Stay-Green Trait: A Prospective Approach for Yield Potential, and Drought and Heat Stress Adaptation in Globally Important Cereals
Abstract
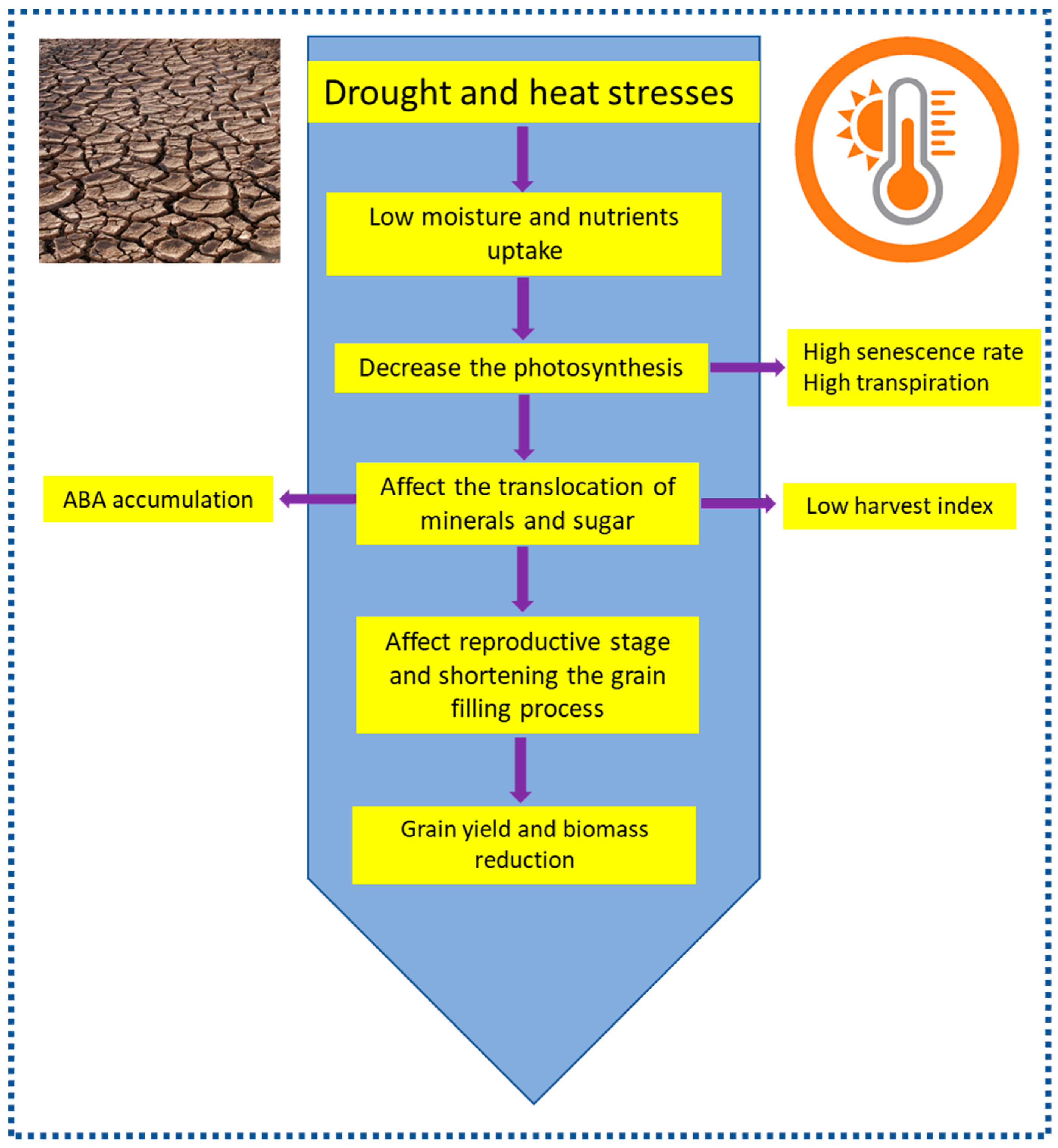
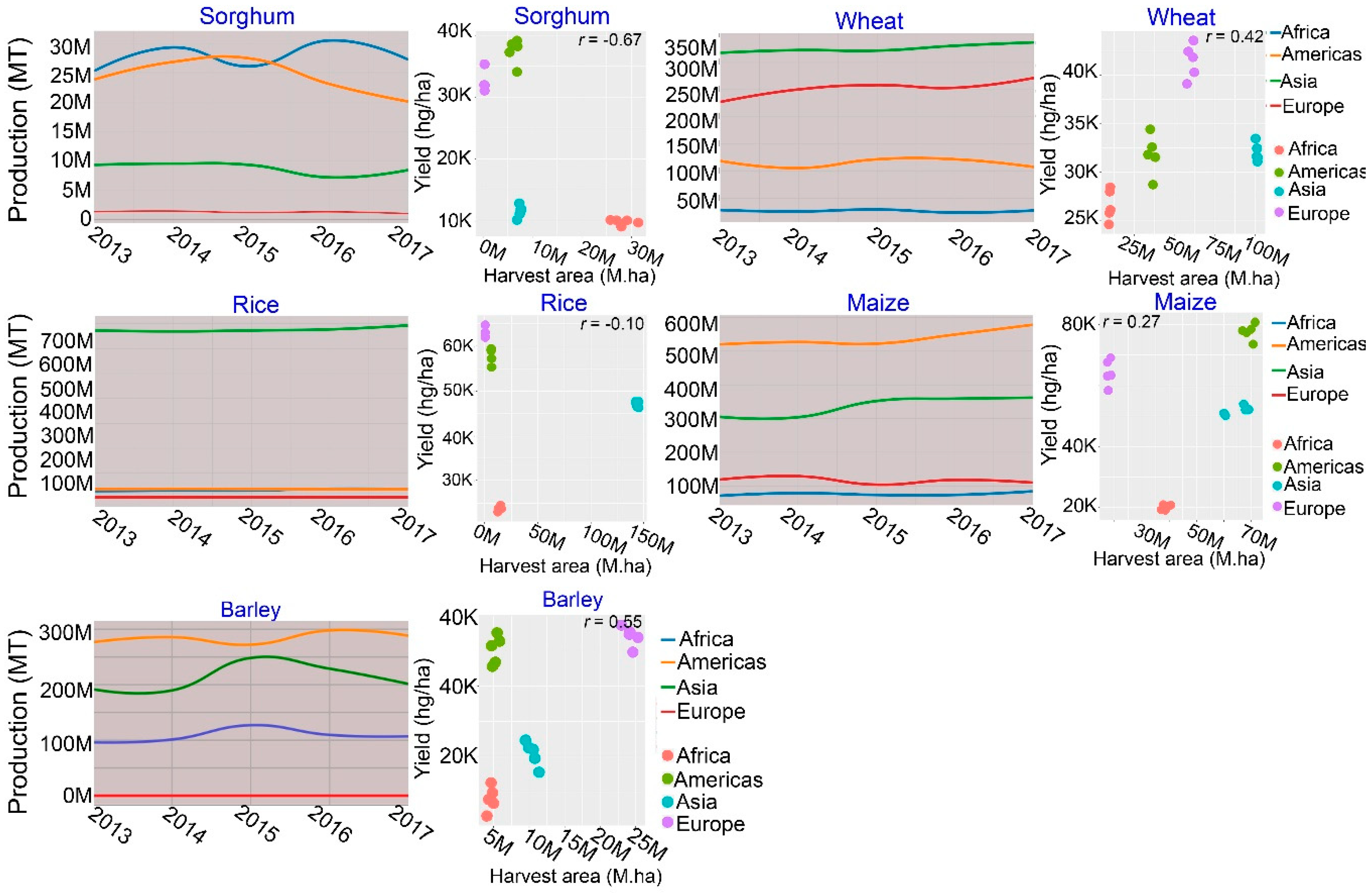
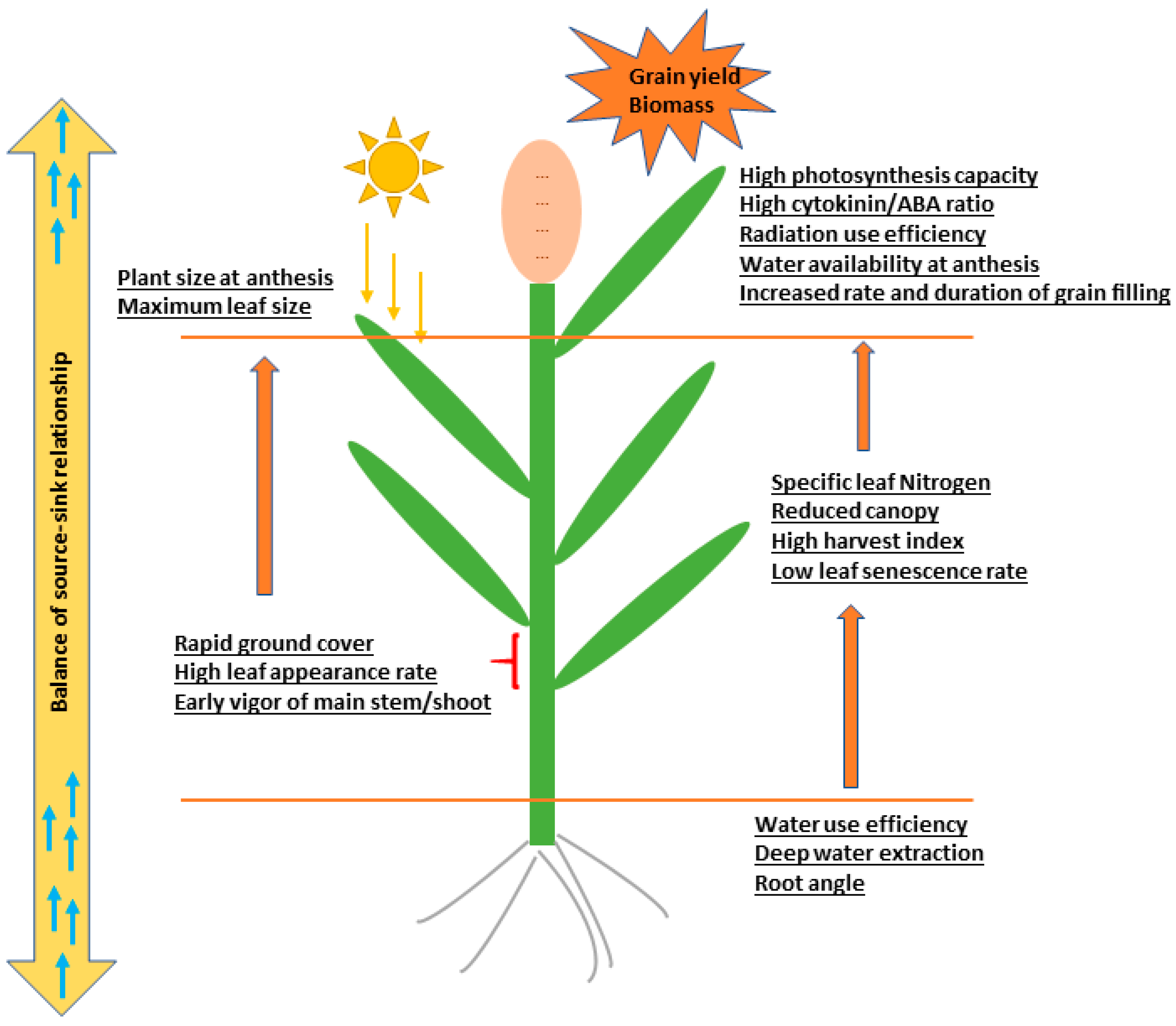
1. Introduction
2. Stay-Green in Sorghum
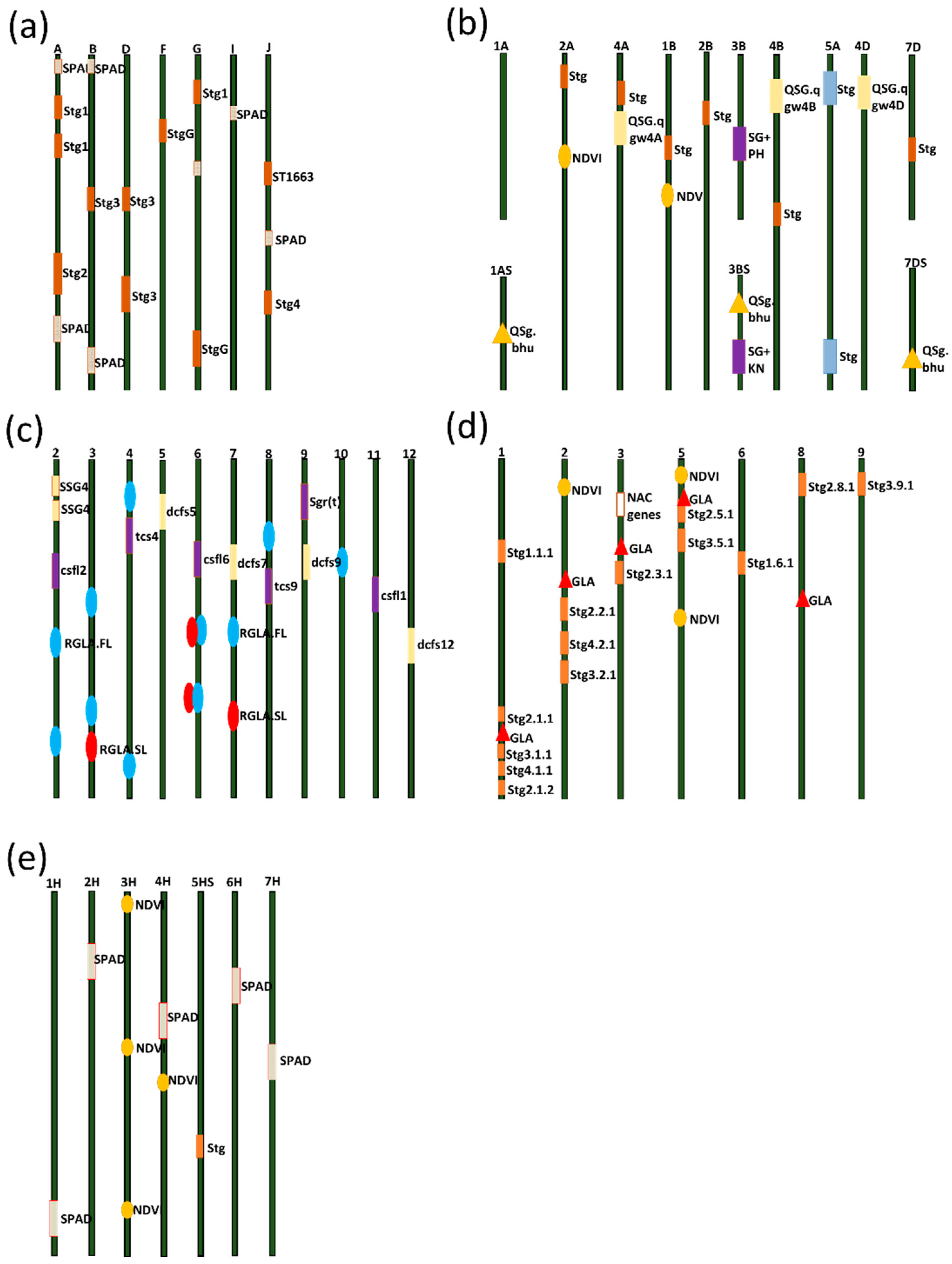
2.1. Stay-Green QTLs in Sorghum
2.2. The Physiology of Stay-Green in Sorghum
3. Stay-Green in Wheat
3.1. Stay-Green QTLs in Wheat
3.2. The Physiology of Stay-Green in Wheat
4. Stay-Green in Rice
4.1. QTLs for the Stay-Green Trait in Rice
4.2. The Physiology of Stay-Green in Rice
5. Stay-Green in Maize
5.1. QTLs for the Stay-Green Trait in Maize
5.2. The Physiology of Stay-Green in Maize
6. Stay-Green in Barley
6.1. QTLs for the Stay-Green Trait in Barley
6.2. The Physiology of Stay-Green in Barley
7. Stay-Green and Grain or End-Use Quality
8. Conclusions and Future Prospective
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SG | Stay-green |
| Chl | Chlorophyll |
| NDVI | Normalized difference vegetation index |
| CTD | Canopy temperature depression |
| MAS | Marker-assisted selection |
References
- FAO Statistics 2019. Available online: http://www.fao.org/faostat/en/#data (accessed on 20 November 2019).
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Global synthesis of drought effects on maize and wheat production. PLoS ONE 2016, 11, e0156362. [Google Scholar] [CrossRef] [PubMed]
- Viola, D.; Daniel, K.Y.; Tan, I.D. Impact of High Temperature and Drought Stresses on Chickpea Production. Agronomy 2018, 8, 145. [Google Scholar]
- Barnabas, B.; Jäger, K.; Fehér, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; Burritt, D.J.; Gupta, A.; Tsujimoto, H.; Tran, L. Heat stress effects on source–sink relationships and metabolome dynamics in wheat. J. Exp. Bot. 2019. [CrossRef] [PubMed]
- Abdelrahman, M.; El-Sayed, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.S.P. The “STAY-GREEN” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 2017, 36, 1009–1025. [Google Scholar] [CrossRef] [PubMed]
- Rosenow, D.T. Breeding for Resistance to Root and Stalk Rots in Texas. In Sorghum Root and Stalk Rots, A Critical Review; ICRISTAT: Patancheru, AP, India, 1983; pp. 209–217. [Google Scholar]
- Peingao, L. Structural and biochemical mechanism responsible for the stay-green phenotype in common wheat. Chi. Sci. Bull. 2013, 51, 2595–2603. [Google Scholar]
- Gregersen, N.; Bross, P.; Vang, S.; Christensen, J.H. Protein misfolding and human disease. Genomics Hum. Genet. 2006, 7, 103–124. [Google Scholar] [CrossRef]
- Borrell, A.K.; Mullet, J.E.; George-Jaeggli, B.; van Oosterom, E.J.; Hammer, G.L.; Klein, P.E.; Jordan, D.R. Drought adaptation of stay-green cereals associated with canopy development, leaf anatomy, root growth and water uptake. J. Exp. Bot. 2014, 65, 6251–6263. [Google Scholar] [CrossRef]
- Jaegglia, B.G.; Mortlockb, M.Y.; Borrell, A. Bigger is not always better: Reducing leaf area helps stay-green sorghum use soil water more slowly. Environ. Exp. Bot. 2017, 138, 119–129. [Google Scholar] [CrossRef]
- Kamal, N.M.; Gorafi, Y.S.A.; Tsujimoto, H.; Ghanim, A.M.A. Stay-green QTLs response in adaptation to post-flowering drought depends on the drought severity. BioMed Res. Int. 2018, 2018, 7082095. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fengler, K.A.; John, L.; Hemert, V.; Gupta, R.; Mongar, N.; Sun, J.; Allen, W.B.; Wang, Y.; Weers, B.; et al. Identification and characterization of a novel stay-green QTL that increases yield in maize. Plant Biot. J. 2019, 17, 2272–2285. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Ougham, H. The stay-green trait. J. Exp. Bot. 2014, 65, 3889–3900. [Google Scholar] [CrossRef]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, Non-Yellow Coloring 1 and NYC1-LIKE, are required for Chl b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hörtensteiner, S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in Chl breakdown during leaf senescence in Arabidopsis. Plant Cell 2009, 21, 767–785. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis stay-green, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chenggen, Q.; Wang, X.; Chen, Y.; Jinqiang, D.; Jiang, C.; Sun, X.; Chen, H.; Li, J.; Piao, W.; et al. New alleles for Chl content and stay-green traits revealed by a genome wide association study in rice (Oryza sativa). Sci. Rep. 2019, 9, 2541. [Google Scholar]
- Sjödin, J. Induced morphological variation in Vicia faba L. Hereditas 1971, 67, 155–179. [Google Scholar] [CrossRef]
- Kassahun, B.; Bidinger, F.; Hash, C.; Kuruvinashetti, M. Stay-green expression in early generation sorghum [Sorghum bicolor (L.) Moench] QTL introgression lines. Euphytica 2010, 172, 351–362. [Google Scholar] [CrossRef]
- Luche, H.S.; Silva, J.A.G.; Nörnberg, R.; Silveira, F.S.; Baretta, D.; Groli, E.L.; Maia, L.C.; Oliveira, A.C. Stay-green: A potentiality in plant breeding. Ciência Rural 2015, 45, 1755–1760. [Google Scholar] [CrossRef]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The Submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol. 2012, 160, 1795–1807. [Google Scholar] [CrossRef] [PubMed]
- Christopher, J.T.; Christopher, M.J.; Borrell, A.K.; Fletcher, S.; Chenu, K. Stay-green traits to improve wheat adaptation in well-watered and water-limited environments. J. Exp. Bot. 2016, 67, 5159–5172. [Google Scholar] [CrossRef] [PubMed]
- Antonietta, M.; Acciaresi, H.A.; Guiamet, J.J. Responses to N deficiency in stay green and non-stay green Argentinean hybrids of maize. J. Agron. Crop Sci. 2015, 202, 231–242. [Google Scholar] [CrossRef]
- Adeyanju, A.; Yu, J.M.; Little, C.; Rooney, W.; Klein, P.; Burke, J.; Tesso, T. Sorghum RILs segregating for stay-green QTL and leaf dhurrin content show differential reaction to stalk rot diseases. Crop Sci. 2016, 56, 2895–2903. [Google Scholar] [CrossRef]
- Distelefeld, A.; Avni, R.; Fishcher, A.M. Senescence, nutrient remobilization, and yield in wheat and barley. J. Exp. Bot. 2014, 65, 3783–3798. [Google Scholar] [CrossRef]
- Joshi, A.K.; Kumari, M.; Singh, V.P.; Reddy, C.M.; Kumar, S.; Rane, J.; Chand, R. Stay green trait: Variation, inheritance and its association with spot blotch resistance in spring wheat (Triticum aestivum L.). Euphytica 2007, 153, 59–71. [Google Scholar] [CrossRef]
- Spano, G.; Di, F.N.; Perrotta, C.; Platani, C.; Ronga, G.; Lawlor, D.W.; Napier, J.A.; Shewry, P.R. Physiological characterization of stay green mutants in durum wheat. J. Exp. Bot. 2003, 54, 1415–1420. [Google Scholar] [CrossRef]
- Veyradier, M.; Christopher, J.; Chenu, K. Quantifying the potential yield benefit of root traits. In Proceedings of the 7th International Conference on Functional-Structural Plant Models, Saariselkä, Finland, 9–14 June 2013; Sievänen, R., Nikinmaa, E., Godin, C., Lintunen, A., Nygren, P., Eds.; Finnish Society of Forest Science: Vantaa, Finland; MELTA: Helsinki, Finland; pp. 317–319.
- Kumar, R.R.; Goswami, S.; Shamim, M. Biochemical defense response: Characterizing the plasticity of source and sink in spring wheat under terminal heat stress. Front. Plant Sci. 2017, 8, 1603. [Google Scholar] [CrossRef]
- Kumar, U.; Joshi, A.K.; Kumari, M.; Paliwal, R.; Kumar, S.; Röder, M.S. Identification of QTLs for stay green trait in wheat (Triticum aestivum L.) in the ‘Chirya 3′ 3 ‘Sonalika’ population. Euphytica 2010, 174, 437–445. [Google Scholar] [CrossRef]
- Vijayalakshmi, K.; Fritz, A.K.; Paulsen, G.M.; Bai, G.H.; Pandravada, S.; Gill, B.S. Modelling and mapping QTL for senescence-related traits in winter wheat under high temperature. Mol. Breed. 2010, 26, 163–175. [Google Scholar] [CrossRef]
- Zheng, H.J.; Wu, A.Z.; Zheng, C.C.; Wang, Y.F.; Cai, R.; Shen, X.F.; Xu, R.R.; Liu, P.; Kong, L.J.; Dong, S.T. QTL mapping of maize (Zea mays) stay-green traits and their relationship to yield. Plant Breed. 2009, 128, 54–62. [Google Scholar] [CrossRef]
- Ribeiro, T.; Alves da Silva, D.; Fátima Esteves, J.A.; Azevedo, C.V.G.; Gonçalves João, G.R.; Carbonell, S.A.M.; Chiorato, A.F. Evaluation of common bean genotypes for drought tolerance. Bragantia 2019, 78. [Google Scholar] [CrossRef]
- Yue, B.; Wei-Ya, X.; Li-Jun, L.; Yong-Zhong, X. QTL analysis for flag leaf characteristics and their relationships with yield and yield traits in rice. Acta Genetica Sin. 2006, 33, 824–832. [Google Scholar] [CrossRef]
- He, Y.; Li, L.; Zhang, Z.; Wu, J. Identification and comparative analysis of premature, senescence leaf mutants in rice (Oryza sativa L.). Int. J. Mol. Sci. 2018, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.; Subudhi, P.K.; Borrell, A.; Jordan, D.; Rosenow, D.; Nguyen, H.; Klein, P.; Klein, R.; Mullet, J. Sorghum stay-green QTL individually reduce post-flowering drought-induced leaf senescence. J. Exp. Bot. 2007, 58, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.Z.; Henzell, R.G.; Jordan, D.R.; Butler, D.G.; Kelly, A.M.; McIntyre, C.L. Identification of genomic regions associated with stay green in sorghum by testing RILs in multiple environments. Theor. Appl. Gen. 2000, 100, 1225–1232. [Google Scholar] [CrossRef]
- Kholová, J.; Tharanya, M.; Sivasakthi, K.; Srikanth, M.; Rekha, B.; Hammer, G.L.; McLean, G.; Deshpande, S.; Hash, C.T.; Craufurd, P.; et al. Modelling the effect of plant water use traits on yield and stay-green expression in sorghum. Funct. Plant Biol. 2014, 41, 1019–1034. [Google Scholar] [CrossRef]
- Borrell, A.K.; Hammer, G.L.; Douglas, A.C.L. Does maintaining green leaf area in sorghum improve yield under drought? I. Leaf growth and senescence. Crop Sci. 2000, 40, 1026–1037. [Google Scholar] [CrossRef]
- Borrell, A.K.; Hammer, G.L.; van Oosterom, E. Stay-green: A consequence of the balance between supply and demand for nitrogen during grain filling? Ann. Appl. Biol. 2001, 138, 91–95. [Google Scholar] [CrossRef]
- Jordan, D.R.; Tao, Y.; Godwin, I.D.; Henzell, R.G.; Cooper, M.; McIntyre, C.L. Prediction of hybrid performance in grain sorghum using RFLP markers. Theor. App. Genet. 2003, 106, 559–567. [Google Scholar] [CrossRef]
- Xu, W.W.; Subudhi, P.K.; Crasta, O.R.; Rosenow, D.T.; Mullet, J.E.; Nguyen, H.T. Molecular mapping of QTLs conferring stay-green in grain sorghum (Sorghum bicolor L. Moench). Genome 2000, 43, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Subudhi, P.K.; Rosenow, D.T.; Nguyen, H.T. Quantitative trait loci for the stay green trait in sorghum (Sorghum bicolor L. Moench): Consistency across genetic backgrounds and environments. Theor. App. Genet. 2000, 101, 733–741. [Google Scholar] [CrossRef]
- Kebede, H.; Subudhi, P.K.; Rosenow, D.T.; Nguyen, H.T. Quantitative trait loci influencing drought tolerance in grain sorghum (Sorghum bicolor L. Moench). Theor. App. Genet. 2001, 103, 266–276. [Google Scholar] [CrossRef]
- Sanchez, A.C.; Subudhi, P.K.; Rosenow, D.T.; Nguyen, H.T. Mapping QTLs associated with drought resistance in sorghum (Sorghum bicolor L. Moench). Plant Mol. Biol. 2002, 48, 713–726. [Google Scholar] [CrossRef]
- Hash, C.T.; Bhasker, A.G.; Lindup, S. Opportunities for marker assisted selection (MAS) to improve the feed quality of crop residues in pearl millet and sorghum. Field Crops Res. 2003, 84, 79–88. [Google Scholar] [CrossRef]
- Haussmann, B.I.G.; Mahalakshmi, V.; Reddy, B.V.S.; Seetharama, N.; Hash, C.T.; Geiger, H.H. QTL mapping of stay-green in two sorghum recombinant inbred populations. Theor. Appl. Genet. 2002, 106, 133–142. [Google Scholar] [CrossRef]
- Reddy, N.R.R.; Ragimasalawada, M.; Sabbavarapu, M.M.; Nadoor, S.; Patil, J.V. Detection and validation of stay-green QTL in post-rainy sorghum involving widely adapted cultivar, M35-1 and a popular stay-green genotype B35. BMC Genom. 2014, 15, 909. [Google Scholar] [CrossRef]
- Kamal, N.M.; Gorafi, Y.S.A.; Ghanim, A.M.A. Performance of sorghum stay-green introgression lines under post-flowering drought. Int. J. Plant Res. 2017, 7, 65–74. [Google Scholar]
- Ngugi, K.W.; Kimani, D.K. Introgression of stay-green trait into a Kenyan farmer preferred sorghum variety. Afr. Crop Sci. J. 2010, 18, 141–146. [Google Scholar]
- Vadez, V.; Deshpande, S.P.; Kholova, J.; Hammer, G.L.; Borrell, A.K.; Talwar, H.S.; Hash, C.T. Stay-green quantitative trait loci’s effects on water extraction, transpiration efficiency and seed yield depend on recipient parent background. Funct. Plant Biol. 2011, 38, 553–566. [Google Scholar] [CrossRef]
- Emebiri, L.C. QTL dissection of the loss of green colour during post anthesis grain maturation in two-rowed barley. Theor. Appl. Genet. 2013, 126, 1873–1884. [Google Scholar] [CrossRef] [PubMed]
- Mace, E.; Singh, V.; van Oosterom, E.; Hammer, G.; Hunt, C.; Jordan, D. QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theor. Appl. Genet. 2012, 124, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Yan, F.; Lee, W. Physiological characteristics of a functional stay-green rice “SNU-SG1” during grain-filling period. J. Crop Sci. Biotechnol. 2009, 12, 47–52. [Google Scholar] [CrossRef]
- Manschadi, A.M.; Christopher, J.; Devoil, P.; Hammer, G.L. The role of root architectural traits in adaptation of wheat to water-limited environments. Funct. Plant Biol. 2006, 33, 823–837. [Google Scholar] [CrossRef]
- Borrell, A.K.; van Oosterom, E.J.; Mullet, J.E.; George-Jaeggli, B.; Jordan, D.R.; Klein, P.E.; Hammer, G.L. Stay-green alleles enhance grain yield in sorghum under drought by modifying canopy development and enhancing water uptake. New Phytol. 2014, 203, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Hammer, G.; Cooper, M.; Tardieu, F.; Weich, S.; Walsh, B.; van Eeuwijk, F.; Chapman, S.; Podlich, D. Models for navigating biological complexity in breeding improved crop plants. Trends Plant Sci. 2006, 11, 587–593. [Google Scholar] [CrossRef] [PubMed]
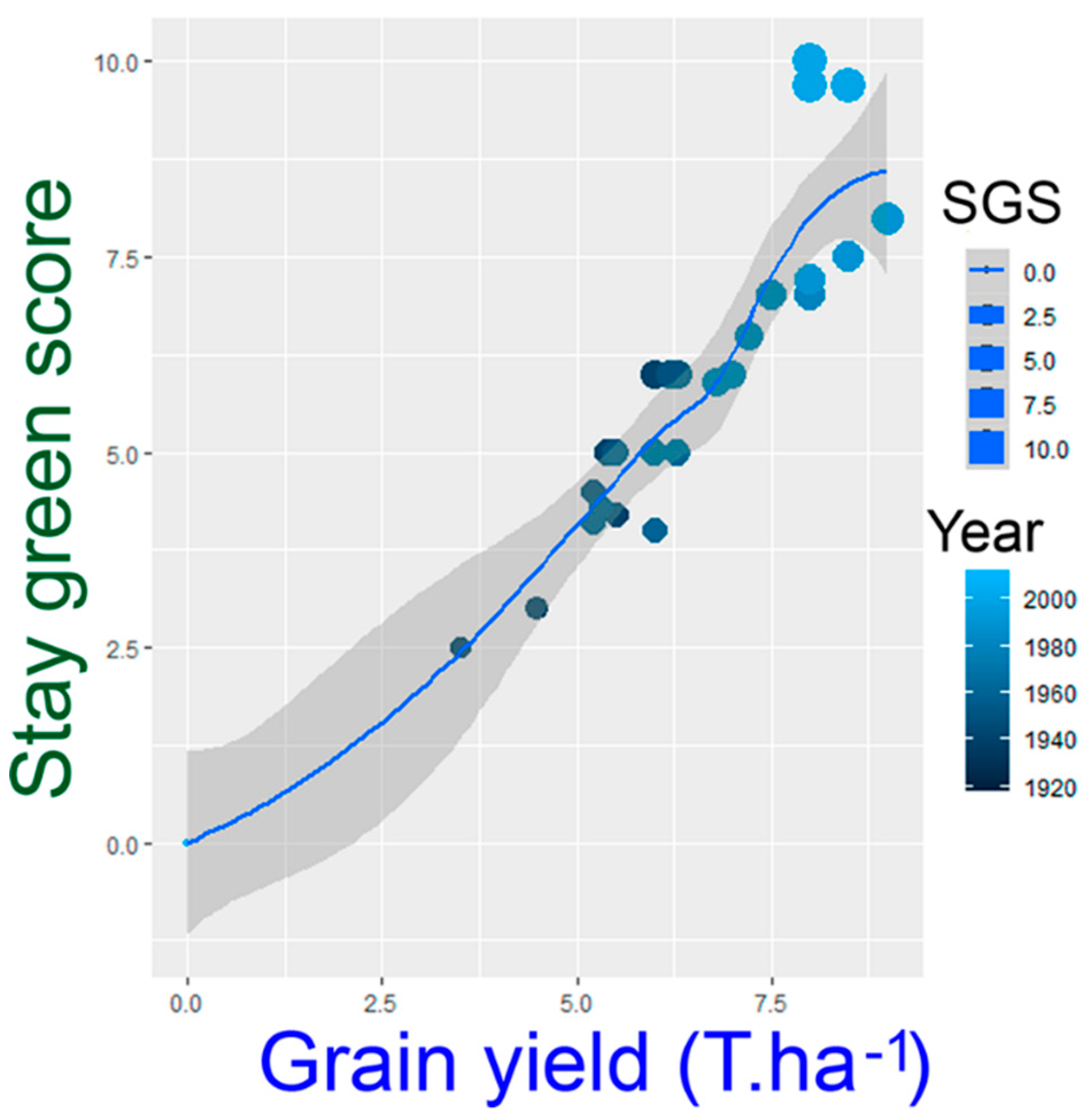
- Jordan, D.R.; Hunt, C.H.; Cruickshank, A.W.; Borrell, A.K.; Henzell, R.G. The relationship between the stay-green trait and grain yield in elite sorghum hybrids grown in a range of environments. Crop Sci. 2012, 52, 1153–1161. [Google Scholar] [CrossRef]
- Borrell, A.K.; Hammer, G.L.; Henzel, R.G. Does maintaining green leaf area in sorghum improve yield under drought? II. Dry matter production and yield. Crop Sci. 2000, 40, 1037–1048. [Google Scholar] [CrossRef]
- Johnson, S.M.; Cummins, I.; Lim, F.L.; Slabas, A.R.; Knight, M.R. Transcriptomic analysis comparing stay-green and senescent Sorghum bicolor lines identifies a role for proline biosynthesis in the stay-green trait. J. Exp. Bot. 2015, 66, 7061–7073. [Google Scholar] [CrossRef]
- Nanjundaswamy, A.; Vadlani, P.V.; Prasad, P.V.V. Evaluation of drought and heat stressed grain sorghum (Sorghum bicolor) for ethanol production. Ind. Crops Prod. 2011, 33, 779–782. [Google Scholar]
- Tacka, J.; Lingenfelserb, J.; Krishna, J.S.V. Disaggregating sorghum yield reductions under warming scenarios exposes narrow genetic diversity in US breeding programs. PNAS 2017, 114, 9296–9301. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, C.D.; Lim, S.; Salzman, R.A.; Kagiampakis, I.; Morishige, D.T.; Weers, B.D.; Klein, R.R.; Pratt, L.H.; Cordonnier-Pratt, M.M.; Klein, P.; et al. Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Mol. Biol. 2005, 58, 699–720. [Google Scholar] [CrossRef] [PubMed]
- Dugas, D.V.; Monaco, M.K.; Olesen, A.; Klein, R.R.; Kumari, S.; Ware, D.; Klein, P.E. Functional annotation of the transcriptome of Sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genom. 2011, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Azam, F.; Li, H.; Chang, X.; Li, B.; Jing, R. Mapping QTL for stay-green and agronomic traits in wheat under diverse water regimes. Euphytica 2017, 2017, 213–246. [Google Scholar] [CrossRef]
- Huang, X.Q.; Kempf, H.; Ganal, M.W.; Röder, M.S. Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 933–943. [Google Scholar] [CrossRef]
- Marza, F.; Bai, G.H.; Carver, B.F.; Zhou, W.C. Quantitative trait loci for yield and related traits in the wheat population Ning7840 9 Clark. Theor. Appl. Genet. 2006, 112, 688–698. [Google Scholar] [CrossRef]
- Pinto, R.S.; Reynolds, M.P.; Mathews, K.L.; McIntyre, C.L.; Olivares-Villegas, J.J.; Chapman, S.C. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor. Appl. Genet. 2010, 121, 1001–1021. [Google Scholar] [CrossRef]
- Wang, A.-Y.; Li, Y.; Zhang, C.-Q. QTL mapping for stay-green in maize (Zea mays). Can. J. Plant Sci. 2012, 92, 249–256. [Google Scholar] [CrossRef]
- Sourdille, P.; Singh, S.; Cadalen, T.; Brown-Guedira, G.L.; Gay, G.; Qi, L.; Gill, B.S.; Dufour, P.; Murigneux, A.; Bernard, M. Microsatellite based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genomics 2004, 4, 12–25. [Google Scholar] [CrossRef]
- Christopher, J.T.; Manschadi, A.M.; Hammer, G.L.; Borrell, A.K. Developmental and physiological traits associated with high yield and stay-green phenotype in wheat. Aust. J. Agric. Res. 2008, 59, 354–364. [Google Scholar] [CrossRef]
- Lopes, M.S.; Reynolds, M.P. Stay-green in spring wheat can be determined by spectral reflectance measurements (normalized difference vegetation index) independently from phenology. J. Exp. Bot. 2012, 63, 3789–3798. [Google Scholar] [CrossRef] [PubMed]
- Bogard, M.; Jourdan, M.; Allard, V.; Martre, P.; Perretant, M.R.; Ravel, C.; Heumez, E.; Orford, S.; Snape, J.; Griffiths, S. Anthesis date mainly explained correlations between post-anthesis leaf senescence, grain yield, and grain protein concentration in a winter wheat population segregating for flowering time QTLs. J. Exp. Bot. 2011, 62, 3621–3636. [Google Scholar] [CrossRef] [PubMed]
- Pask, A.; Pietragalla, J. Leaf area, green crop area and senescence. In Physiological Breeding II: A Field Guide to Wheat Phenotyping; Pask, A., Pietragalla, J., Mullan, D., Reynolds, M., Eds.; International Maize and Wheat Improvement Center (CIMMYT): Mexico City, Mexico, 2012; pp. 58–62. [Google Scholar]
- Kichey, T.; Hirel, B.; Heumez, E.; Dubois, F.; Le Gouis, J. In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilization to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crops Res. 2007, 102, 22–32. [Google Scholar] [CrossRef]
- Derkx, A.; Orford, S.; Griffiths, S.; Foulkes, J.; Hawkesford, M.J.I. dentification of differentially senescing mutants of wheat and impacts on yield, biomass and nitrogen partitioning. J. Integ. Plant Biol. 2012, 54, 555–566. [Google Scholar] [CrossRef]
- Christopher, J.T.; Veyradier, M.; Borrell, A.K.; Harvey, G.; Fletche, S.; Chenu, K. Phenotyping novel stay-green traits to capture genetic variation in senescence dynamics. Funct. Plant Biol. 2014, 41, 1035–1048. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Balota, M.; Delgado, M.I.B.; Amani, I.; Fischer, R.A. Physiological and morphological traits associated with spring wheat yield under hot irrigated conditions. Aust. J. Plan Physiol. 1994, 21, 717–730. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Singh, R.P.; Ibrahim, A.; Ageeb, O.A.A.; Larque Saavedra, A.; Quick, J.S. Evaluating Physiological traits to complement empirical selection for wheat in warm environments. Euphytica 1998, 100, 84–95. [Google Scholar] [CrossRef]
- Gorny, A.G.; Garczynski, S. Genotypic and nutrition-dependent variation in water use efficiency and photosynthetic activity of leaves in winter wheat (Triticum aestivum L.). J. Appl. Genet. 2002, 43, 145–160. [Google Scholar]
- Montazeaud, G.; Karatogma, H.; Ozturk, I.; Roumet, P.; Ecarnot, M.; Crossa, J.; Lopes, M.S. Predicting wheat maturity and stay-green parameters by modeling spectral reflectance measurements and their contribution to grain yield under rainfed conditions. Field Crop Res. 2016, 196, 191–198. [Google Scholar] [CrossRef]
- Gizaw, S.A.; Garland-Campbell, K.; Carter, A.H. Evaluation of agronomic traits and spectral reflectance in Pacific Northwest wheat under rain-fed and irrigated conditions. Field Crop Res. 2016, 196, 168–179. [Google Scholar] [CrossRef]
- Foulkes, M.J.; Sylvester-Bradley, R.; Weightman, R.; Snape, J.W. Identifying physiological traits associated with improved drought resistance in winter wheat. Field Crops Res. 2007, 103, 11–24. [Google Scholar] [CrossRef]
- Fischer, R.A. The importance of grain or kernel number in wheat: A reply to Sinclair and Jamieson. Field Crops Res. 2008, 105, 15–21. [Google Scholar] [CrossRef]
- Liang, X.; Liu, Y.; Chen, J.; Adams, C. Late-season photosynthetic rate and senescence were associated with grain yield in winter wheat of diverse origins. J. Agron. Crop Sci. 2018, 204, 1–12. [Google Scholar] [CrossRef]
- Lopes, M.S.; Reynolds, M.P.; Manes, Y.; Singh, R.P.; Crossa, J.; Braun, H.J. Genetic yield gains and changes in associated traits of CIMMYT spring bread wheat in a “Historic” set representing 30 years of breeding. Crop Sci. 2012, 52, 1123–1131. [Google Scholar] [CrossRef]
- Kipp, S.; Mistele, B.; Schmidhalter, U. Identification of stay-green and early senescence phenotypes in high-yielding winter wheat, and their relationship to grain yield and grain protein concentration using high-throughput phenotyping techniques. Funct. Plant Biol. 2013, 41, 227–235. [Google Scholar] [CrossRef]
- João, P.; Pennacchi, D.; Elizabete, C.D.; Andralojc, P.J.; Feuerhelm, D.; Powers, S.J.; Martin Parry, A.J. Dissecting wheat grain yield drivers in a mapping population in the UK. Agronomy 2018, 8, 94. [Google Scholar]
- Rebetzke, G.J.; Jimenez-Berni, J.A.; Bovill, W.D.; Deery, D.M.; James, R.A. High-throughput phenotyping technologies allow accurate selection of stay-green. J. Exp. Bot. 2016, 67, 4919–4924. [Google Scholar] [CrossRef]
- Fu, J.D.; Yan, Y.F.; Kim, M.Y.; Lee, S.H.; Lee, B.W. Population-specific quantitative trait loci mapping for functional stay-green trait in rice (Oryza sativa L.). Genome 2011, 5, 235. [Google Scholar] [CrossRef]
- Ishimaru, K.; Yano, M.; Aoki, N.; Ono, K.; Hirose, T. Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor. Appl. Genet. 2004, 102, 793–800. [Google Scholar] [CrossRef]
- Teng, S.; Qian, Q.; Zeng, D.; Kunihiro, Y.; Fujimoto, K.; Huang, D.; Zhu, L. QTL analysis of leaf photosynthetic rate and related physiological traits in rice (Oryza sativa L.). Euphytica 2004, 135, 1–7. [Google Scholar] [CrossRef]
- Jiang, G.H.; He, Y.Q.; Xu, C.G.; Li, X.H.; Zhang, Q. The genetic basis of stay-green in rice analyzed in a population of doubled haploid lines derived from an indica by japonica cross. Theor. Appl. Genet. 2004, 108, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Yang, H.J.; Jung, K.H.; Yoo, S.C.; Paek, N.C. Quantitative trait locus mapping and candidate gene analysis for plant architecture traits using whole genome re-sequencing in rice. Mol. Cells 2014, 37, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Cha, K.W.; Koh, H.J.; Lee, Y.J.; Lee, B.M.; Nam, Y.W.; Paek, N.C. Isolation, characterization, and mapping of the stay-green mutant in rice. Theor. Appl. Genet. 2002, 104, 526–532. [Google Scholar] [CrossRef]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice non-yellow coloring 1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Sato, Y.; Yu, M.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Li, M.; Liang, N.; Yan, H.; Wei, Y.; Xu, X.; Liu, J.; Xu, Z.; Chen, F.; Wu, G. Molecular cloning and function analysis of the stay green gene in rice. Plant J. 2007, 52, 197–209. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanaka, R. Chlorophyll metabolism. Curr. Opin. Plant Biol. 2006, 9, 248–255. [Google Scholar] [CrossRef]
- Liu, X.; Li, Z.; Jiang, Z.; Zhao, Y.; Peng, J.; Jin, J.; Guo, H.; Luo, J. LSD: A leaf senescence database. Nucl. Acid. Res. 2011, 39, D1103–D1107. [Google Scholar] [CrossRef]
- Wu, H.; Wang, B.; Chen, Y.; Liu, Y.; Chen, L. Characterization and fine mapping of the rice premature senescence mutant ospse1. Theor. Appl. Genet. 2013, 126, 1897–1907. [Google Scholar] [CrossRef]
- Lin, A.; Wang, Y.; Tang, J.; Xue, P.; Li, C.; Liu, L.; Hu, B.; Yang, F.; Loake, G.J.; Chu, C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012, 158, 451–464. [Google Scholar] [CrossRef]
- Huang, Q.; Shi, Y.; Zhang, X.; Song, L.; Feng, B.; Wang, H.; Xu, X.; Li, X.; Guo, D.; Wu, J. Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J. Integr. Plant Biol. 2016, 58, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Yang, Y.; Xu, J.; Li, X.; Leng, Y.; Dai, L.; Huang, L.; Shao, G.; Ren, D.; Hu, J.; et al. EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol. 2015, 169, 1225–1239. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Ren, D.; Hu, S.; Li, G.; Dong, G.; Jiang, L.; Hu, X.; Ye, W.; Cui, Y.; Zhu, L.; et al. Down-regulation of a nicotinate phosphoribosyl transferase gene, OsNaPRT1, leads to withered leaf tips. Plant Physiol. 2016, 171, 1085–1098. [Google Scholar] [PubMed]
- Jiao, B.; Wang, J.; Zhu, X.; Zeng, L.; Li, Q.; He, Z. A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol. Plant 2012, 5, 205–217. [Google Scholar] [CrossRef]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. PNAS 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Hoang, T.B.; Kobata, T. Stay-green in Rice (Oryza sativa L.) of drought prone areas in desiccated soils. Plant Prod. Sci. 2009, 12, 397–408. [Google Scholar] [CrossRef]
- Adams, W.W.; Winter, K.; Schreiber, U.; Schramel, P. Photosynthesis and Chl fluorescence characteristics in relationship to changes in pigment and element composition of leaves of Platanus occidentalis L. during autumnal leaf senescence. Plant Physiol. 1990, 92, 1184–1190. [Google Scholar] [CrossRef]
- Weng, X.Y.; Xu, H.X.; Jiang, D.A. Characteristics of gas exchange, Chl fluorescence and expression of key enzymes in photosynthesis during leaf senescence in rice plants. J. Integr. Plant Biol. 2005, 47, 560–566. [Google Scholar] [CrossRef]
- Zhang, C.J.; Chen, G.X.; Gao, X.X.; Chu, C.J. Photosynthetic decline in flag leaves of two field-grown spring wheat cultivars with different senescence properties. S. Afr. J. Bot. 2006, 72, 15–23. [Google Scholar] [CrossRef]
- Hörtensteiner, S.; Kräutler, B. Chl breakdown in higher plants. Biochim. Biophys. Acta 2011, 1807, 977–988. [Google Scholar] [CrossRef]
- Kusaba, M.; Tanaka, A.; Tanaka, R. Stay-green plants: What do they tell us about the molecular mechanism of leaf senescence. Photosynth Res. 2013, 117, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; Tang, Y.; Zhang, H.; Wu, P.; Chen, Y.; Li, M.; Wu, G.; Jiang, H. The Stay-green rice like (SGRL) gene regulates Chl degradation in rice. J. Plant Physiol. 2013, 170, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, B.W. Photosynthetic characteristics of rice cultivars with depending on leaf senescence during grain filling. Korean J. Crop Sci. 2003, 48, 216–223. [Google Scholar]
- Agrama, H.A.S.; Moussa, M.E. Mapping QTLs in breeding for drought tolerance in maize (Zea mays L.). Euphytica 1996, 91, 89–97. [Google Scholar] [CrossRef]
- Ribaut, J.M.; Jiang, C.; Gonzalez-de-Leon, D.; Edmeades, G.O.; Hoisington, D.A. Identification of quantitative trait loci under drought conditions in tropical maize: Part 2. Yield components and marker-assisted selection strategies. Theor. Appl. Genet. 1997, 94, 887–896. [Google Scholar] [CrossRef]
- Tuberosa, R.; Salvi, S.; Sanguineti, M.C.; Landi, P.; Maccaferri, M.; Conti, S. Mapping QTLs regulating morpho-physiological traits and yield: Case studies, shortcomings and perspectives in drought-stressed maize. Ann. Bot. 2002, 89, 941–963. [Google Scholar] [CrossRef]
- Beavis, W.D.; Smith, O.S.; Grant, D.; Fincher, R. Identification of quantitative trait loci using a small sample of top crossed and F4 progeny from maize. Crop Sci. 1994, 34, 882–896. [Google Scholar] [CrossRef]
- Câmara, T.M.M. Mapping QTL for Traits Related to Drought Stress Tolerance in Tropical Maize (In Portuguese with English Abstract). Ph.D. Thesis, Agriculture College Luiz de Queiroz, University of São Paulo, Piracicaba, SP, Brazil, 2006. [Google Scholar]
- Yang, Z.; Li, X.; Zhang, N.; Wang, X.; Zhang, Y.; Ding, Y.; Kuai, B.; Huang, X. Mapping and validation of the quantitative trait loci for leaf stay-green-associated parameters in maize. Plant Breed. 2017, 136, 188–196. [Google Scholar] [CrossRef]
- Sekhon, R.D.; Saski, C.; Kumar, R.; Flinn, B.S.; Luo, F.; Beissinger, T.M.; Ackerman, A.J.; Breitzman, M.M.W.; Bridges, W.C.; de Leon, N.; et al. Integrated genome-scale analysis identifies novel genes and networks underlying senescence in maize. Plant Cell 2019, 31, 1968–1989. [Google Scholar] [CrossRef]
- Benchimol, L.L.; de Souza, C.L., Jr.; de Souza, A.P. Microsatellite-assisted backcross selection in maize. Genet. Mol. Biol. 2005, 28, 789–797. [Google Scholar] [CrossRef]
- Neereja, C.N.; Maghirang-Rodriguez, R.; Pamplona, A.; Heuer, S.; Collard, B.C.Y.; Sptiningsih, E.M.; Vergara, G.; Sanchez, D.; Xu, K.; Ismail, A.M.; et al. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor. Appl. Genet. 2007, 115, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Garzón, L.N.; Ligarreto, G.A.; Blair, M.W. Molecular marker-assisted backcrossing of anthracnose resistance into Andean climbing beans Phaseolus vulgaris L. Crop Sci. 2008, 48, 562–570. [Google Scholar] [CrossRef]
- Chen, L.; An, Y.; Li, Y.; Li, C.; Shi, Y.; Song, Y.; Zhang, D.; Wang, T.; Li, Y. Candidate loci for yield-related traits in maize revealed by a combination of meta QTL analysis and regional association mapping. Front. Plant Sci. 2017, 8, 2190. [Google Scholar] [CrossRef] [PubMed]
- Swanckaert, J.; Pannecocque, J.; Vanwaes, J.; Steppee, K.; Vanlabeke, M.-C.; Rehul, D. Stay-green characterization in Belgian forage maize. J. Agric. Sci. 2017, 155, 766–776. [Google Scholar] [CrossRef]
- Hay, R.K.M.; Porter, J.R. The Physiology of Crop Yield; Blackwell Publishing: Oxford, UK, 2006; ISBN 9781405108591. [Google Scholar]
- Pennisi, E. Plant genetics: The blue revolution, drop by drop, gene by gene. Science 2008, 320, 171–173. [Google Scholar] [CrossRef]
- Ludlow, M.M.; Muchow, R.C. A critical evaluation of traits for improving crop yields in water-limited environments. Adv. Agron. 1990, 43, 107–153. [Google Scholar]
- He, Y.; Liu, Y.; Cao, W.; Huai, M.; Xu, B.; Huang, B. Effects of salicylic acid on heat tolerance associated with antioxidant metabolism in Kentucky Bluegrass. Am. J. Crop Sci. 2005, 45, 988–995. [Google Scholar] [CrossRef]
- Cai, H.; Chu, Q.; Yuan, L.; Liu, J.; Chen, X.; Chen, F.; Mi, G.; Zhang, F. Identification of quantitative trait loci for leaf area and Chl content in maize (Zea mays L.) under low nitrogen and low phosphorus supply. Mol. Breed. 2012, 30, 251–266. [Google Scholar] [CrossRef]
- Cai, H.; Chu, Q.; Yuan, L.; Liu, J.; Chen, X.; Chen, F.; Mi, G.; Zhang, F. Identification of QTL for plant height, ear height and grain yield in maize (Zea mays L.) in response to nitrogen and phosphorus supply. Plant Breed. 2012, 131, 502–510. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D.A. Review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Trachsel, S.; Sun, D.; San Vicente, F.M.; Zheng, H.; Atlin, G.N.; Suarez, E.A. Identification of QTL for early vigor and stay-green conferring tolerance to drought in two connected advanced backcross populations in tropical maize (Zea mays L.). PLoS ONE 2016, 11, e0149636. [Google Scholar]
- Cerrudo, D.; Cao, S.; Yuan, Y.; Martinez, C.; Suarez, E.A.; Babu, R.; Zhang, X.; Trachsel, S. Genomic selection outperforms marker assisted selection for grain yield and physiological traits in a maize doubled haploid population across water treatments. Front. Plant Sci. 2018, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Duvick, D.N.; Smith, J.S.C.; Cooper, M. Long-term selection in a commercial hybrid maize breeding program. Plant Breed Rev. 2004, 24, 109–151. [Google Scholar]
- Crosbie, T.M.; Pearce, R.B.; Mock, J.J. Selection for high CO2 exchange rate among inbred lines of maize. Crop Sci. 1981, 21, 629–631. [Google Scholar] [CrossRef]
- Valentinuz, O.R.; Tollenaar, M. Effect of genotype, nitrogen, plant density, and row spacing on the area-per leaf profile in maize. Agron. J. 2006, 98, 94–99. [Google Scholar] [CrossRef]
- Cairns, J.E.; Sanchez, C.; Vargas, M.; Ordoñez, R.; Araus, J.L. Dissecting maize productivity: Ideotypes associated with grain yield under drought stress and well-watered conditions. J. Integr. Plant Biol. 2012, 54, 1007–1020. [Google Scholar] [CrossRef]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Börner, A. Drought Stress Tolerance in Wheat and Barley: Advances in Physiology, Breeding and Genetics Research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef]
- Obsa, B.T.; Eglinton, J.; Coventry, S.; March, T.; Langridge, P.; Fleury, D. Genetic analysis of developmental and adaptive traits in three doubled haploid populations of barley (Hordeum vulgare L.). Theor. Appl. Genet. 2016, 129, 1139–1151. [Google Scholar] [CrossRef]
- Fox, G.P. Discovery of QTL for stay-green and heat-stress in barley (Hordeum vulgare) grown under simulated abiotic stress conditions. Euphytica 2015, 207, 2. [Google Scholar]
- Gonzalez, A.; Martin, I.; Ayerbe, L. Response of barley genotypes to terminal soil moisture stress: Phenology, growth, and yield. Aust. J. Agric. Res. 2007, 58, 29–37. [Google Scholar] [CrossRef]
- Maydup, M.L.; Antonietta, M.; Buiamet, J.J.; Graciano, C.; Lopez, J.R.; Tambussi, E.A. The contribution of ear photosynthesis to grain filling in bread wheat (Triticum aestivum L.). Field Crops Res. 2010, 119, 48–58. [Google Scholar] [CrossRef]
- Vaezi1, B.; Bavei1, V.; Shiran, B. Screening of barley genotypes for drought tolerance by agro-physiological traits in field condition. Afr. J. Agric. Res. 2010, 5, 881–892. [Google Scholar]
- Seiler, C.; Harshavardhan, V.T.; Reddy, P.S.; Hensel, G.; Kumlehn, J.; Eschen-Lippold, L.; Rajesh, K.; Korzun, V.; Wobus, U.; Lee, J.; et al. Abscisic acid flux alterations result in differential ABA signaling responses and impact assimilation efficiency in barley under terminal drought stress. Plant Physiol. 2014, 164, 1677–1696. [Google Scholar] [CrossRef] [PubMed]
- Shirdelmoghanloo, H.; Paynter, B.; Chen, K.; D’Antuono, M.; Balfour, C.A.; Angesa, T.; Westcott, S.; Li, C. Grain plumpness in barley under grain filling heat stress: Association with grain growth components and stay-green. In Proceedings of the 19th Australian Barley Technical Symposium, Perth, WA, Australia, 9–12 September 2019. [Google Scholar]
- Magney, T.S.; Eitel, J.U.H.; Huggins, D.R.; Viering, L.A. Proximal NDVI derived phenology improves in-season predictions of wheat quality and quantity. Agric. Forest Meteorol. 2015, 217, 46–60. [Google Scholar] [CrossRef]
- Gous, P.W.; Hasjim, J.; Franckowiak, J.; Fox, G.P.; Gilbert, R. Barley genotype expressing “SG”-like characteristics maintains starch quality of the grain during water stress condition. J. Cereal Sci. 2013, 58, 414–419. [Google Scholar] [CrossRef]
- Zhou, C.; Han, L.; Pislariu, C.; Nakashima, J.; Fu, C.; Jiang, Q.; Quan, L.; Blancaflor, E.B.; Tang, Y.; Bouton, J.H. From model to crop: Functional analysis of a Stay-green gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 2011, 157, 1483–1496. [Google Scholar] [CrossRef]
- Patro, L.; Mohapatra, P.K.; Biswal, U.C.; Biswal, B. Dehydration induced loss of photosynthesis in Arabidopsis leaves during senescence is accompanied by the reversible enhancement in the activity of cell wall β-glucosidase. J. Photochem. Photobiol. 2014, 137, 49–54. [Google Scholar] [CrossRef]
- Bromley, J.R.; Busse-Wicher, M.; Tryfona, T.; Mortimer, J.C.; Zhang, Z.; Brown, D.M.; Dupree, P. GUX1 and GUX2 glucuronyl transferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 2013, 74, 423–434. [Google Scholar] [CrossRef]
- Rennie, E.A.; Hansen, S.F.; Baidoo, E.E.; Hadi, M.Z.; Keasling, J.D.; Scheller, H.V. Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyl transferases. Plant Physiol. 2012, 159, 1408–1417. [Google Scholar] [CrossRef]
- Zeng, W.; Chatterjee, M.; Faik, A. UDP-Xylose-stimulated glucuronyl transferase activity in wheat microsomal membranes: Characterization and role in glucuronol (arabino) xylan biosynthesis. Plant Physiol. 2008, 147, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, Y.; Zhang, Y.; Chai, C.; Gang, W.; Wei, X.; Xu, H.; Wang, M.; Ouwerkerk, P.B.F.; Zhu, Z. Molecular cloning, functional characterization and expression analysis of a novel monosaccharide transporter gene OsMST6 from rice (Oryza sativa L.). Planta 2008, 228, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.A.; Mahé, A.; Guy, S.; Brangeona, J.; Rochea, O.; Chourey, P.S.; Priou, J.L. Characterization of two members of the maize gene family, Incw3andIncw4, encoding cell-wall invertases. Gene 2000, 245, 89–102. [Google Scholar] [CrossRef]
- Gal, C.; Moore, K.M.; Paszkiewicz, K.; Kent, N.A.; Whitehall, S.K. The impact of the HIRA histone chaperone upon global nucleosome architecture. Cell Cycle 2015, 14, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Yang, Y.; Feng, X.; Zhang, M.; Song, R. Mitochondrial function and maize kernel development requires Dek2, a pentatricopeptide repeat protein involved in nad1 mRNA splicing. Genetics 2017, 205, 239–249. [Google Scholar] [CrossRef]
- Abdi Woldesemayat, A.; Modise, D.M.; Gemeildien, J.; Ndimba, B.K.; Christoffels, A. Cross-species multiple environmental stress responses: An integrated approach to identify candidate genes for multiple stress tolerance in sorghum (Sorghum bicolor (L.) Moench) and related model species. PLoS ONE 2018, 13, e0192678. [Google Scholar]
- Moore, G. Cereal genome evolution: Pastoral pursuits with Lego’ genomes. Curr. Opin. Genet. Dev. 1995, 5, 717–724. [Google Scholar] [CrossRef]
- Foote, T.; Roberts, M.; Kurata, N.; Sasaki, T.; Moore, G. Detailed comparative mapping of cereal chromosome regions corresponding to the Phl locus in wheat. Genetics 1997, 147, 801–807. [Google Scholar]
| Genes | Function | Cereals | References | |||
|---|---|---|---|---|---|---|
| Rice | Wheat | Maize | Sorghum | |||
| Stay-Green Rice like (SGRL) | Affect Chlorophyll (Chl) degradation during natural and dark-induced leaf senescence | √ | - | - | 0 | [116,154] |
| Glucuronic acid substitution of xylan1 (GUX1) | Required for substitution of the xylan backbone with 4-O-methylglucuronic acid [Me]GlcA | - | 0 | √ | 0 | [125,155,156,157,158] |
| β-Glucosidase (BGLU42) | The exact role remains to be determined | √ | - | √ | 0 | [125] |
| Trehalose-6-phospate synthase13(trps13) | Sugar-mediated signaling in stay-green | - | - | √ | 0 | [125] |
| Monosaccharide transporter (MST4) | Associated with senescence and nitrogen use efficiency, transport of Sucrose out of leaf cells by sugar transporters to alternative sinks | √ | - | √ | - | [125,159] |
| Sugar will Eventually be exported transporters (SWEET) | Transport of Sucrose out of leaf cells by sugar transporters to alternative sinks | √ | - | √ | - | [125,160] |
| Cell wall invertase (incw4) | Sinks hydrolyzation | - | - | √ | 0 | [125,161] |
| Mitochondrial pentatricopeptide repeat protein (dek10) | Activation sugar-sinks | - | - | √ | - | [125,162,163] |
| Sb09g004170 and Sb09g022580 | DRGs (Associated with Stg1 QTL) | - | - | - | √ | [164] |
| NAC-transcription factor 9(nactf9) | Appears to be one of the master regulators of stay-green, acting in conjunction with ZmIRX15-L, ZmGUX1, mlg3 | - | - | √ | - | [125] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamal, N.M.; Gorafi, Y.S.A.; Abdelrahman, M.; Abdellatef, E.; Tsujimoto, H. Stay-Green Trait: A Prospective Approach for Yield Potential, and Drought and Heat Stress Adaptation in Globally Important Cereals. Int. J. Mol. Sci. 2019, 20, 5837. https://doi.org/10.3390/ijms20235837
Kamal NM, Gorafi YSA, Abdelrahman M, Abdellatef E, Tsujimoto H. Stay-Green Trait: A Prospective Approach for Yield Potential, and Drought and Heat Stress Adaptation in Globally Important Cereals. International Journal of Molecular Sciences. 2019; 20(23):5837. https://doi.org/10.3390/ijms20235837
Chicago/Turabian StyleKamal, Nasrein Mohamed, Yasir Serag Alnor Gorafi, Mostafa Abdelrahman, Eltayb Abdellatef, and Hisashi Tsujimoto. 2019. "Stay-Green Trait: A Prospective Approach for Yield Potential, and Drought and Heat Stress Adaptation in Globally Important Cereals" International Journal of Molecular Sciences 20, no. 23: 5837. https://doi.org/10.3390/ijms20235837
APA StyleKamal, N. M., Gorafi, Y. S. A., Abdelrahman, M., Abdellatef, E., & Tsujimoto, H. (2019). Stay-Green Trait: A Prospective Approach for Yield Potential, and Drought and Heat Stress Adaptation in Globally Important Cereals. International Journal of Molecular Sciences, 20(23), 5837. https://doi.org/10.3390/ijms20235837