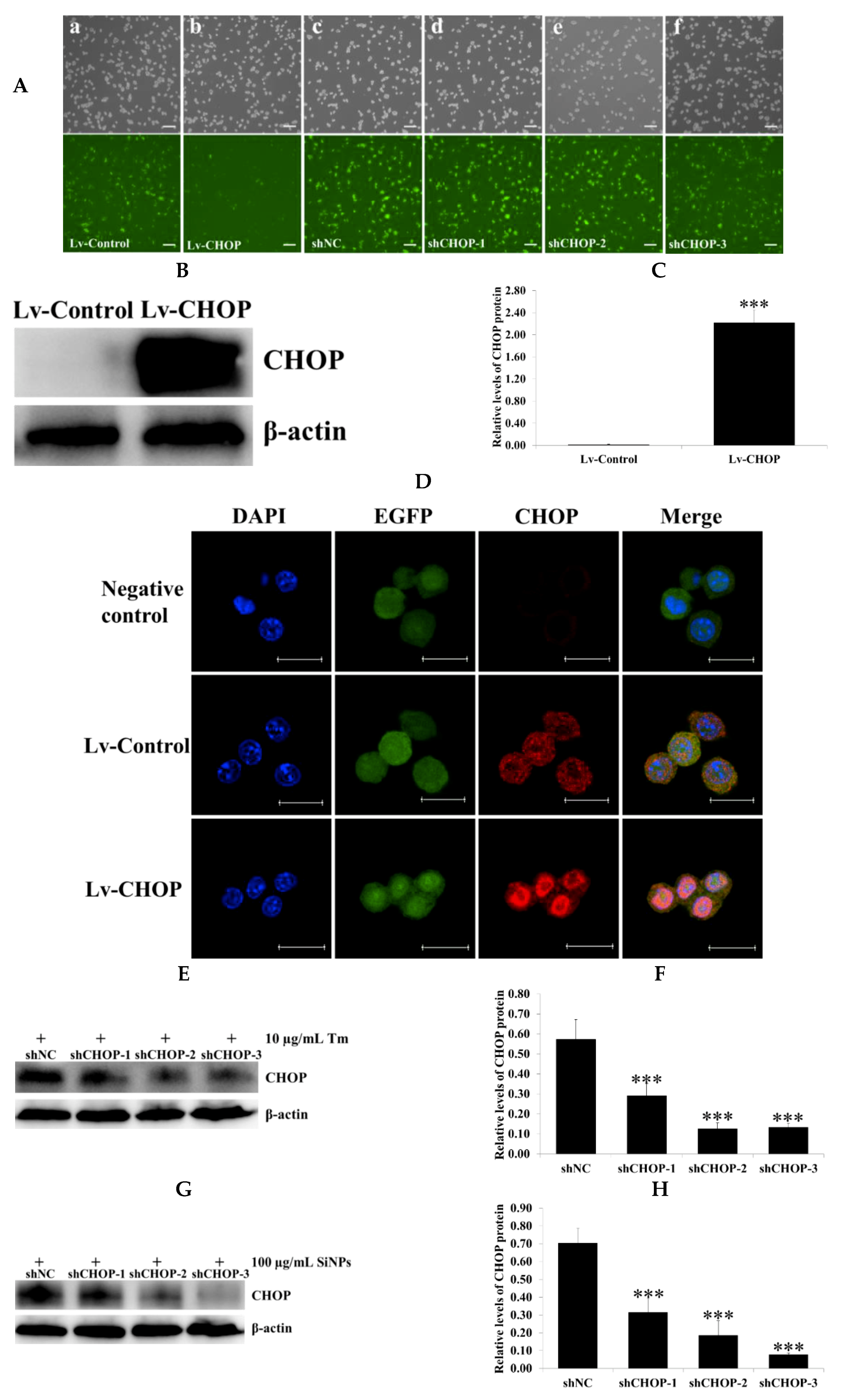
2.5. Verification of the Overexpression and Knockdown Efficiency of Recombinant CCAAT/Enhancer Binding Protein Homologous (CHOP) Lentivirus Vectors in RAW 264.7 Macrophage Cells
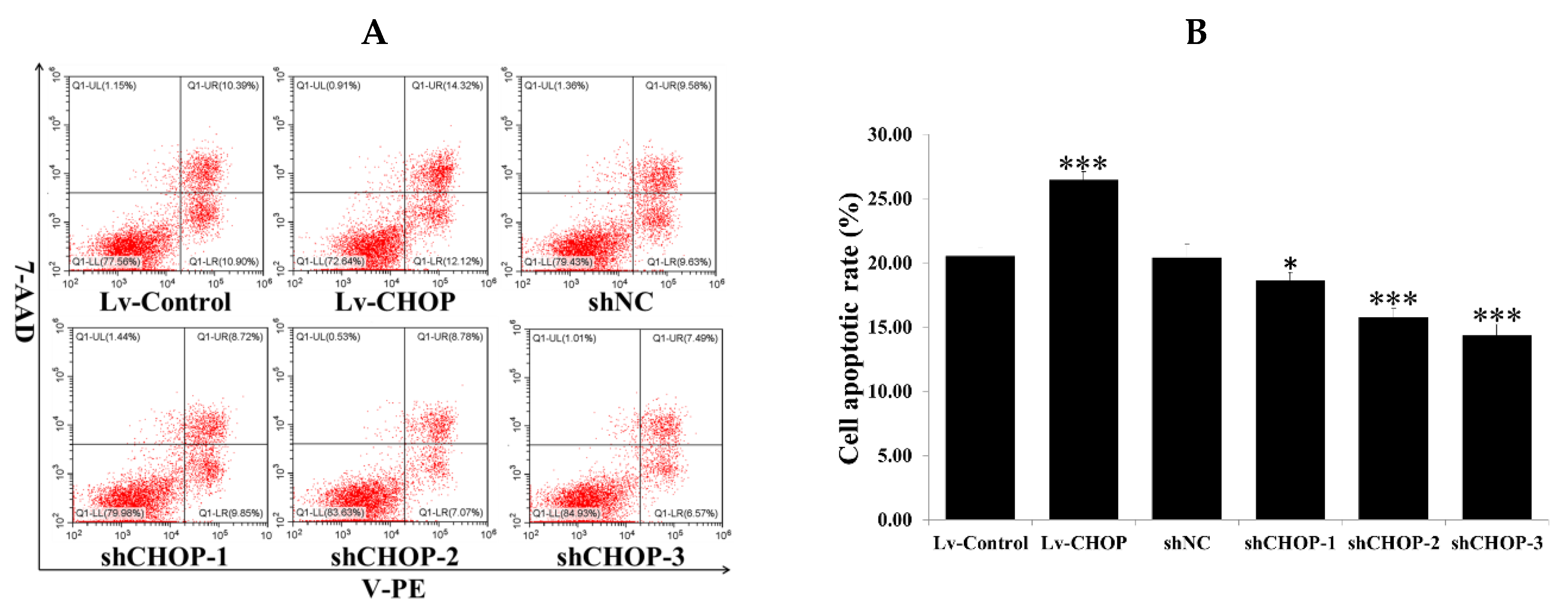
2.6. Regulation of CHOP on SiNP-Induced Cell Apoptosis in RAW 264.7 Macrophage Cells
2.7. Effect of CHOP Overexpression and Knockdown on the Expression of ER Stress and Apoptosis-Related Proteins in RAW 264.7 Macrophage Cells
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Characterization of SiNPs
4.3. Cell Line Culture and Treatment
4.4. Measurement of Cell Viability
4.5. Cell Apoptosis Assay
4.6. Immunofluorescence Staining
4.7. Construction of Recombinant CHOP Overexpression and Short Hairpin Interfering RNA (shRNA) Lentivirus Plasmid and Cell Transduction
4.8. Western Blotting
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SiNPs | Silica nanoparticles |
| ER | Endoplasmic reticulum |
| NPs | Nanoparticles |
| OECD | Organization for Economic Cooperation and Development |
| ROS | Reactive oxygen species |
| GRP78 | Glucose-regulated protein 78 |
| CHOP | CCAAT/enhancer binding protein homologous protein |
| ERO1α | ER oxidoreduclin 1α |
| PI | Propidium iodide |
| PVDF | Polyvinylidene difluoride |
| MOI | Multiplicity of infection |
| ECL | Enhanced chemiluminescence |
| BCL-2 | B-cell lymphoma 2 |
| BAD | BCL-2-associated death promoter |
| BAX | BCL-2-associated X protein |
| Tm | Tunicamycin |
| UPRmt | Mitochondrial unfolded protein response |
References
- Brumfiel, G. Nanotechnology: A little knowledge. Nature 2003, 424, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Service, R.F. Nanotoxicology: Nanotechnology grows up. Science 2004, 304, 1732–1734. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, J. Lipid, protein and poly(NIPAM) coated mesoporous silica nanoparticles for biomedical applications. Adv. Colloid Interface Sci. 2014, 207, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Mai, W.X.; Meng, H. Mesoporous silica nanoparticles: A multifunctional nano therapeutic system. Integr. Biol. 2013, 5, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhang, G.; Mai, J.; Deng, X.; Segura-Ibarra, V.; Wu, S.; Shen, J.; Liu, H.; Hu, Z.; Chen, L.; et al. An injectable nanoparticle generator enhances delivery of cancer therapeutics. Nat. Biotechnol. 2016, 34, 414–418. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, H.; Shi, Y.; Yang, X.; Cao, L.; Wu, J.; Asweto, C.O.; Feng, L.; Duan, J.; Sun, Z. (1)H NMR-based metabolomics study on repeat dose toxicity of fine particulate matter in rats after intratracheal instillation. Sci. Total Environ. 2017, 589, 212–221. [Google Scholar] [CrossRef]
- Oberdorster, G.; Oberdorster, E.; Oberdorster, J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 2005, 113, 823–839. [Google Scholar] [CrossRef]
- Du, Z.; Zhao, D.; Jing, L.; Cui, G.; Jin, M.; Li, Y.; Liu, X.; Liu, Y.; Du, H.; Guo, C.; et al. Cardiovascular toxicity of different sizes amorphous silica nanoparticles in rats after intratracheal instillation. Cardiovasc. Toxicol. 2013, 13, 194–207. [Google Scholar] [CrossRef]
- Matsuo, K.; Hirobe, S.; Okada, N.; Nakagawa, S. Analysis of Skin Permeability and Toxicological Properties of Amorphous Silica Particles. Biol. Pharm. Bull. 2016, 39, 1201–1205. [Google Scholar] [CrossRef]
- Guo, C.X.; Ma, R.; Liu, X.Y.; Xia, Y.Y.; Niu, P.Y.; Ma, J.X.; Zhou, X.Q.; Li, Y.B.; Sun, Z.W. Silica nanoparticles induced endothelial apoptosis via endoplasmic reticulum stress-mitochondrial apoptotic signaling pathway. Chemosphere 2018, 210, 183–192. [Google Scholar] [CrossRef]
- Liang, H.; Jin, C.; Tang, Y.; Wang, F.D.; Ma, C.W.; Yang, Y.J. Cytotoxicity of silica nanoparticles on HaCaT cells. J. Appl. Toxicol. 2014, 34, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.S.; Huang, Y.W.; Zhou, X.D.; Ma, Y.F. In vitro toxicity of silica nanoparticles in human lung cancer cells. Toxicol. Appl. Pharm. 2006, 217, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Park, K. Oxidative stress and pro-inflammatory responses induced by silica nanoparticles in vivo and in vitro. Toxicol. Lett. 2009, 184, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Eom, H.J.; Choi, J. Oxidative stress of silica nanoparticles in human bronchial epithelial cell, Beas-2B. Toxicol. Vitr. 2009, 23, 1326–1332. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Liu, J.; Chen, M.; Sun, L.; Lan, M. In vitro toxicity of silica nanoparticles in myocardial cells. Environ. Toxicol. Pharm. 2010, 29, 131–137. [Google Scholar] [CrossRef]
- Eom, H.J.; Choi, J. SiO(2) Nanoparticles Induced Cytotoxicity by Oxidative Stress in Human Bronchial Epithelial Cell, Beas-2B. Environ. Health Toxicol. 2011, 26, e2011013. [Google Scholar] [CrossRef]
- Passagne, I.; Morille, M.; Rousset, M.; Pujalte, I.; L’Azou, B. Implication of oxidative stress in size-dependent toxicity of silica nanoparticles in kidney cells. Toxicology 2012, 299, 112–124. [Google Scholar] [CrossRef]
- Sun, L.; Li, Y.; Liu, X.M.; Jin, M.H.; Zhang, L.; Du, Z.J.; Guo, C.X.; Huang, P.L.; Sun, Z.W. Cytotoxicity and mitochondrial damage caused by silica nanoparticles. Toxicol. Vitr. 2011, 25, 1619–1629. [Google Scholar] [CrossRef]
- Ahmad, J.; Ahamed, M.; Akhtar, M.J.; Alrokayan, S.A.; Siddiqui, M.A.; Musarrat, J.; Al-Khedhairy, A.A. Apoptosis induction by silica nanoparticles mediated through reactive oxygen species in human liver cell line HepG2. Toxicol. Appl. Pharm. 2012, 259, 160–168. [Google Scholar] [CrossRef]
- Yu, Y.B.; Duan, J.C.; Yu, Y.; Li, Y.; Liu, X.M.; Zhou, X.Q.; Huang, P.L.; Sun, Z.W. Amorphous silica nanoparticles induce autophagic cell death in HepG2 cells triggered by reactive oxygen species. Toxicol. Lett. 2013, 221, S239. [Google Scholar] [CrossRef]
- Rao, R.V.; Peel, A.; Logvinova, A.; del Rio, G.; Hermel, E.; Yokota, T.; Goldsmith, P.C.; Ellerby, L.M.; Ellerby, H.M.; Bredesen, D.E. Coupling endoplasmic reticulum stress to the cell death program: Role of the ER chaperone GRP78. FEBS Lett. 2002, 514, 122–128. [Google Scholar] [CrossRef]
- Oyadomari, S.; Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004, 11, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Christen, V.; Fent, K. Silica nanoparticles and silver-doped silica nanoparticles induce endoplasmatic reticulum stress response and alter cytochrome P4501A activity. Chemosphere 2012, 87, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Yu, Y.B.; Wang, J.H.; Li, Y.B.; Li, Y.; Wei, J.; Zheng, T.; Jin, M.H.; Sun, Z.W. Silica nanoparticles induced intrinsic apoptosis in neuroblastoma SH-SY5Y cells via CytC/Apaf-1 pathway. Environ. Toxicol. Pharmacol. 2017, 52, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.X.; Ma, R.; Liu, X.Y.; Chen, T.; Li, Y.; Yu, Y.; Duan, J.C.; Zhou, X.Q.; Li, Y.B.; Sun, Z.W. Silica nanoparticles promote oxLDL-induced macrophage lipid accumulation and apoptosis via endoplasmic reticulum stress signaling. Sci. Total Environ. 2018, 631–632, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Duan, J.; Yang, M.; Yu, Y.; Feng, L.; Yang, X.; Zhou, X.; Zhao, Z.; Sun, Z. Silica nanoparticles induce autophagosome accumulation via activation of the EIF2AK3 and ATF6 UPR pathways in hepatocytes. Autophagy 2018, 14, 1185–1200. [Google Scholar] [CrossRef]
- Wu, T.S.; Zhang, S.H.; Liang, X.; He, K.Y.; Wei, T.T.; Wang, Y.; Zou, L.Y.; Zhang, T.; Xue, Y.Y.; Tang, M. The apoptosis induced by silica nanoparticle through endoplasmic reticulum stress response in human pulmonary alveolar epithelial cells. Toxicol. Vitr. 2019, 56, 126–132. [Google Scholar] [CrossRef]
- Lee, K.; Lee, J.; Kwak, M.; Cho, Y.L.; Hwang, B.; Cho, M.J.; Lee, N.G.; Park, J.; Lee, S.H.; Park, J.G.; et al. Two distinct cellular pathways leading to endothelial cell cytotoxicity by silica nanoparticle size. J. Nanobiotechnol. 2019, 17, 24. [Google Scholar] [CrossRef]
- Yang, X.; Feng, L.; Zhang, Y.; Hu, H.; Shi, Y.; Liang, S.; Zhao, T.; Cao, L.; Duan, J.; Sun, Z. Co-exposure of silica nanoparticles and methylmercury induced cardiac toxicity in vitro and in vivo. Sci. Total Environ. 2018, 631–632, 811–821. [Google Scholar] [CrossRef]
- Liu, J.; Yang, M.; Jing, L.; Ren, L.; Wei, J.; Zhang, J.; Zhang, F.; Duan, J.; Zhou, X.; Sun, Z. Silica nanoparticle exposure inducing granulosa cell apoptosis and follicular atresia in female Balb/c mice. Environ. Sci. Pollut. Res. Int. 2018, 25, 3423–3434. [Google Scholar] [CrossRef]
- Han, J.W.; Jeong, J.K.; Gurunathan, S.; Choi, Y.J.; Das, J.; Kwon, D.N.; Cho, S.G.; Park, C.; Seo, H.G.; Park, J.K.; et al. Male- and female-derived somatic and germ cell-specific toxicity of silver nanoparticles in mouse. Nanotoxicology 2016, 10, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Hirai, T.; Yoshioka, Y.; Takahashi, H.; Ichihashi, K.; Udaka, A.; Mori, T.; Nishijima, N.; Yoshida, T.; Nagano, K.; Kamada, H.; et al. Cutaneous exposure to agglomerates of silica nanoparticles and allergen results in IgE-biased immune response and increased sensitivity to anaphylaxis in mice. Part. Fibre Toxicol. 2015, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Higashisaka, K.; Kunieda, A.; Iwahara, Y.; Tanaka, K.; Tsunoda, S.; Yoshioka, Y.; Tsutsumi, Y. The contribution of silica nanoparticles-induced neutrophilia to pregnancy complications in mice. Toxicol. Lett. 2014, 229, S193. [Google Scholar] [CrossRef]
- Li, J.; He, X.; Yang, Y.; Li, M.; Xu, C.; Yu, R. Risk assessment of silica nanoparticles on liver injury in metabolic syndrome mice induced by fructose. Sci. Total Environ. 2018, 628–629, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K.; Nishimori, H.; Kondoh, M.; Isoda, K.; Tsunoda, S.; Tsutsumi, Y. Hepatotoxicity of silica nanoparticles in mice. Toxicol. Lett. 2009, 189, S184. [Google Scholar] [CrossRef]
- Marquardt, C.; Fritsch-Decker, S.; Al-Rawi, M.; Diabate, S.; Weiss, C. Autophagy induced by silica nanoparticles protects RAW264.7 macrophages from cell death. Toxicology 2017, 379, 40–47. [Google Scholar] [CrossRef]
- Xi, C.; Wang, Z.; Zhou, J.; Shen, F.; Huang, Z. Activation of autophagy protects against mesoporous silica nanoparticles-induced NF-kappa B dependent inflammation in macrophagy. Toxicol. Lett. 2016, 258, S266–S267. [Google Scholar] [CrossRef]
- Paschen, W.; Doutheil, J. Disturbances of the functioning of endoplasmic reticulum: A key mechanism underlying neuronal cell injury? J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 1999, 19, 1–18. [Google Scholar] [CrossRef]
- Nakka, V.P.; Prakash-babu, P.; Vemuganti, R. Crosstalk Between Endoplasmic Reticulum Stress, Oxidative Stress, and Autophagy: Potential Therapeutic Targets for Acute CNS Injuries. Mol. Neurobiol. 2016, 53, 532–544. [Google Scholar] [CrossRef]
- Li, G.; Mongillo, M.; Chin, K.T.; Harding, H.; Ron, D.; Marks, A.R.; Tabas, I. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J. Cell Biol. 2009, 186, 783–792. [Google Scholar] [CrossRef]
- Rao, J.; Zhang, C.; Wang, P.; Lu, L.; Qian, X.; Qin, J.; Pan, X.; Li, G.; Wang, X.; Zhang, F. C/EBP homologous protein (CHOP) contributes to hepatocyte death via the promotion of ERO1alpha signalling in acute liver failure. Biochem. J. 2015, 466, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, S.J.; Yun, C.Y.; Oyadomari, S.; Novoa, I.; Zhang, Y.H.; Jungreis, R.; Nagata, K.; Harding, H.P.; Ron, D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004, 18, 3066–3077. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Zhang, Y.; Huang, G.; Zhang, Q.; Wang, C.C.; Wang, L.; Lu, D. AtERO1 and AtERO2 Exhibit Differences in Catalyzing Oxidative Protein Folding in the Endoplasmic Reticulum. Plant Physiol. 2019, 180, 2022–2033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Niu, Y.; Zhu, L.; Fang, J.; Wang, X.; Wang, L.; Wang, C.C. Different interaction modes for protein-disulfide isomerase (PDI) as an efficient regulator and a specific substrate of endoplasmic reticulum oxidoreductin-1alpha (Ero1alpha). J. Biol. Chem. 2014, 289, 31188–31199. [Google Scholar] [CrossRef] [PubMed]
- Quiros, P.M.; Prado, M.A.; Zamboni, N.; D’Amico, D.; Williams, R.W.; Finley, D.; Gygi, S.P.; Auwerx, J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017, 216, 2027–2045. [Google Scholar] [CrossRef]
- Fiorese, C.J.; Schulz, A.M.; Lin, Y.F.; Rosin, N.; Pellegrino, M.W.; Haynes, C.M. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr. Biol. CB 2016, 26, 2037–2043. [Google Scholar] [CrossRef]
- Qureshi, M.A.; Haynes, C.M.; Pellegrino, M.W. The mitochondrial unfolded protein response: Signaling from the powerhouse. J. Biol. Chem. 2017, 292, 13500–13506. [Google Scholar] [CrossRef]
- Lu, C.F.; Li, L.Z.; Zhou, W.; Zhao, J.; Wang, Y.M.; Peng, S.Q. Silica nanoparticles and lead acetate co-exposure triggered synergistic cytotoxicity in A549 cells through potentiation of mitochondria-dependent apoptosis induction. Environ. Toxicol. Pharmacol. 2017, 52, 114–120. [Google Scholar] [CrossRef]
- Liu, N.; Tang, M. Toxic effects and involved molecular pathways of nanoparticles on cells and subcellular organelles. J. Appl. Toxicol. 2019. [Google Scholar] [CrossRef]
- Cao, G.D.; Minami, M.; Pei, W.; Yan, C.H.; Chen, D.X.; O’Horo, C.; Graham, S.H.; Chen, J. Intracellular Bax translocation after transient cerebral ischemia: Implications for a role of the mitochondrial apoptotic signaling pathway in ischemic neuronal death. J. Cereb. Blood Flow Metab. 2001, 21, 321–333. [Google Scholar] [CrossRef]
- Gogvadze, V.; Orrenius, S.; Zhivotovsky, B. Multiple pathways of cytochrome c release from mitochondria in apoptosis. BBA-Bioenerg. 2006, 1757, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Vosler, P.S.; Graham, S.H.; Wechsler, L.R.; Chen, J. Mitochondrial Targets for Stroke Focusing Basic Science Research Toward Development of Clinically Translatable Therapeutics. Stroke 2009, 40, 3149–3155. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Cuffe, L.; Szegezdi, E.; Logue, S.E.; Neary, C.; Healy, S.; Samali, A. Mechanisms of ER Stress-Mediated Mitochondrial Membrane Permeabilization. Int. J. Cell Biol. 2010, 2010, 170215. [Google Scholar] [CrossRef] [PubMed]
- Klee, M.; Pallauf, K.; Alcala, S.; Fleischer, A.; Pimentel-Muinos, F.X. Mitochondrial apoptosis induced by BH3-only molecules in the exclusive presence of endoplasmic reticular Bak. EMBO J. 2009, 28, 1757–1768. [Google Scholar] [CrossRef]
- Chen, F.; Lin, P.; Wang, N.; Yang, D.; Wen, X.; Zhou, D.; Wang, A.; Jin, Y. Herp depletion inhibits zearalenone-induced cell death in RAW 264.7 macrophages. Toxicol. Vitr. 2016, 32, 115–122. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, F.; Jin, J.; Hu, J.; Wang, Y.; Ma, Z.; Zhang, J. Endoplasmic Reticulum Stress Cooperates in Silica Nanoparticles-Induced Macrophage Apoptosis via Activation of CHOP-Mediated Apoptotic Signaling Pathway. Int. J. Mol. Sci. 2019, 20, 5846. https://doi.org/10.3390/ijms20235846
Chen F, Jin J, Hu J, Wang Y, Ma Z, Zhang J. Endoplasmic Reticulum Stress Cooperates in Silica Nanoparticles-Induced Macrophage Apoptosis via Activation of CHOP-Mediated Apoptotic Signaling Pathway. International Journal of Molecular Sciences. 2019; 20(23):5846. https://doi.org/10.3390/ijms20235846
Chicago/Turabian StyleChen, Fenglei, Jiaqi Jin, Jiahui Hu, Yujing Wang, Zhiyu Ma, and Jinlong Zhang. 2019. "Endoplasmic Reticulum Stress Cooperates in Silica Nanoparticles-Induced Macrophage Apoptosis via Activation of CHOP-Mediated Apoptotic Signaling Pathway" International Journal of Molecular Sciences 20, no. 23: 5846. https://doi.org/10.3390/ijms20235846
APA StyleChen, F., Jin, J., Hu, J., Wang, Y., Ma, Z., & Zhang, J. (2019). Endoplasmic Reticulum Stress Cooperates in Silica Nanoparticles-Induced Macrophage Apoptosis via Activation of CHOP-Mediated Apoptotic Signaling Pathway. International Journal of Molecular Sciences, 20(23), 5846. https://doi.org/10.3390/ijms20235846




