Interspecies Outer Membrane Vesicles (OMVs) Modulate the Sensitivity of Pathogenic Bacteria and Pathogenic Yeasts to Cationic Peptides and Serum Complement
Abstract
1. Introduction
2. Results
2.1. Mc6 OMVs Characteristics
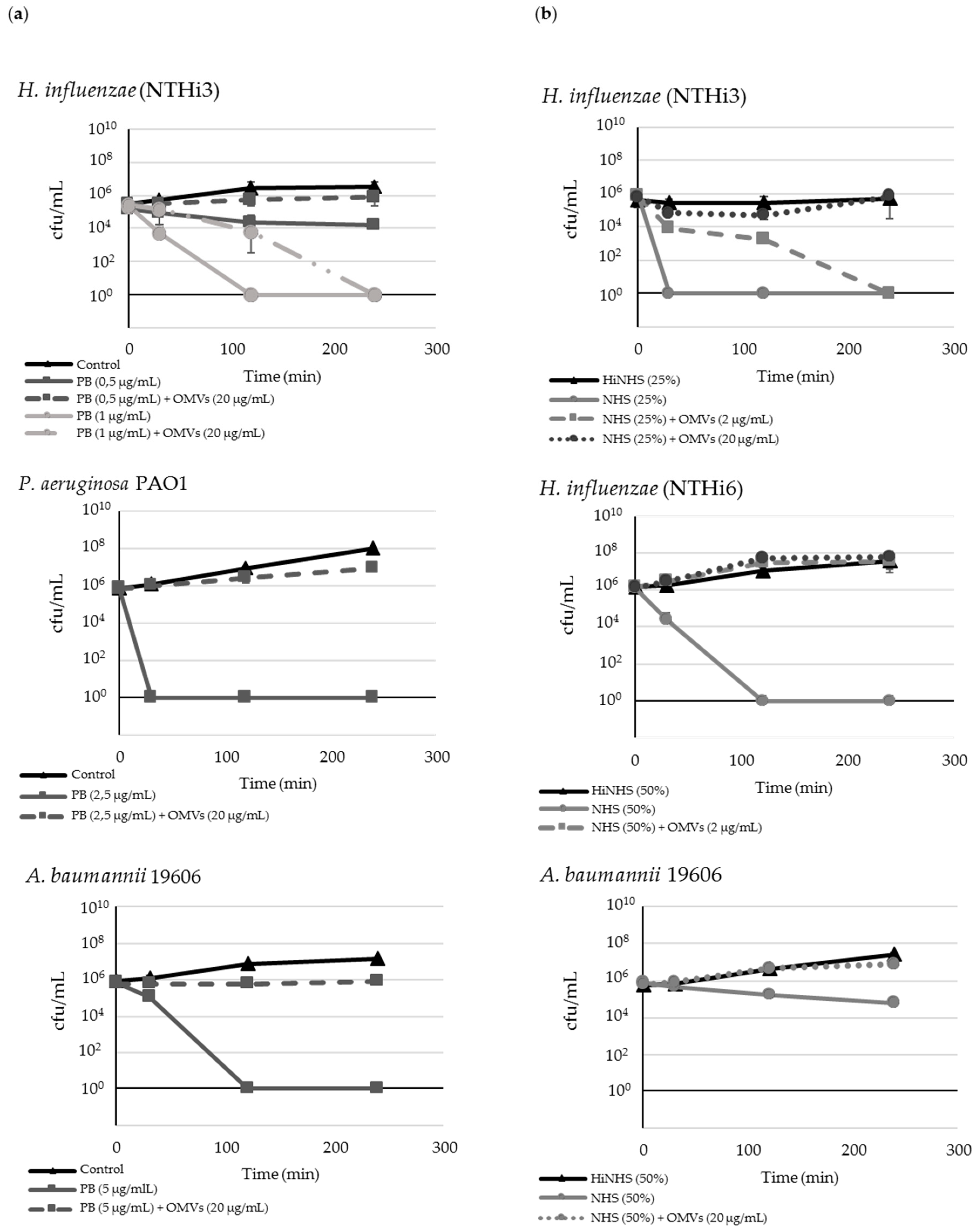
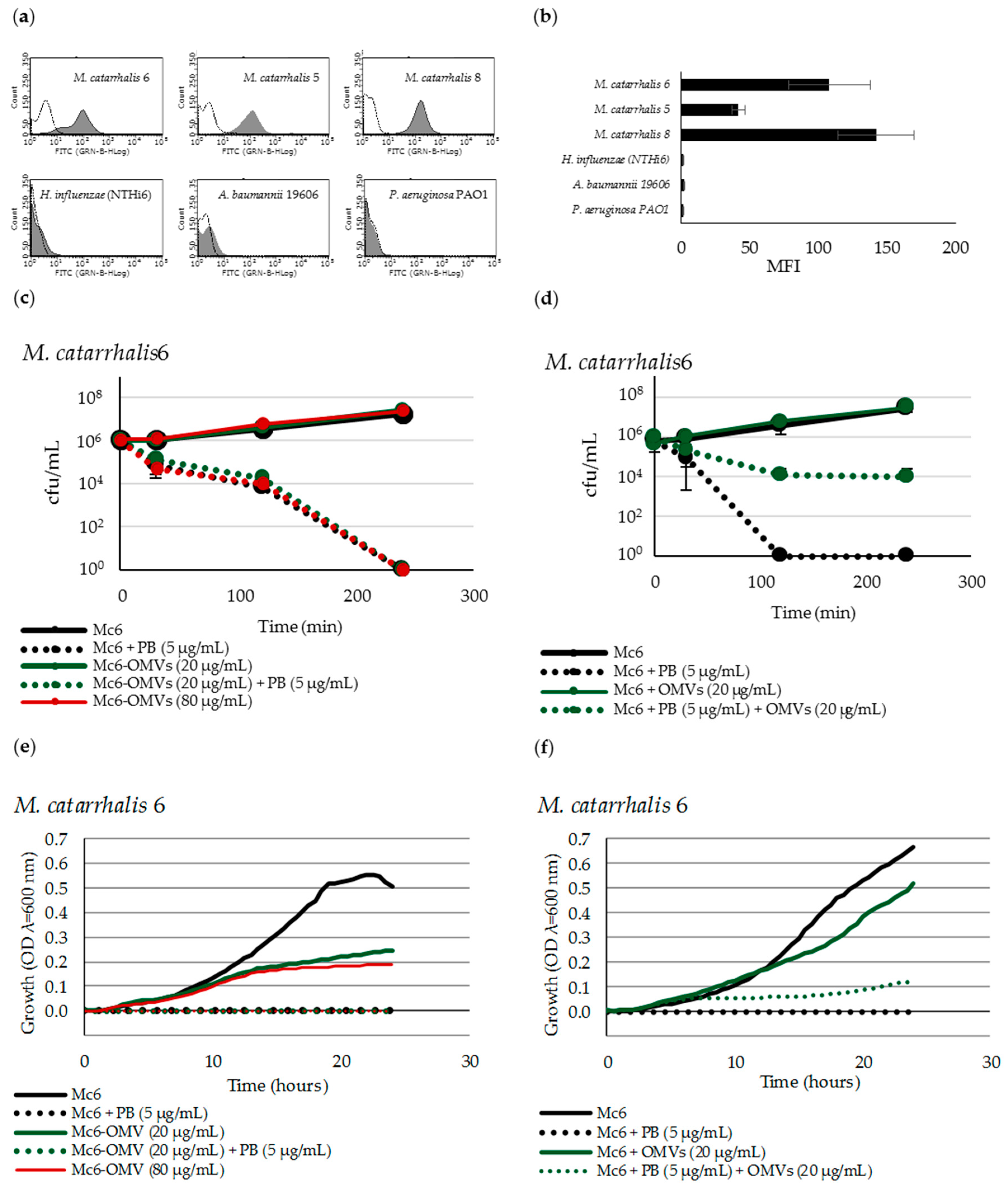
2.2. M. catarrhalis OMVs Passively Protect Cross-Pathogens against Polymyxin B-Dependent Killing
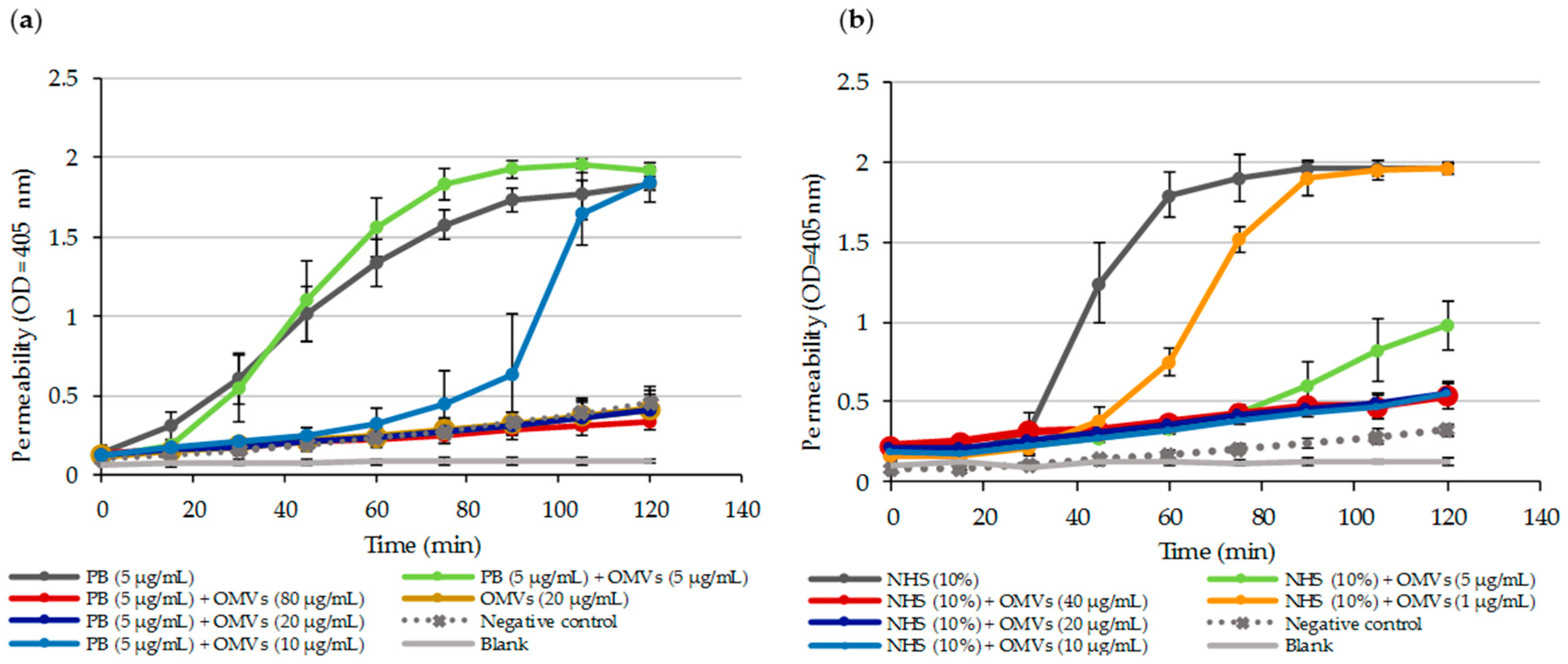
2.3. OMVs Protect Serum-Sensitive Strains and Accelerate Growth of Serum-Resistant Strains against Complement
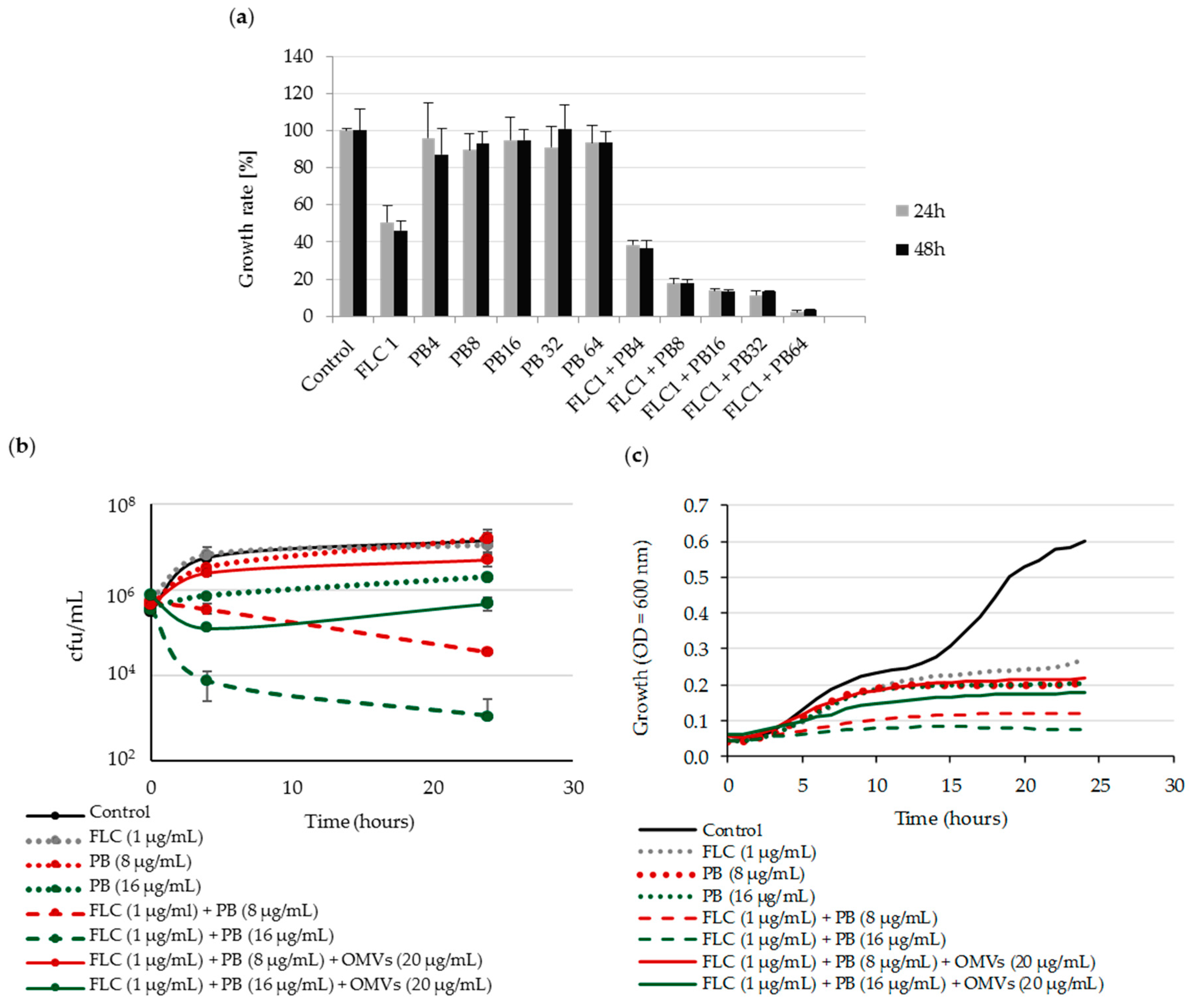
2.4. Mc 6 OMVs Passively Protect Pathogenic Yeasts against Polymyxin B-Dependent Fungicidal Effect in Combination with Fluconazole
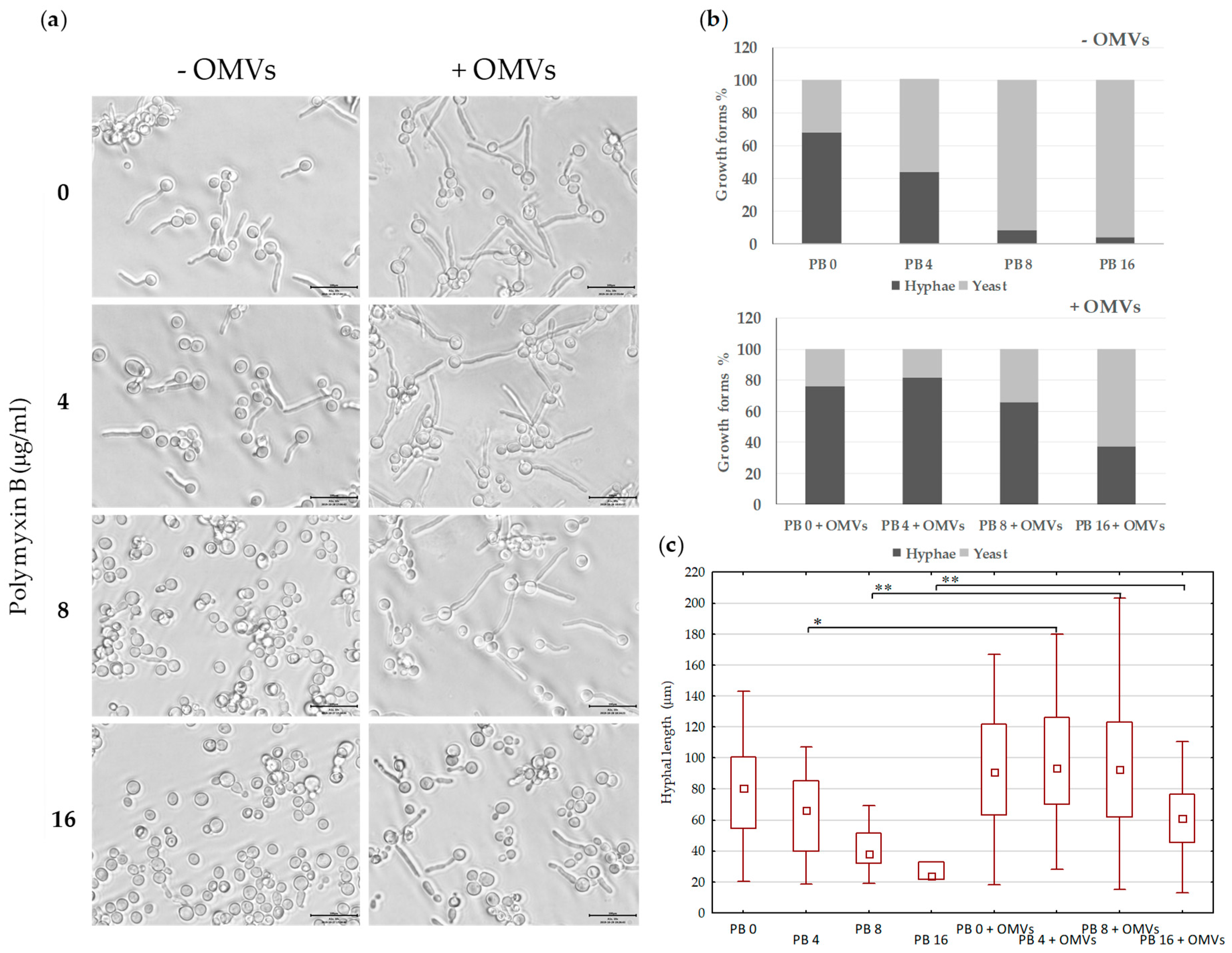
2.5. Mc6 OMVs Passively Enhance the Virulence of Pathogenic Yeasts Facilitating the Formation of Filaments
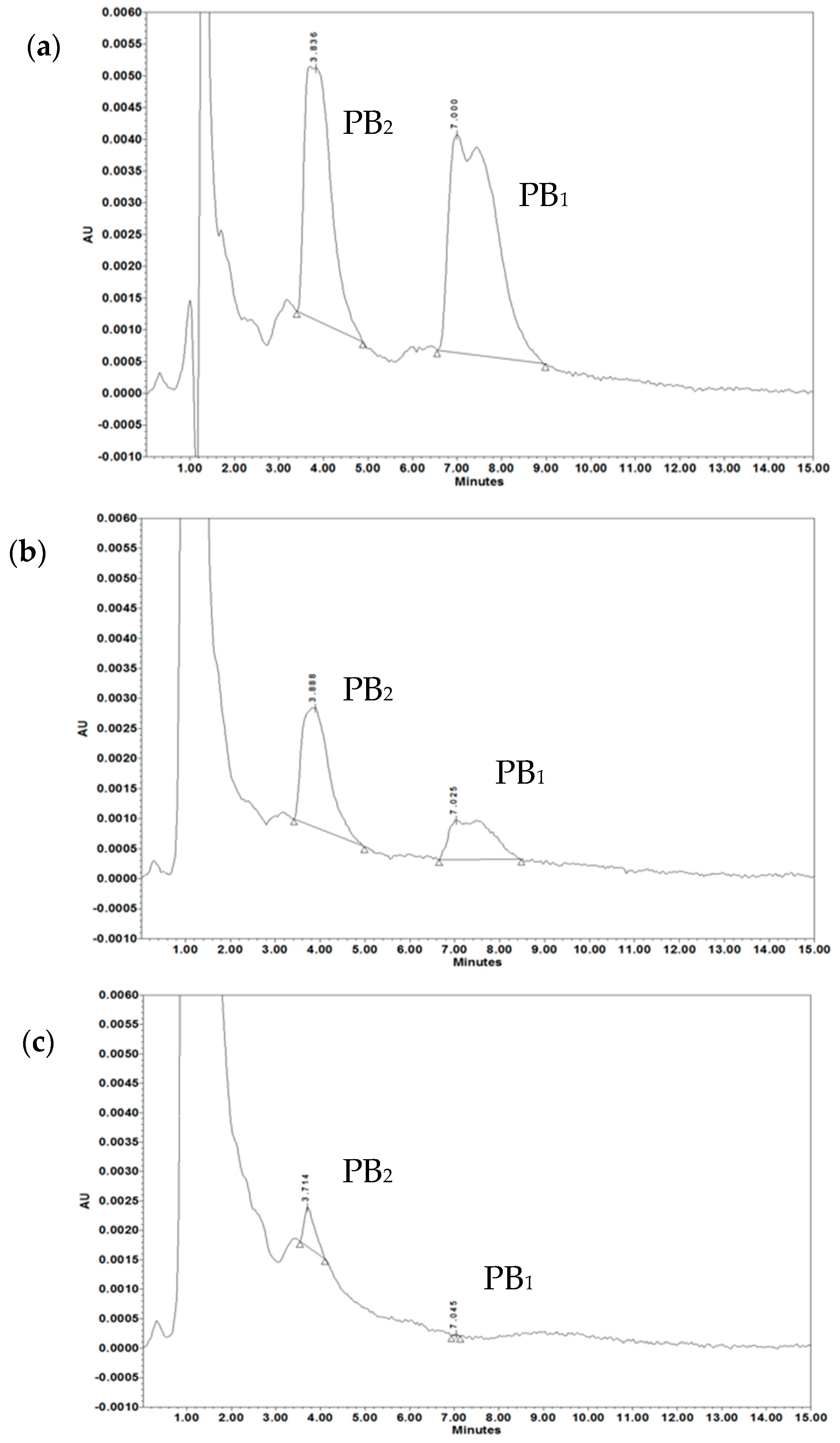
2.6. OMV-Dependent Protection against PB and Complement is the Result of PB Sequestration on Free OMVs
2.7. OMVs Associated with Bacteria do not Protect against PB
3. Discussion
4. Materials and Methods
4.1. Materials
4.1.1. Reagents
4.1.2. Microbial Strains and Growth Condition
4.2. Methods
4.2.1. Outer Membrane Vesicles Isolation
4.2.2. Time-Kill Assay
4.2.3. Growth Kinetics Assay
4.2.4. Checkerboard Microdilution Assay
4.2.5. Induction of Filamentation
4.2.6. Membrane Permeability Assay with ONPG
4.2.7. Complement Activation
4.2.8. OMV Association Assay and Flow Cytometry
4.2.9. Estimation of Zeta Potential
4.2.10. HPLC-UV System and Method
4.2.11. LOS-OMVs Electrophoresis
4.2.12. TEM
4.2.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AMP | Antimicrobial peptide |
| FBS | Fetal bovine serum |
| FLC | Fluconazole |
| HiNHS | Heat-inactivated normal human serum |
| LOS | Lipooligosaccharide |
| MIC | Minimal inhibitory concentration |
| NHS | Normal human serum |
| OMV | Outer membrane vesicle |
| ONPG | o-nitrophenyl-β-D-galactopyranoside |
| PB | Polymyxin B |
References
- Guerrero-Mandujano, A.; Hernández-Cortez, C.; Ibarra, J.A.; Castro-Escarpulli, G. The outer membrane vesicles: Secretion system type zero. Traffic 2017, 18, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Dusane, D.H.; Zinjarde, S.S.; Venugopalan, V.P.; McLean, R.J.C.; Weber, M.M.; Rahman, P.K.S.M. Quorum sensing: Implications on rhamnolipid biosurfactant production. Biotechnol. Genet. Eng. Rev. 2010, 27, 159–184. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T. Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: A Perspective Update. Front. Microbiol. 2017, 8, 1053. [Google Scholar] [CrossRef] [PubMed]
- Augustyniak, D.; Roszkowiak, J.; Wiśniewska, I.; Skała, J.; Gorczyca, D.; Drulis-Kawa, Z. Neuropeptides SP and CGRP Diminish the Moraxella catarrhalis Outer Membrane Vesicle- (OMV-) Triggered Inflammatory Response of Human A549 Epithelial Cells and Neutrophils. Mediat. Inflamm. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Bitto, N.J.; Chapman, R.; Pidot, S.; Costin, A.; Lo, C.; Choi, J.; D’Cruze, T.; Reynolds, E.C.; Dashper, S.G.; Turnbull, L.; et al. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Kulkarni, H.M.; Nagaraj, R.; Jagannadham, M.V. Protective role of E. coli outer membrane vesicles against antibiotics. Microbiol. Res. 2015, 181, 1–7. [Google Scholar] [CrossRef]
- Lee, J.; Lee, E.-Y.; Kim, S.-H.; Kim, D.-K.; Park, K.-S.; Kim, K.P.; Kim, Y.-K.; Roh, T.-Y.; Gho, Y.S. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob. Agents Chemother. 2013, 57, 2589–2595. [Google Scholar] [CrossRef]
- Manning, A.J.; Kuehn, M.J. Contribution of bacterial outer membrane vesicles to innate bacterial defense. Bmc Microbiol. 2011, 11, 258. [Google Scholar] [CrossRef]
- Rumbo, C.; Fernández-Moreira, E.; Merino, M.; Poza, M.; Mendez, J.A.; Soares, N.C.; Mosquera, A.; Chaves, F.; Bou, G. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: A new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3084–3090. [Google Scholar] [CrossRef]
- Chatterjee, S.; Mondal, A.; Mitra, S.; Basu, S. Acinetobacter baumannii transfers the blaNDM-1 gene via outer membrane vesicles. J. Antimicrob. Chemother. 2017, 72, 2201–2207. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K.; Jaganandham, M.V. Vesicles-mediated resistance to antibiotics in bacteria. Front. Microbiol. 2015, 6, 758. [Google Scholar] [CrossRef] [PubMed]
- Kadurugamuwa, J.L.; Mayer, A.; Messner, P.; Sára, M.; Sleytr, U.B.; Beveridge, T.J. S-layered Aneurinibacillus and Bacillus spp. are susceptible to the lytic action of Pseudomonas aeruginosa membrane vesicles. J. Bacteriol. 1998, 180, 2306–2311. [Google Scholar] [PubMed]
- Evans, A.G.L.; Davey, H.M.; Cookson, A.; Currinn, H.; Cooke-Fox, G.; Stanczyk, P.J.; Whitworth, D.E. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 2012, 158, 2742–2752. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, Y.; Yawata, Y.; Toyofuku, M.; Uchiyama, H.; Nomura, N. Interspecies Interaction between Pseudomonas aeruginosa and Other Microorganisms. Microbe. Environ. 2013, 28, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Schaar, V.; Nordström, T.; Mörgelin, M.; Riesbeck, K. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob. Agents Chemother. 2011, 55, 3845–3853. [Google Scholar] [CrossRef]
- Kim, S.W.; Park, S.B.; Im, S.P.; Lee, J.S.; Jung, J.W.; Gong, T.W.; Lazarte, J.M.S.; Kim, J.; Seo, J.-S.; Kim, J.-H.; et al. Outer membrane vesicles from β-lactam-resistant Escherichia coli enable the survival of β-lactam-susceptible E. coli in the presence of β-lactam antibiotics. Sci. Rep. 2018, 8, 5402. [Google Scholar] [CrossRef]
- Kulkarni, H.M.; Jagannadham, M.V. Biogenesis and multifaceted roles of outer membrane vesicles from Gram-negative bacteria. Microbiology 2014, 160, 2109–2121. [Google Scholar] [CrossRef]
- Joo, H.-S.; Fu, C.-I.; Otto, M. Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150292. [Google Scholar] [CrossRef]
- Tashiro, Y.; Ichikawa, S.; Shimizu, M.; Toyofuku, M.; Takaya, N.; Nakajima-Kambe, T.; Uchiyama, H.; Nomura, N. Variation of Physiochemical Properties and Cell Association Activity of Membrane Vesicles with Growth Phase in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2010, 76, 3732–3739. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Futamata, H.; Tashiro, Y. Complexities of cell-to-cell communication through membrane vesicles: Implications for selective interaction of membrane vesicles with microbial cells. Front. Microbiol. 2015, 6, 633. [Google Scholar] [CrossRef]
- Augustyniak, D.; Seredyński, R.; McClean, S.; Roszkowiak, J.; Roszniowski, B.; Smith, D.L.; Drulis-Kawa, Z.; Mackiewicz, P. Virulence factors of Moraxella catarrhalis outer membrane vesicles are major targets for cross-reactive antibodies and have adapted during evolution. Sci. Rep. 2018, 8, 4955. [Google Scholar] [CrossRef] [PubMed]
- Zhai, B.; Zhou, H.; Yang, L.; Zhang, J.; Jung, K.; Giam, C.Z.; Xiang, X.; Lin, X. Polymyxin B, in combination with fluconazole, exerts a potent fungicidal effect. J. Antimicrob. Chemother. 2010, 65, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Ha, K.C.; White, T.C. Effects of Azole Antifungal Drugs on the Transition from Yeast Cells to Hyphae in Susceptible and Resistant Isolates of the Pathogenic Yeast Candida albicans. Antimicrob. agents Chemother. 1999, 43, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Rogers, P.D.; Perlin, D.S. Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 2015, 5. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updat. 2016, 26, 43–57. [Google Scholar] [CrossRef]
- Tashiro, Y.; Hasegawa, Y.; Shintani, M.; Takaki, K.; Ohkuma, M.; Kimbara, K.; Futamata, H. Interaction of Bacterial Membrane Vesicles with Specific Species and Their Potential for Delivery to Target Cells. Front. Microbiol. 2017, 8, 571. [Google Scholar] [CrossRef]
- Yousfi, H.; Ranque, S.; Rolain, J.-M.; Bittar, F. In vitro polymyxin activity against clinical multidrug-resistant fungi. Antimicrob. Resist. Infect. Control. 2019, 8, 66. [Google Scholar] [CrossRef]
- Scorzoni, L.; de Paula e Silva, A.C.A.; Marcos, C.M.; Assato, P.A.; de Melo, W.C.M.A.; de Oliveira, H.C.; Costa-Orlandi, C.B.; Mendes-Giannini, M.J.S.; Fusco-Almeida, A.M. Antifungal Therapy: New Advances in the Understanding and Treatment of Mycosis. Front. Microbiol. 2017, 08, 36. [Google Scholar] [CrossRef]
- Kathiravan, M.K.; Salake, A.B.; Chothe, A.S.; Dudhe, P.B.; Watode, R.P.; Mukta, M.S.; Gadhwe, S. The biology and chemistry of antifungal agents: A review. Bioorg. Med. Chem. 2012, 20, 5678–5698. [Google Scholar] [CrossRef]
- Tatsumi, Y.; Nagashima, M.; Shibanushi, T.; Iwata, A.; Kangawa, Y.; Inui, F.; Siu, W.J.J.; Pillai, R.; Nishiyama, Y. Mechanism of action of efinaconazole, a novel triazole antifungal agent. Antimicrob. Agents Chemother. 2013, 57, 2405–2409. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.A.; Lewis, R.E.; Kontoyiannis, D.P. Direct effects of non-antifungal agents used in cancer chemotherapy and organ transplantation on the development and virulence of Candida and Aspergillus species. Virulence 2011, 2, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.-H.; Wang, H.-F.; Sun, P.-L.; Hu, F.-R.; Chen, Y.-L. The antibiotic polymyxin B exhibits novel antifungal activity against Fusarium species. Int. J. Antimicrob. Agents 2017, 49, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Villar, C.C.; Kashleva, H.; Dongari-Bagtzoglou, A. Role of Candida albicans polymorphism in interactions with oral epithelial cells. Oral Microbiol. Immunol. 2004, 19, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Murzyn, A.; Krasowska, A.; Augustyniak, D.; Majkowska-Skrobek, G.; Łukaszewicz, M.; Dziadkowiec, D. The effect of Saccharomyces boulardii on Candida albicans-infected human intestinal cell lines Caco-2 and Intestin 407. FEMS Microbiol. Lett. 2010, 310, 17–23. [Google Scholar] [CrossRef]
- Allison, D.L.; Willems, H.M.E.; Jayatilake, J.A.M.S.; Bruno, V.M.; Peters, B.M.; Shirtliff, M.E. Candida–Bacteria Interactions: Their Impact on Human Disease. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011, 19, 557–563. [Google Scholar] [CrossRef]
- Kerr, J.R.; Taylor, G.W.; Rutman, A.; Høiby, N.; Cole, P.J.; Wilson, R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 1999, 52, 385–387. [Google Scholar] [CrossRef]
- Gaddy, J.A.; Tomaras, A.P.; Actis, L.A. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 2009, 77, 3150–3160. [Google Scholar] [CrossRef]
- Krüger, W.; Vielreicher, S.; Kapitan, M.; Jacobsen, I.D.; Niemiec, M.J. Fungal-Bacterial Interactions in Health and Disease. Pathogens 2019, 8, 70. [Google Scholar] [CrossRef]
- Roux, D.; Gaudry, S.; Khoy-Ear, L.; Aloulou, M.; Phillips-Houlbracq, M.; Bex, J.; Skurnik, D.; Denamur, E.; Monteiro, R.C.; Dreyfuss, D.; et al. Airway Fungal Colonization Compromises the Immune System Allowing Bacterial Pneumonia to Prevail. Crit. Care Med. 2013, 41, e191–e199. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, M.P.; Rong, L. Human recombinant lactoferrin acts synergistically with antimicrobials commonly used in neonatal practice against coagulase-negative staphylococci and Candida albicans causing neonatal sepsis. J. Med. Microbiol. 2008, 57, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.M.; Gonçalves, S.; Santos, N.C. Defensins: Antifungal lessons from eukaryotes. Front. Microbiol. 2014, 5, 97. [Google Scholar] [CrossRef] [PubMed]
- Swidergall, M.; Ernst, J.F. Interplay between Candida albicans and the antimicrobial peptide armory. Eukaryot. Cell 2014, 13, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Goh, Y.H.; Goode, R.L. State of the Art Review: Current Status of Topical Nasal Antimicrobial Agents. Laryngoscope 2000, 110, 875–880. [Google Scholar] [CrossRef]
- Ragioto, D.A.M.T.; Carrasco, L.D.M.; Carmona-Ribeiro, A.M. Novel gramicidin formulations in cationic lipid as broad-spectrum microbicidal agents. Int. J. Nanomed. 2014, 9, 3183–3192. [Google Scholar]
- Tan, T.T.; Mörgelin, M.; Forsgren, A.; Riesbeck, K. Haemophilus influenzae Survival during Complement-Mediated Attacks Is Promoted by Moraxella catarrhalis Outer Membrane Vesicles. J. Infect. Dis. 2007, 195, 1661–1670. [Google Scholar] [CrossRef]
- Baumgarten, T.; Sperling, S.; Seifert, J.; von Bergen, M.; Steiniger, F.; Wick, L.Y.; Heipieper, H.J. Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl. Environ. Microbiol. 2012, 78, 6217–6224. [Google Scholar] [CrossRef]
- MacDonald, I.A.; Kuehna, M.J. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2971–2981. [Google Scholar] [CrossRef]
- Yun, S.H.; Park, E.C.; Lee, S.Y.; Lee, H.; Choi, C.W.; Yi, Y.S.; Ro, H.J.; Lee, J.C.; Jun, S.; Kim, H.Y.; et al. Antibiotic treatment modulates protein components of cytotoxic outer membrane vesicles of multidrug-resistant clinical strain, Acinetobacter baumannii DU202. Clin. Proteom. 2018, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Choi, K.H.; Kim, Y.S.; Hong, B.S.; Kim, O.Y.; Kim, J.H.; Yoon, C.M.; Koh, G.Y.; Kim, Y.K.; Gho, Y.S. Outer membrane vesicles derived from Escherichia coli induce systemic inflammatory response syndrome. PLoS ONE 2010, 5, e11334. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.; Sullivan, C.J.; Lonergan, N.E.; Stanley, S.; Soult, M.C.; Britt, L.D. Circulating bacterial membrane vesicles cause sepsis in rats. Shock 2012, 37, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Lee, J.; Jang, S.C.; Kim, S.R.; Jang, M.H.; Lötvall, J.; Kim, Y.K.; Gho, Y.S. Pulmonary inflammation induced by bacteria-free outer membrane vesicles from Pseudomonas aeruginosa. Am. J. Respir. Cell Mol. Biol. 2013, 49, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Namork, E.; Brandtzaeg, P. Fatal meningococcal septicaemia with “blebbing” meningococcus. Lancet 2002, 360, 1741. [Google Scholar] [CrossRef]
- Wayne, P.A. Reference method for broth dilution antifungal susceptibility testing of yeasts, Approved standard-second edition. Clsi Doc. M27-A2 2002. [Google Scholar]
- Augustyniak, D.; Jankowski, A.; Mackiewicz, P.; Skowyra, A.; Gutowicz, J.; Drulis-Kawa, Z. Innate immune properties of selected human neuropeptides against Moraxella catarrhalis and nontypeable Haemophilus influenzae. BMC Immunol. 2012, 13, 24. [Google Scholar] [CrossRef]
- Arabski, M.; Lisowska, H.; Lankoff, A.; Davydova, V.N.; Drulis-Kawa, Z.; Augustyniak, D.; Yermak, I.M.; Molinaro, A.; Kaca, W. The properties of chitosan complexes with smooth and rough forms of lipopolysaccharides on CHO-K1 cells. Carbohydr. Polym. 2013, 97, 284–292. [Google Scholar] [CrossRef]
- Orwa, J.A.; Van Gerven, A.; Roets, E.; Hoogmartens, J. Liquid chromatography of polymyxin B sulphate. J. Chromatogr. A 2000, 870, 237–243. [Google Scholar] [CrossRef]
- Inka, B. Glycobiology Protocols; Humana Press: Totowa, NJ, USA, 2006. [Google Scholar]
- Tsai, C.-M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
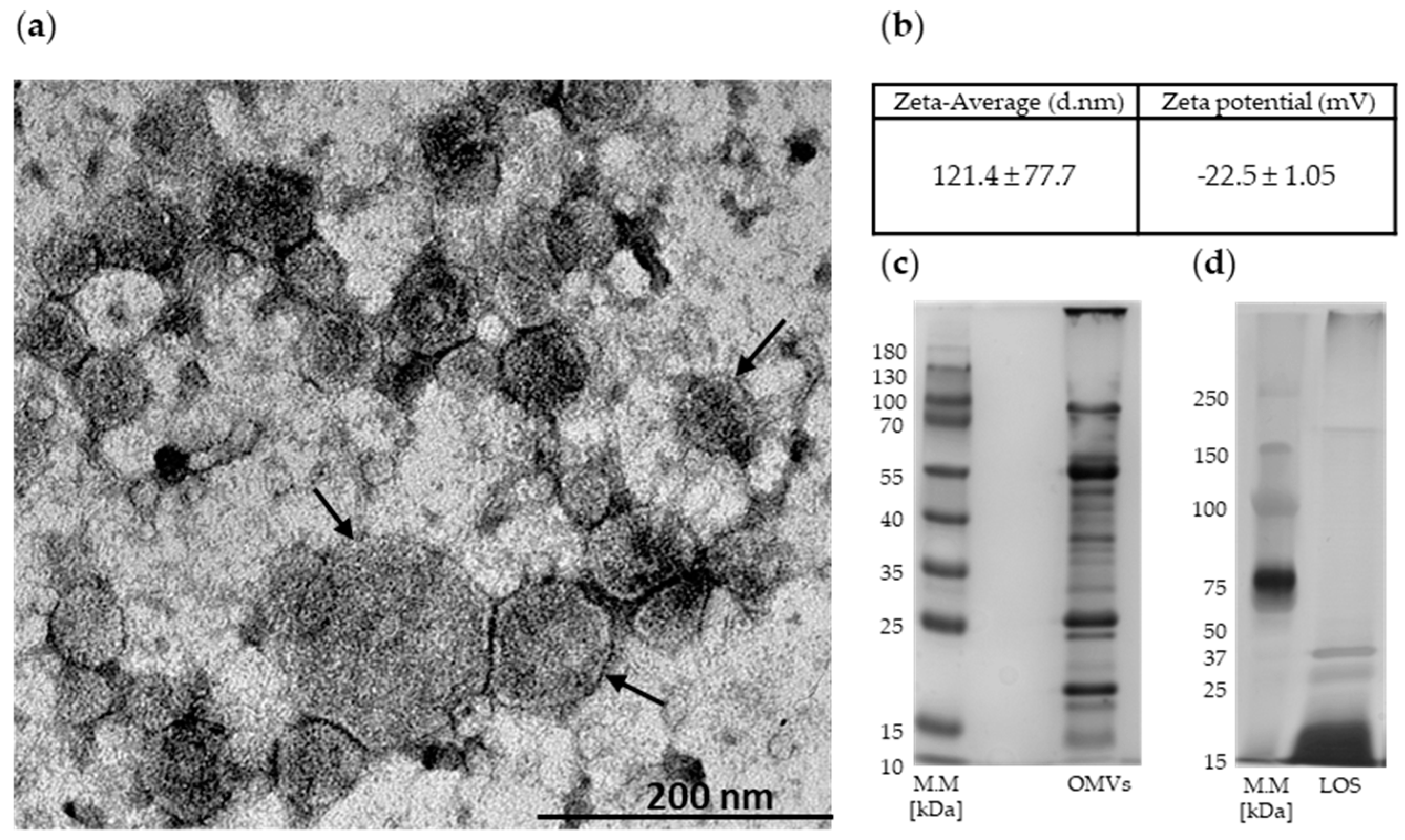
| PB Binding by OMVs in NaPB Buffer | PB Binding by OMVs in miliQ | ||
|---|---|---|---|
| Treatment | Zeta Potential (mV) | Treatment | Zeta Potential (mV) |
| 20 µg/mL OMVs (control) | −22.5 ± 1.05 | 20 µg/mL OMVs (control) | −24.7 ± 0.76 |
| 20 µg/mL OMVs + 5 µg/mL PB | −21.2 ± 1.06 | 20 µg/mL OMVs + 5 µg/mL PB | −25.9 ± 0.97 |
| 20 µg/mL OMVs + 50 µg/mL PB | −16.5 ± 0.76 * | 20 µg/mL OMVs + 50 µg/mL PB | −9.6 ± 0.35 * |
| 20 µg/mL OMVs + 250 µg/mL PB | −10.1 ± 0.46 * | 20 µg/mL OMVs + 250 µg/mL PB | −1.17 ± 0.14 * |
| Treatment | PB2 (µg/mL) | PB1 (µg/mL) | PB2 + PB1 (µg/mL) |
|---|---|---|---|
| Standard PB | 14.38 ± 0.48 | 17.45 ± 3.58 | 31.83 ± 3.47 |
| PB after treatment with 5 µg/mL OMVs | 7.76 ± 1.01 | 5.19 ± 1.05 | 13.67 ± 1.33 * |
| PB after treatment with 20 µg/mL OMVs | 0.96 ± 0.42 | 0.06 ± 0.01 | 1.02 ± 0.42 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roszkowiak, J.; Jajor, P.; Guła, G.; Gubernator, J.; Żak, A.; Drulis-Kawa, Z.; Augustyniak, D. Interspecies Outer Membrane Vesicles (OMVs) Modulate the Sensitivity of Pathogenic Bacteria and Pathogenic Yeasts to Cationic Peptides and Serum Complement. Int. J. Mol. Sci. 2019, 20, 5577. https://doi.org/10.3390/ijms20225577
Roszkowiak J, Jajor P, Guła G, Gubernator J, Żak A, Drulis-Kawa Z, Augustyniak D. Interspecies Outer Membrane Vesicles (OMVs) Modulate the Sensitivity of Pathogenic Bacteria and Pathogenic Yeasts to Cationic Peptides and Serum Complement. International Journal of Molecular Sciences. 2019; 20(22):5577. https://doi.org/10.3390/ijms20225577
Chicago/Turabian StyleRoszkowiak, Justyna, Paweł Jajor, Grzegorz Guła, Jerzy Gubernator, Andrzej Żak, Zuzanna Drulis-Kawa, and Daria Augustyniak. 2019. "Interspecies Outer Membrane Vesicles (OMVs) Modulate the Sensitivity of Pathogenic Bacteria and Pathogenic Yeasts to Cationic Peptides and Serum Complement" International Journal of Molecular Sciences 20, no. 22: 5577. https://doi.org/10.3390/ijms20225577
APA StyleRoszkowiak, J., Jajor, P., Guła, G., Gubernator, J., Żak, A., Drulis-Kawa, Z., & Augustyniak, D. (2019). Interspecies Outer Membrane Vesicles (OMVs) Modulate the Sensitivity of Pathogenic Bacteria and Pathogenic Yeasts to Cationic Peptides and Serum Complement. International Journal of Molecular Sciences, 20(22), 5577. https://doi.org/10.3390/ijms20225577






