Acute Liver Injury after CCl4 Administration Is Independent of Smad7 Expression in Myeloid Cells
Abstract
1. Introduction
2. Results
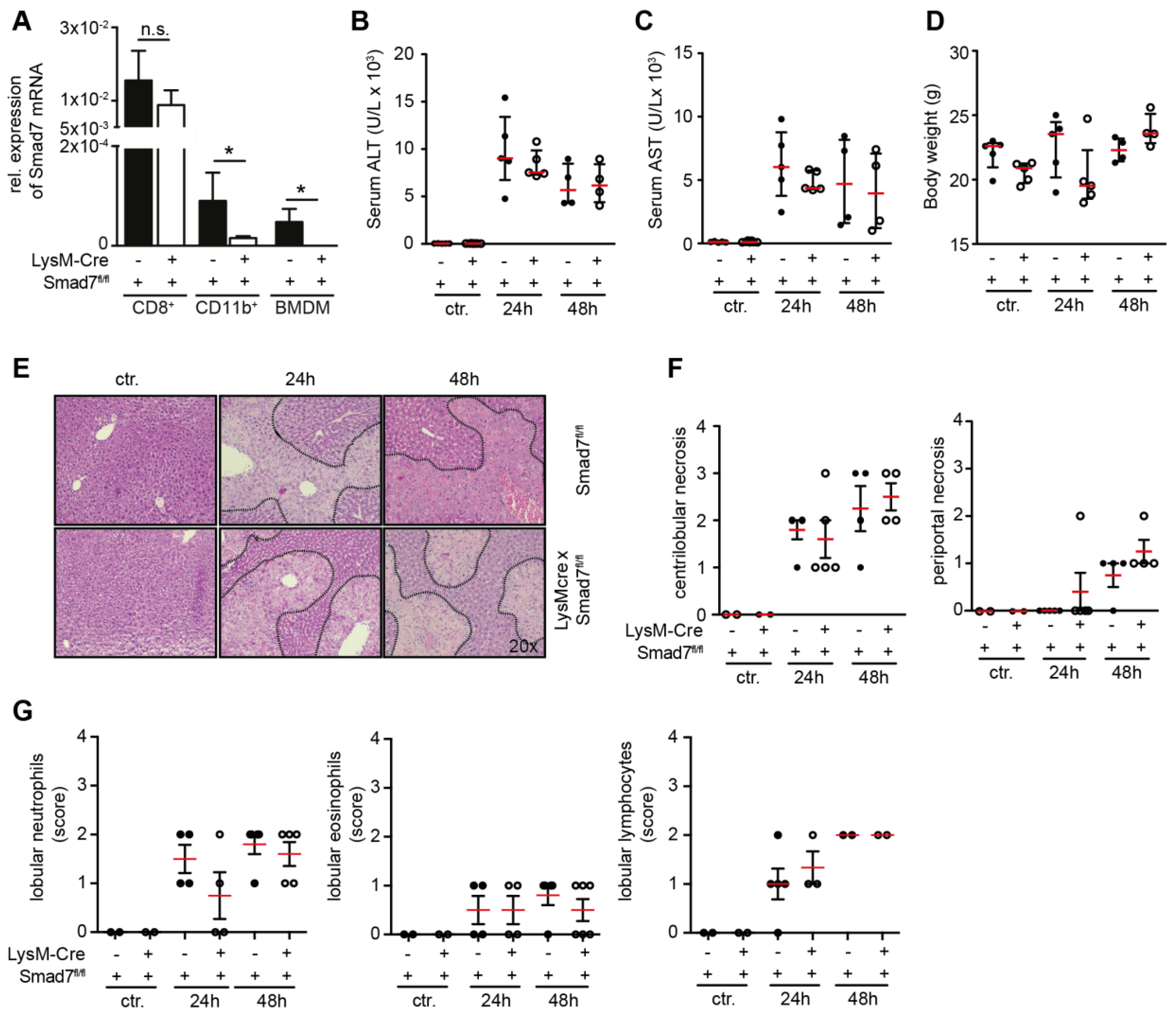
2.1. Liver Injury after CCl4 Administration is Similar in Myeloid-Specific Smad7 Knockdown and Wild-Type Controls
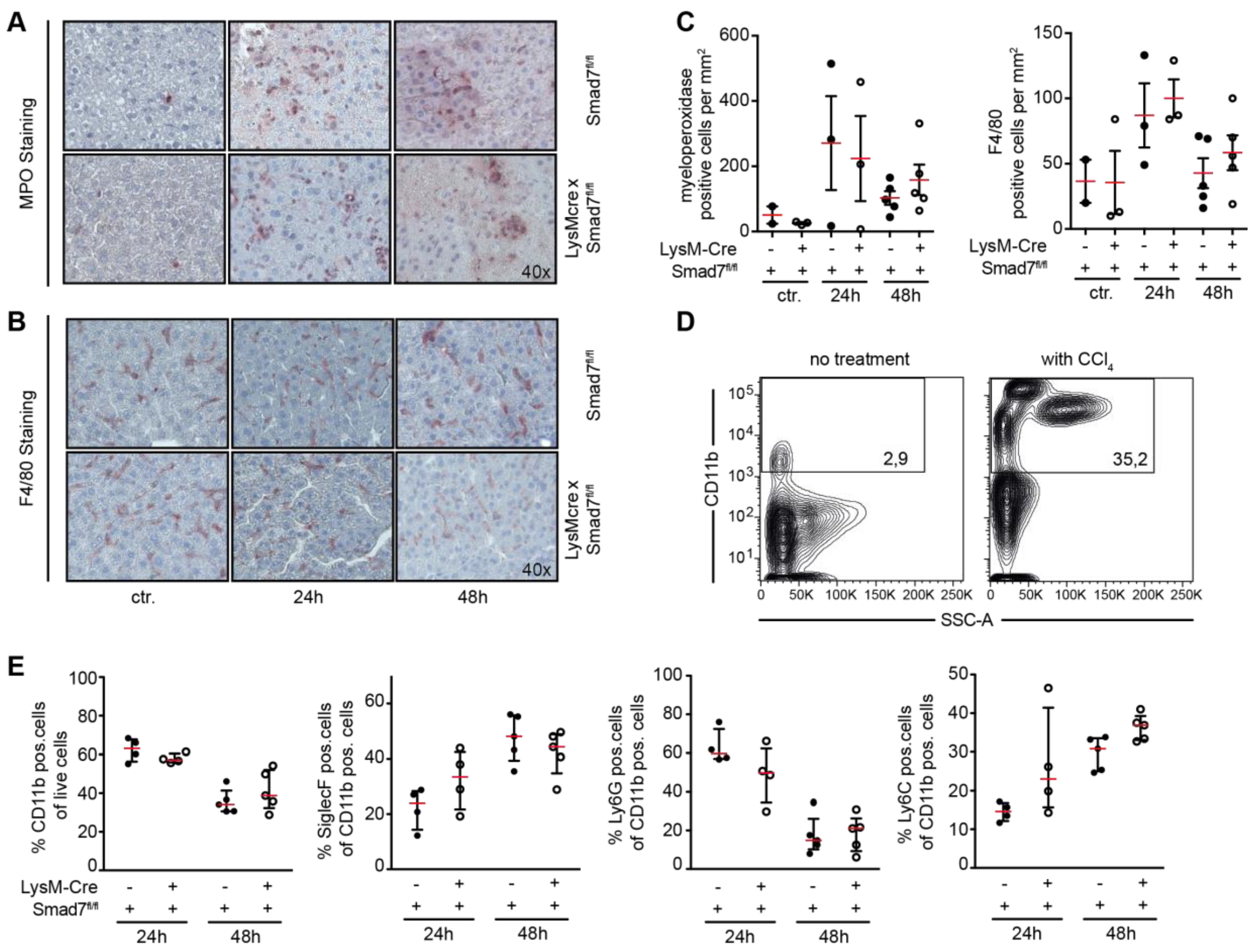
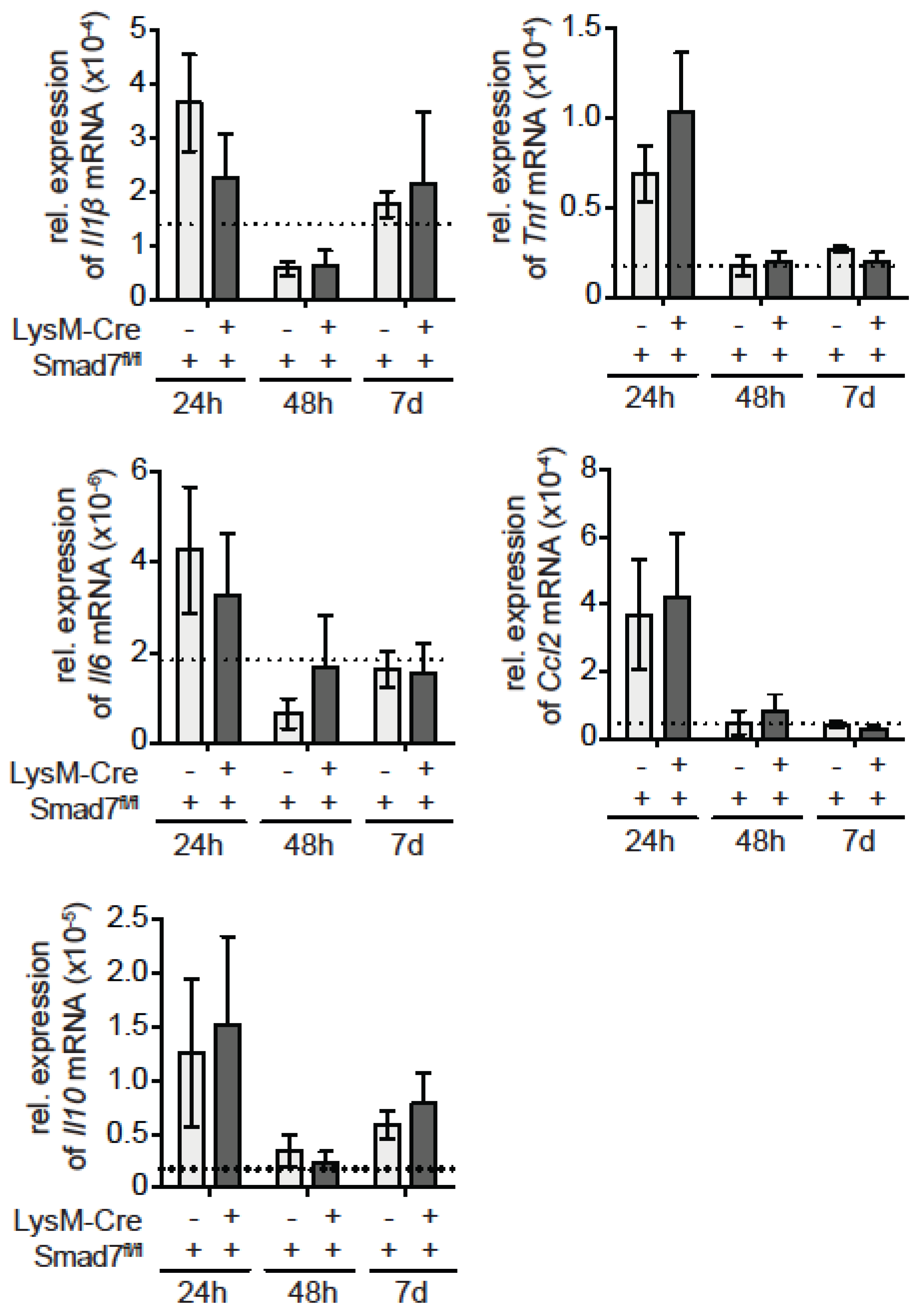
2.2. Neither Immune Cell Infiltration nor Inflammatory Gene Expression is Affected by the Loss of Smad7 Expression in Myeloid Cells after CCl4 Administration
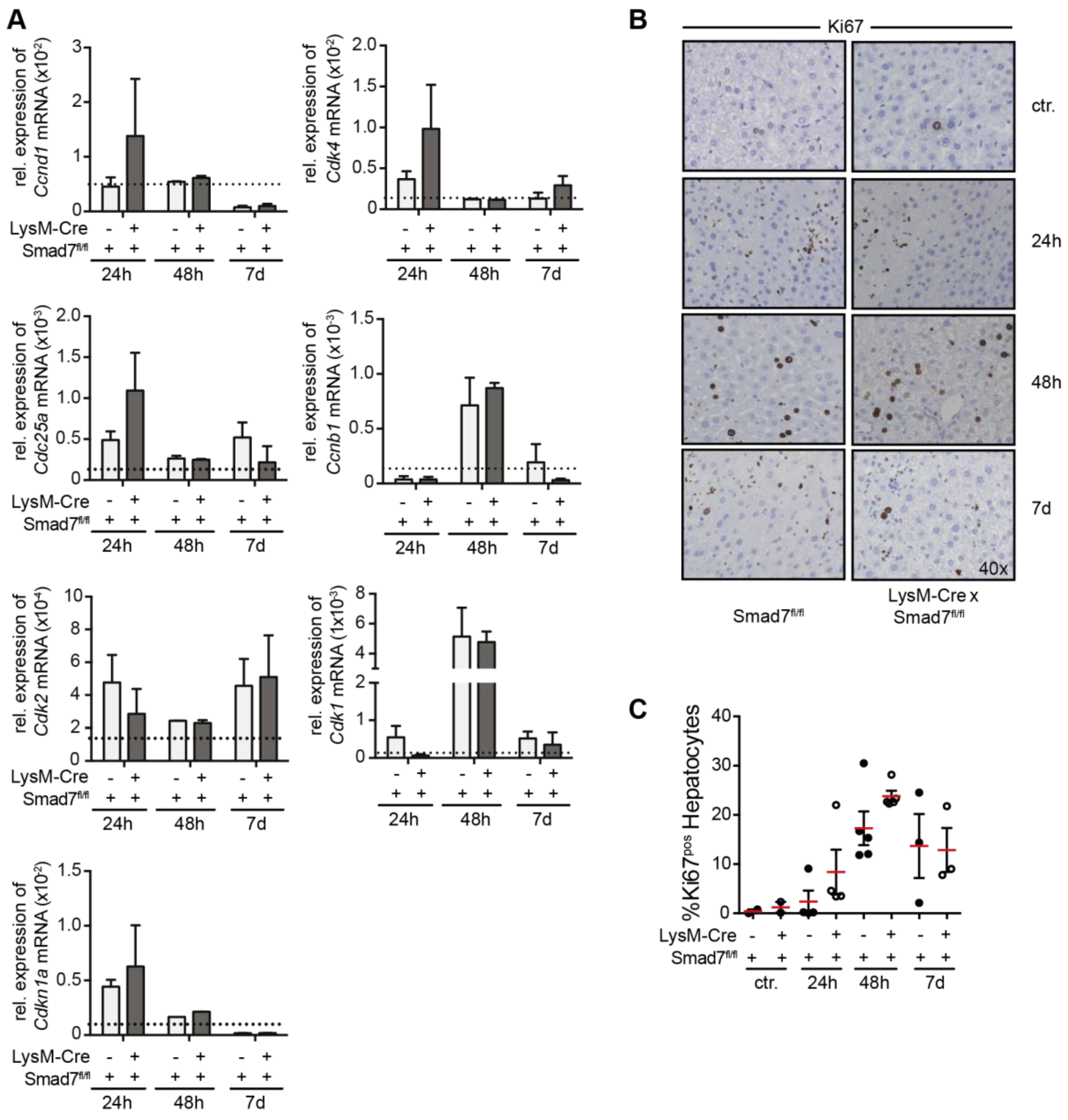
2.3. Smad7 Expressing Myeloid Cells Do not Affect the Regenerative Capacity of the Liver after CCl4 Mediated Injury
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Induction of Acute Hepatitis
4.3. Isolation of Non-Parenchymal Liver Cells
4.4. Fluorescence-Activated Cell Sorting
4.5. mRNA Isolation and Quantitative RT-PCR
4.6. Histology
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dooley, S.; ten Dijke, P. TGF-β in progression of liver disease. Cell Tissue Res. 2012, 347, 245–256. [Google Scholar] [CrossRef]
- Yan, X.; Liu, Z.; Chen, Y. Regulation of TGF- signaling by Smad7. Acta Biochim. Biophys. Sin. 2009, 41, 263–272. [Google Scholar] [CrossRef]
- Zhang, Y.; Alexander, P.B.; Wang, X.-F. TGF-β Family signaling in the control of cell proliferation and survival. Cold Spring Harb. Perspect. Biol. 2017, 9, a022145. [Google Scholar] [CrossRef] [PubMed]
- Puche, J.E.; Lee, Y.A.; Jiao, J.; Aloman, C.; Fiel, M.I.; Muñoz, U.; Kraus, T.; Lee, T.; Yee, H.F.; Friedman, S.L. A novel murine model to deplete hepatic stellate cells uncovers their role in amplifying liver damage in mice. Hepatology 2013, 57, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Duffield, J.S.; Forbes, S.J.; Constandinou, C.M.; Clay, S.; Partolina, M.; Vuthoori, S.; Wu, S.; Lang, R.; Iredale, J.P. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J. Clin. Investig. 2005, 115, 56–65. [Google Scholar] [CrossRef]
- Karkampouna, S.; Goumans, M.-J.; ten Dijke, P.; Dooley, S.; Kruithof-de Julio, M. Inhibition of TGFβ type I receptor activity facilitates liver regeneration upon acute CCl4 intoxication in mice. Arch. Toxicol. 2016, 90, 347–357. [Google Scholar] [CrossRef]
- Zhu, L.; Wang, L.; Wang, X.; Luo, X.; Yang, L.; Zhang, R.; Yin, H.; Xie, D.; Pan, Y.; Chen, Y. Hepatic deletion of Smad7 in mouse leads to spontaneous liver dysfunction and aggravates alcoholic liver injury. PLoS ONE 2011, 6, e17415. [Google Scholar] [CrossRef]
- Hamzavi, J.; Ehnert, S.; Godoy, P.; Ciuclan, L.; Weng, H.; Mertens, P.R.; Heuchel, R.; Dooley, S. Disruption of the Smad7 gene enhances CCI4-dependent liver damage and fibrogenesis in mice. J. Cell. Mol. Med. 2008, 12, 2130–2144. [Google Scholar] [CrossRef]
- Feng, T.; Dzieran, J.; Yuan, X.; Dropmann, A.; Maass, T.; Teufel, A.; Marhenke, S.; Gaiser, T.; Rückert, F.; Kleiter, I.; et al. Hepatocyte-specific Smad7 deletion accelerates DEN-induced HCC via activation of STAT3 signaling in mice. Oncogenesis 2017, 6, e294. [Google Scholar] [CrossRef] [PubMed]
- Dooley, S.; Hamzavi, J.; Ciuclan, L.; Godoy, P.; Ilkavets, I.; Ehnert, S.; Ueberham, E.; Gebhardt, R.; Kanzler, S.; Geier, A.; et al. Hepatocyte-specific Smad7 expression attenuates TGF-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology 2008, 135, 642–659. [Google Scholar] [CrossRef] [PubMed]
- MohanKumar, K.; Namachivayam, K.; Chapalamadugu, K.C.; Garzon, S.A.; Premkumar, M.H.; Tipparaju, S.M.; Maheshwari, A. Smad7 interrupts TGF-β signaling in intestinal macrophages and promotes inflammatory activation of these cells during necrotizing enterocolitis. Pediatr. Res. 2016, 79, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F. Current and emerging therapeutic targets for IBD. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, I.; Moreno-Càceres, J.; Sánchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; ten Dijke, P. IT-LIVER Consortium TGF-β signaling and liver disease. FEBS J. 2016, 283, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Kleiter, I.; Song, J.; Lukas, D.; Hasan, M.; Neumann, B.; Croxford, A.L.; Pedre, X.; Hovelmeyer, N.; Yogev, N.; Mildner, A.; et al. Smad7 in T cells drives T helper 1 responses in multiple sclerosis and experimental autoimmune encephalomyelitis | Brain. Brain 2010, 133, 1067–1081. [Google Scholar] [CrossRef]
- Manibusan, M.K.; Odin, M.; Eastmond, D.A. Postulated carbon tetrachloride mode of action: A review. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2007, 25, 185–209. [Google Scholar] [CrossRef]
- Liedtke, C.; Luedde, T.; Sauerbruch, T.; Scholten, D.; Streetz, K.; Tacke, F.; Tolba, R.; Trautwein, C.; Trebicka, J.; Weiskirchen, R. Experimental liver fibrosis research: Update on animal models, legal issues and translational aspects. Fibrogenesis Tissue Repair 2013, 6, 19. [Google Scholar] [CrossRef]
- Gong, D.; Shi, W.; Yi, S.-J.; Chen, H.; Groffen, J.; Heisterkamp, N. TGFβ signaling plays a critical role in promoting alternative macrophage activation. BMC Immunol. 2012, 13, 31. [Google Scholar] [CrossRef]
- Troncone, E.; Marafini, I.; Stolfi, C.; Monteleone, G. Transforming Growth Factor-β1/Smad7 in Intestinal Immunity, Inflammation, and Cancer. Front. Immunol. 2018, 9, 1407. [Google Scholar] [CrossRef]
- Sedda, S.; De Simone, V.; Marafini, I.; Bevivino, G.; Izzo, R.; Paoluzi, O.A.; Colantoni, A.; Ortenzi, A.; Giuffrida, P.; Corazza, G.R.; et al. High Smad7 sustains inflammatory cytokine response in refractory coeliac disease. Immunology 2016, 150, 356–363. [Google Scholar] [CrossRef]
- Ka, S.-M.; Huang, X.-R.; Lan, H.Y.; Tsai, P.-Y.; Yang, S.-M.; Shui, H.-A.; Chen, A. Smad7 gene therapy ameliorates an autoimmune crescentic glomerulonephritis in mice. J. Am. Soc. Nephrol. 2007, 18, 1777–1788. [Google Scholar] [CrossRef]
- Rizzo, A.; Waldner, M.J.; Stolfi, C.; Sarra, M.; Fina, D.; Becker, C.; Neurath, M.F.; MacDonald, T.T.; Pallone, F.; Monteleone, G.; et al. Smad7 expression in T cells prevents colitis-associated cancer. Cancer Res. 2011, 71, 7423–7432. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Endig, J.; Unrau, L.; Sprezyna, P.; Rading, S.; Karsak, M.; Goltz, D.; Heukamp, L.C.; Tiegs, G.; Diehl, L. Acute Liver Injury after CCl4 Administration Is Independent of Smad7 Expression in Myeloid Cells. Int. J. Mol. Sci. 2019, 20, 5528. https://doi.org/10.3390/ijms20225528
Endig J, Unrau L, Sprezyna P, Rading S, Karsak M, Goltz D, Heukamp LC, Tiegs G, Diehl L. Acute Liver Injury after CCl4 Administration Is Independent of Smad7 Expression in Myeloid Cells. International Journal of Molecular Sciences. 2019; 20(22):5528. https://doi.org/10.3390/ijms20225528
Chicago/Turabian StyleEndig, Jessica, Ludmilla Unrau, Paulina Sprezyna, Sebasting Rading, Meliha Karsak, Diane Goltz, Lukas C. Heukamp, Gisa Tiegs, and Linda Diehl. 2019. "Acute Liver Injury after CCl4 Administration Is Independent of Smad7 Expression in Myeloid Cells" International Journal of Molecular Sciences 20, no. 22: 5528. https://doi.org/10.3390/ijms20225528
APA StyleEndig, J., Unrau, L., Sprezyna, P., Rading, S., Karsak, M., Goltz, D., Heukamp, L. C., Tiegs, G., & Diehl, L. (2019). Acute Liver Injury after CCl4 Administration Is Independent of Smad7 Expression in Myeloid Cells. International Journal of Molecular Sciences, 20(22), 5528. https://doi.org/10.3390/ijms20225528



