Cloning and Expression Analysis of the BocMBF1c Gene Involved in Heat Tolerance in Chinese Kale
Abstract
1. Introduction
2. Results
2.1. Cloning and Analysis of the BocMBF1c Gene
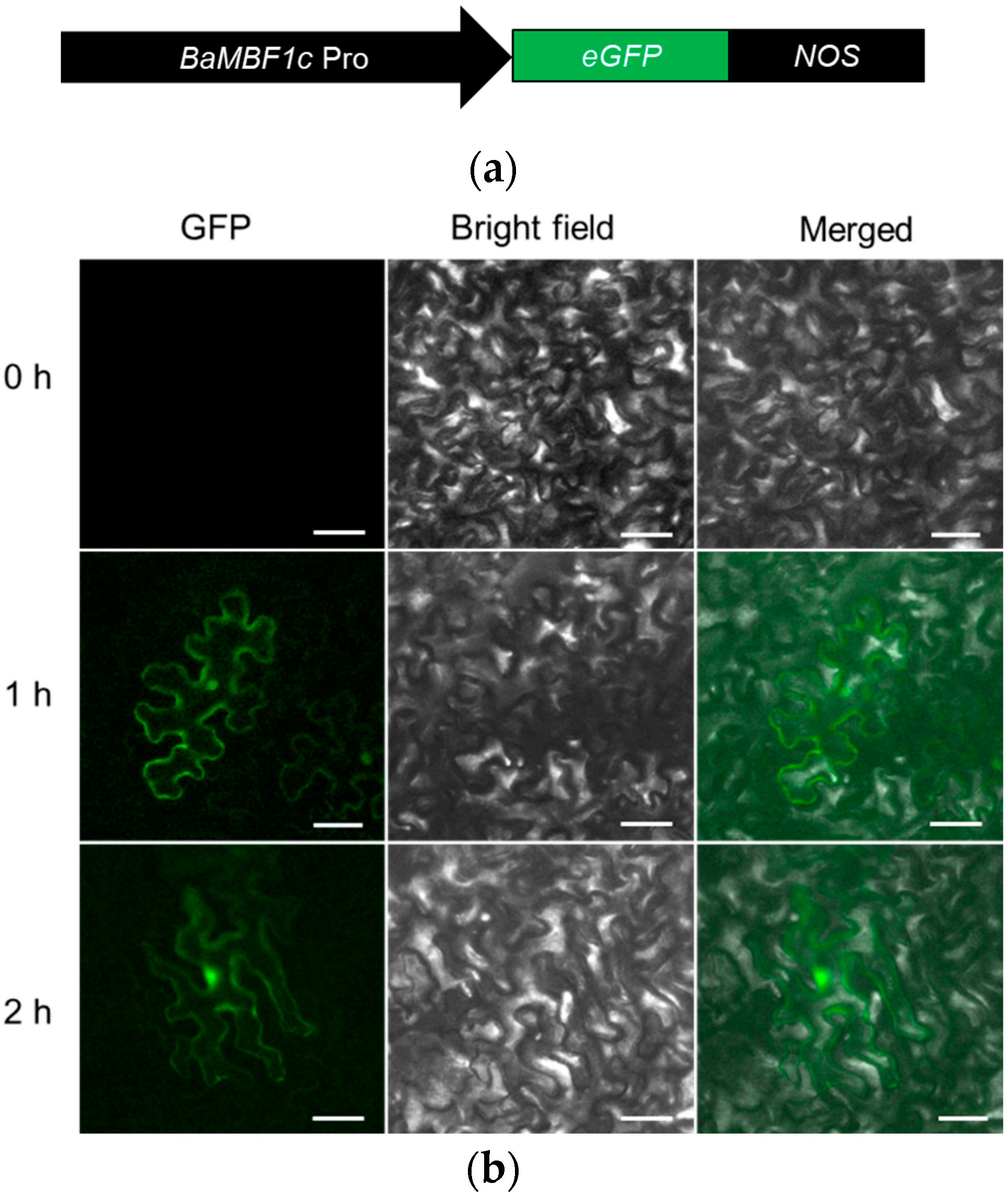
2.2. Transient Expression Analysis Showed BocMBF1c Promoter Activity Can Be Induced by Heat Stress
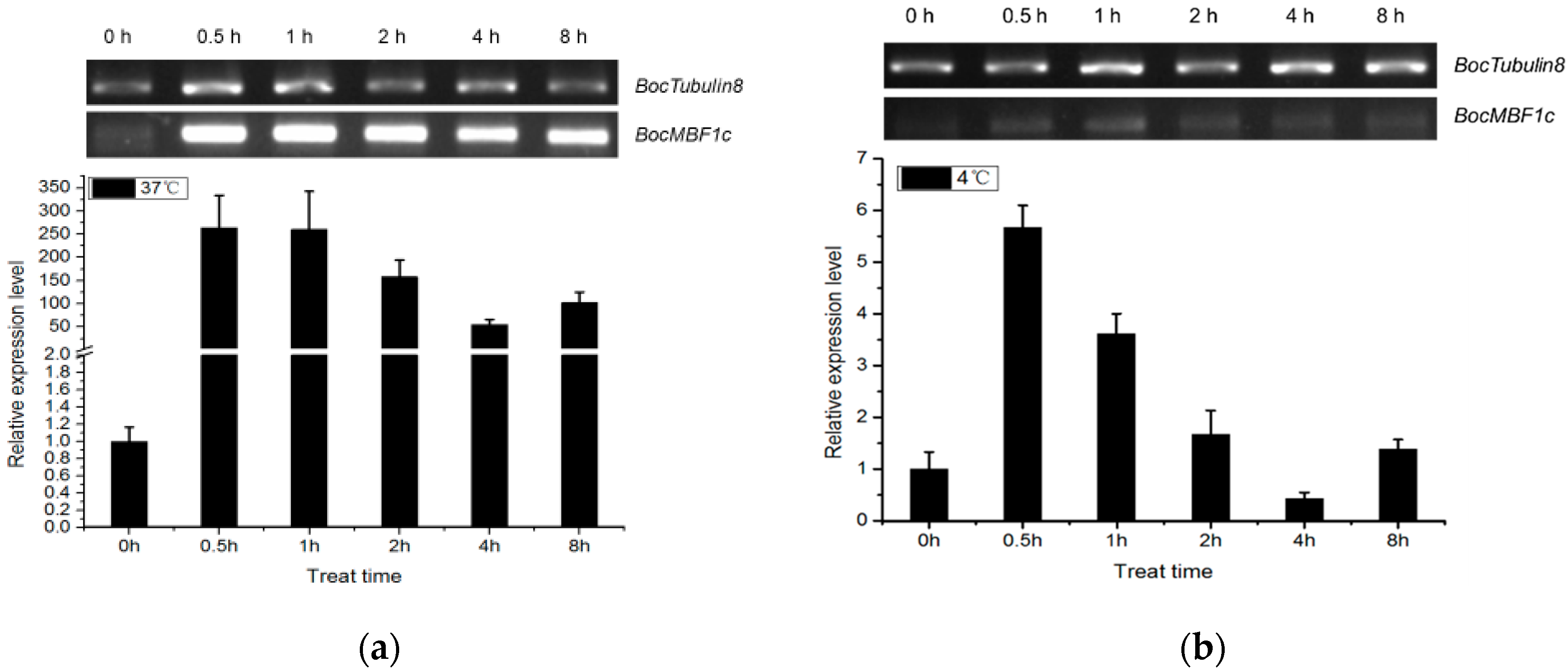
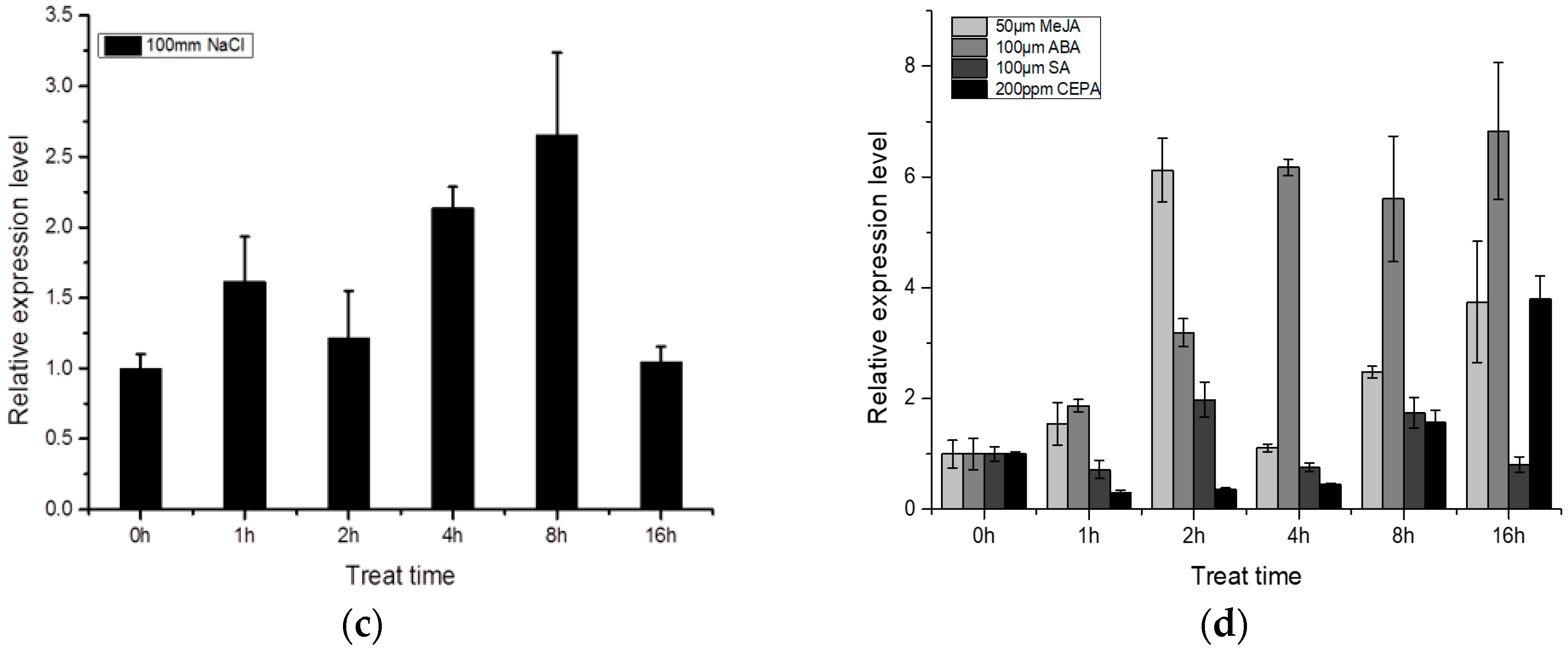
2.3. Expression Pattern of BocMBF1c
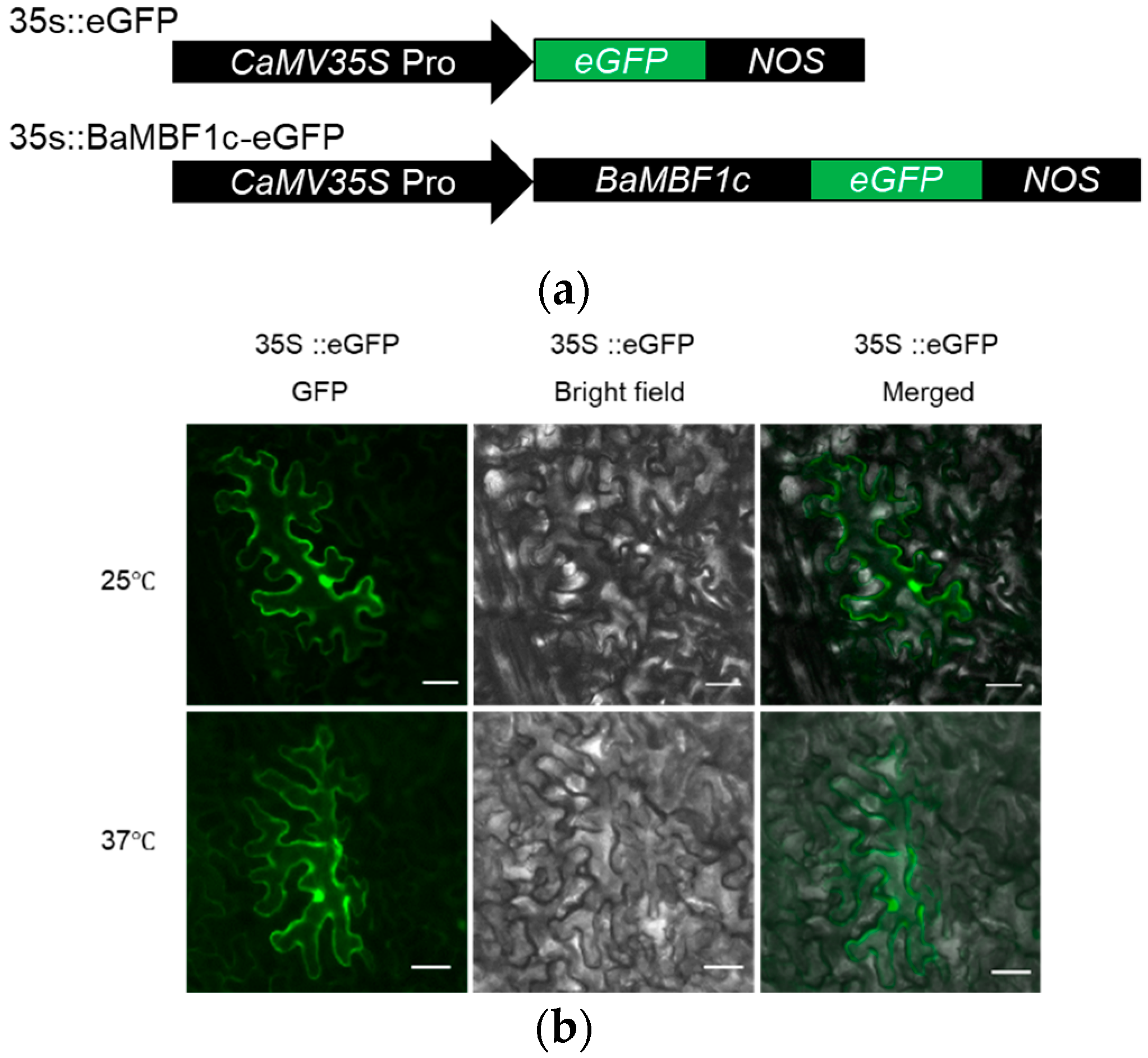
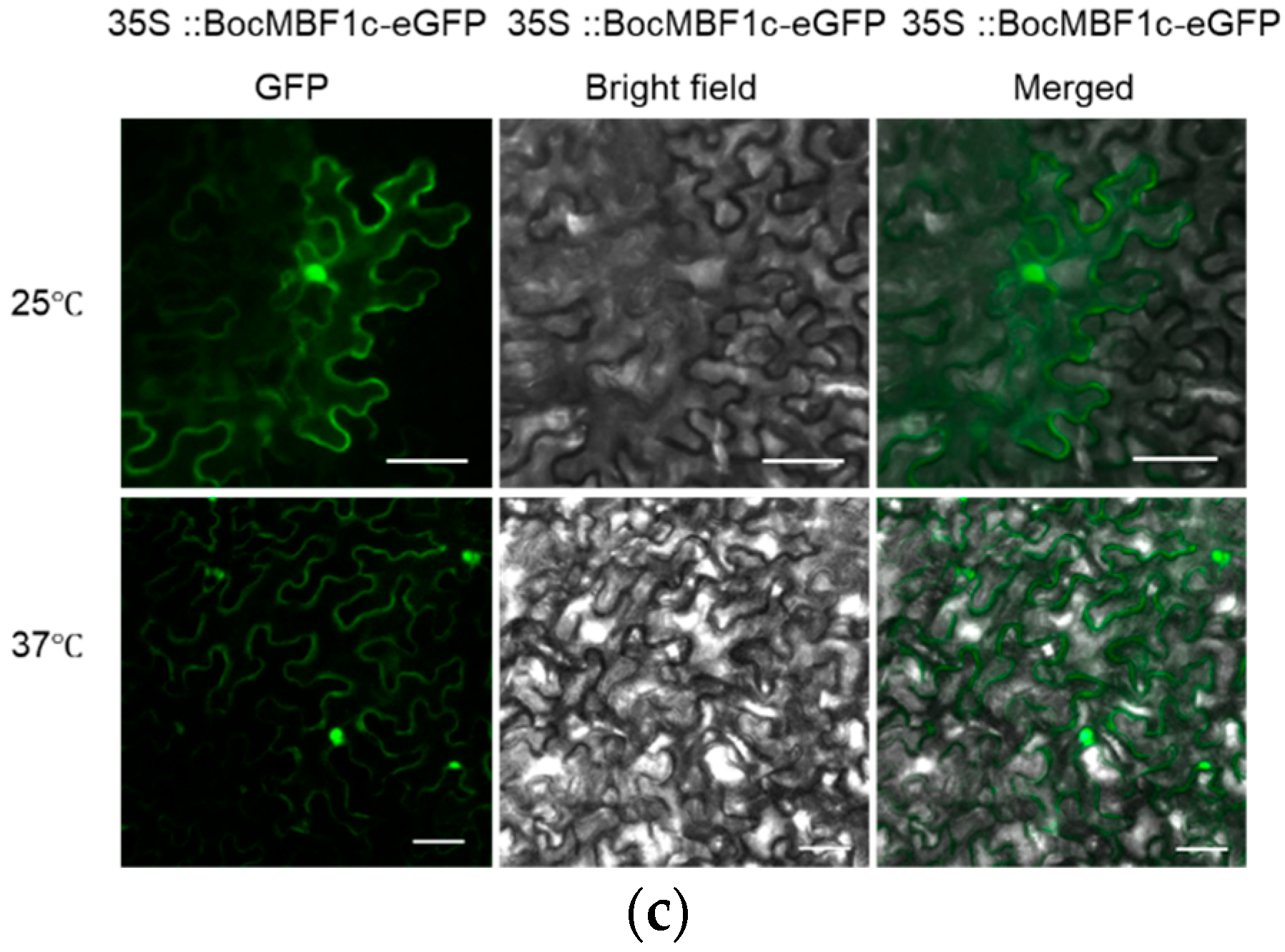
2.4. BocMBF1c Protein Localizes to the Nucleus under Heat Stress
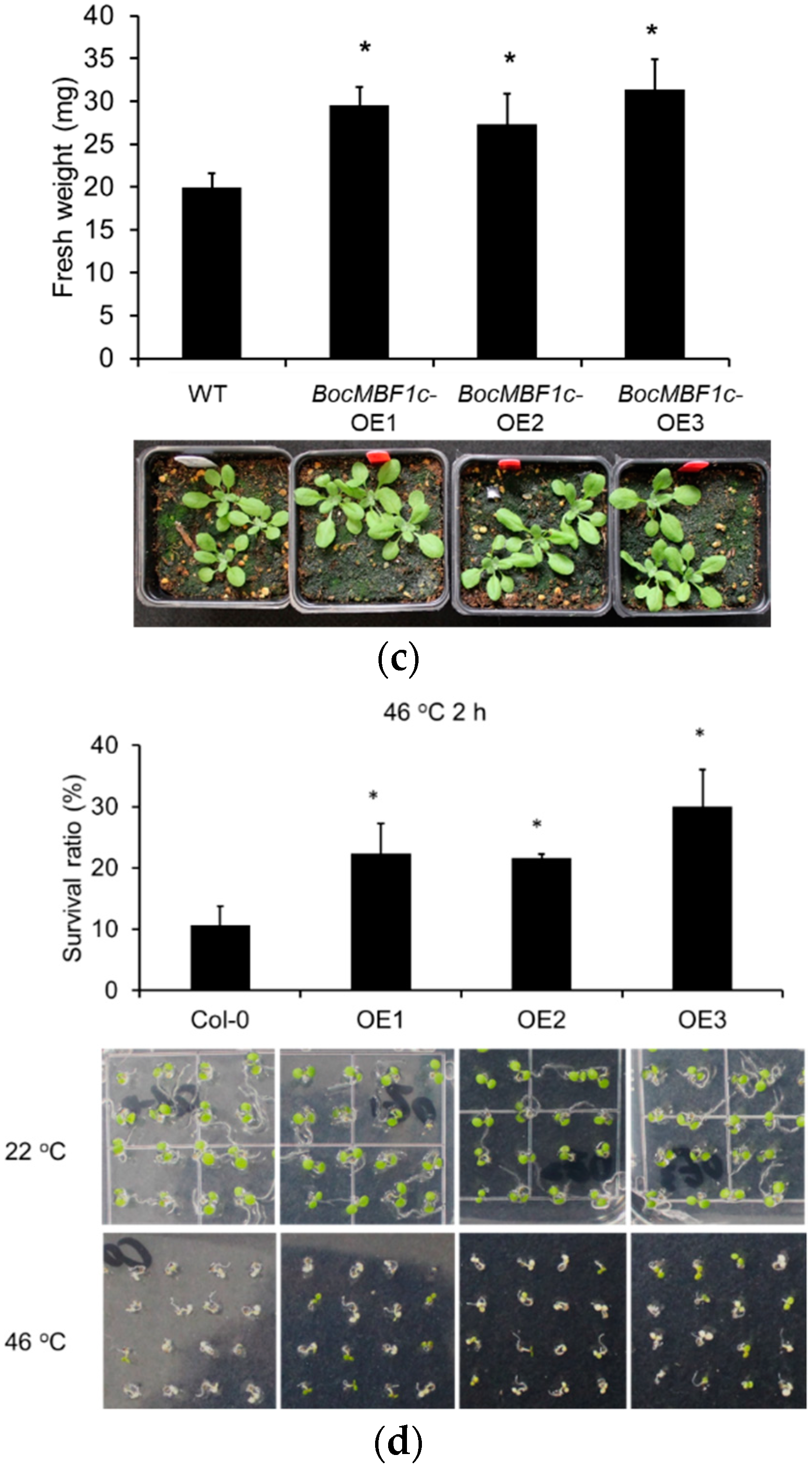
2.5. Phenotype and Thermotolerance Analysis of BocMBF1c Overexpression Lines
3. Discussion
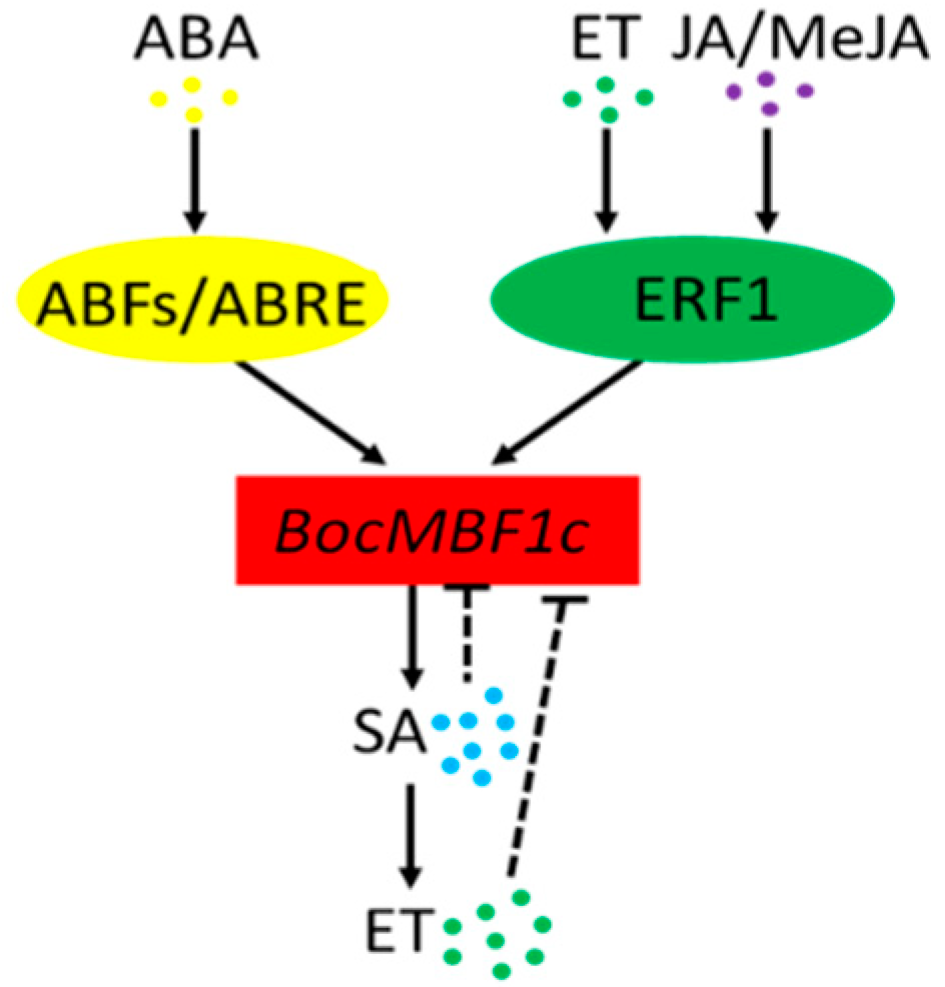
3.1. BocMBF1c Can Respond to Multiple Stresses and Hormone Treatment in Addition to Heat Stress in Chinese Kale
3.2. BocMBF1c Can be Applied to Improve the Productivity of Chinese Kale
4. Materials and Methods
4.1. Plant Growth and Abiotic Treatment Conditions
4.2. Cloning of the BocMBF1c CDS and Promoter
4.3. Sequence Analysis of the BocMBF1c Gene
4.4. Promoter Activity and Subcellular Localization
4.5. Gene Expression Patterns of BocMBF1c by qPCR and Semi-Quantitative RT-PCR
4.6. Arabidopsis Transformation and Phenotypic Analysis of Transgenic Plants
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MBF | Multi-protein bridging factor |
| HTH | Helix-turn-helix |
| HSEs | Heat shock elements |
| GFP | Green fluorescent protein |
| eGFP | Enhanced green fluorescent protein |
| ABA | Abscisic acid |
| ABRFs | ABA responsive transcription factors |
| HSF | Heat stress transcription factor |
| HSPs | Heat shock proteins |
| MeJA | Methyl jasmonate |
| SA | Salicylic acid |
| ET | Ethylene |
| JA | Jasmonic acid |
| ERF | Ethylene response factor |
| DRE | Dehydration-responsive element |
| ORF | Open reading frame |
| CDS | Coding sequence |
| ERE | Ethylene-responsive elements |
| ABRE | Abscisic acid-responsive element |
| MBS | MYB binding site |
| N. | Nicotiana |
| qPCR | Quantitative PCR |
| CEPA | Ethephon |
| RT-PCR | Reverse transcription PCR |
| GUS | β-Glucuronidase |
| CaMV | Cauliflower mosaic virus |
| WT | Wild type |
| ABFs | ABRE binding factors |
References
- Lei, J.; Chen, G.; Chen, C.; Cao, B. Germplasm Diversity of Chinese Kale in China. Hortic. Plant J. 2017, 3, 101–104. [Google Scholar] [CrossRef]
- Wu, S.; Lei, J.; Chen, G.; Chen, H.; Cao, B.; Chen, C. De novo Transcriptome Assembly of Chinese Kale and Global Expression Analysis of Genes Involved in Glucosinolate Metabolism in Multiple Tissues. Front. Plant Sci. 2017, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Chen, G.; Lei, J.; Cao, B.; Feng, E. Analysis on Composition and Content of Glucosinolates in Different Genotypes of Chinese Kale. China Veg. 2009, 6, 7–13. [Google Scholar]
- Yang, X.; Yang, Y. The Effects of Temperature on Flower Bud Differentiation, Yield and Quality Formation in Chinese Kale (Brassica alboglabra Bailey). J. S. China Agric. Univ. 2002, 23, 5–7. [Google Scholar]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhuang, L.; Shi, Y.; Huang, B. Up-Regulation of HSFA2c and HSPs by ABA Contributing to Improved Heat Tolerance in Tall Fescue and Arabidopsis. Int. J. Mol. Sci. 2017, 18, 1981. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.-H.; Guo, J.; Ding, K.; Wang, S.-J.; Zhang, H.; Dai, X.-W.; Chen, Y.-Y.; Govers, F.; Huang, L.-L.; Kang, Z.-S. Characterization of a wheat HSP70 gene and its expression in response to stripe rust infection and abiotic stresses. Mol. Biol. Rep. 2011, 38, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Iqbal, N.; Masood, A.; Per, T.S.; Khan, N.A. Salicylic acid alleviates adverse effects of heat stress on photosynthesis through changes in proline production and ethylene formation. Plant Signal. Behav. 2013, 8, e26374. [Google Scholar] [CrossRef] [PubMed]
- Qu, A.-L.; Ding, Y.-F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Bajad, S.; Shuman, J.; Shulaev, V.; Mittler, R. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 2008, 283, 9269–9275. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Sejima, H.; Tam, R.; Schlauch, K.; Mittler, R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 2011, 66, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Alavilli, H.; Lee, H.; Park, M.; Lee, B. Antarctic Moss Multiprotein Bridging Factor 1c Overexpression in Arabidopsis Resulted in Enhanced Tolerance to Salt Stress. Front. Plant Sci. 2017, 8, 1206. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Yamazaki, K.I. Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim. Biophys. Acta—Gene Struct. Expr. 2004, 1680, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N. Enhanced Tolerance to Environmental Stress in Transgenic Plants Expressing the Transcriptional Coactivator Multiprotein Bridging Factor 1c. Plant Physiol. 2005, 139, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, G.; Chen, C.; Cao, B.; Zou, L.; Lei, J. Identification of Heat Tolerance in Chinese Kale and the Expression Analysis of Heat Tolerance Transcription Factor MBF1c. China Veg. 2017, 2, 30–37. [Google Scholar]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-C.; Liao, P.-M.; Kuo, W.-W.; Lin, T.-P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 Regulates Abiotic Stress-Responsive Gene Expression by Binding to Different cis-Acting Elements in Response to Different Stress Signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Toufighi, K.; Brady, S.M.; Austin, R.; Ly, E.; Provart, N.J. The botany array resource: E-Northerns, expression angling, and promoter analyses. Plant J. 2005, 43, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Sun, B.; Xu, X.; Chen, H.; Zou, L.; Chen, G.; Cao, B.; Chen, C.; Lei, J. Overexpression of AtEDT1/HDG11 in Chinese Kale (Brassica oleracea var. alboglabra) Enhances Drought and Osmotic Stress Tolerance. Front. Plant Sci. 2016, 7, 1285. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Wang, F.; Geng, X.; Zhang, L.; Yao, Y.; Ni, Z.; Peng, H.; Sun, Q. Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) Multiprotein Bridging Factor, confers heat tolerance in both yeast and rice. Plant Mol. Biol. 2015, 87, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Goodin, M.M.; Zaitlin, D.; Naidu, R.A.; Lommel, S.A. Nicotiana benthamiana: Its History and Future as a Model for Plant–Pathogen Interactions. Mol. Plant-Microbe Interact. 2008, 21, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, L.; Yu, B.; Ma, X.-L.; Cao, B.; Chen, G.; Chen, C.; Lei, J. Cloning and Expression Analysis of the BocMBF1c Gene Involved in Heat Tolerance in Chinese Kale. Int. J. Mol. Sci. 2019, 20, 5637. https://doi.org/10.3390/ijms20225637
Zou L, Yu B, Ma X-L, Cao B, Chen G, Chen C, Lei J. Cloning and Expression Analysis of the BocMBF1c Gene Involved in Heat Tolerance in Chinese Kale. International Journal of Molecular Sciences. 2019; 20(22):5637. https://doi.org/10.3390/ijms20225637
Chicago/Turabian StyleZou, Lifang, Bingwei Yu, Xing-Liang Ma, Bihao Cao, Guoju Chen, Changming Chen, and Jianjun Lei. 2019. "Cloning and Expression Analysis of the BocMBF1c Gene Involved in Heat Tolerance in Chinese Kale" International Journal of Molecular Sciences 20, no. 22: 5637. https://doi.org/10.3390/ijms20225637
APA StyleZou, L., Yu, B., Ma, X.-L., Cao, B., Chen, G., Chen, C., & Lei, J. (2019). Cloning and Expression Analysis of the BocMBF1c Gene Involved in Heat Tolerance in Chinese Kale. International Journal of Molecular Sciences, 20(22), 5637. https://doi.org/10.3390/ijms20225637



