Allelic Variants for Candidate Nitrogen Fixation Genes Revealed by Sequencing in Red Clover (Trifolium pratense L.)
Abstract
1. Introduction
2. Results
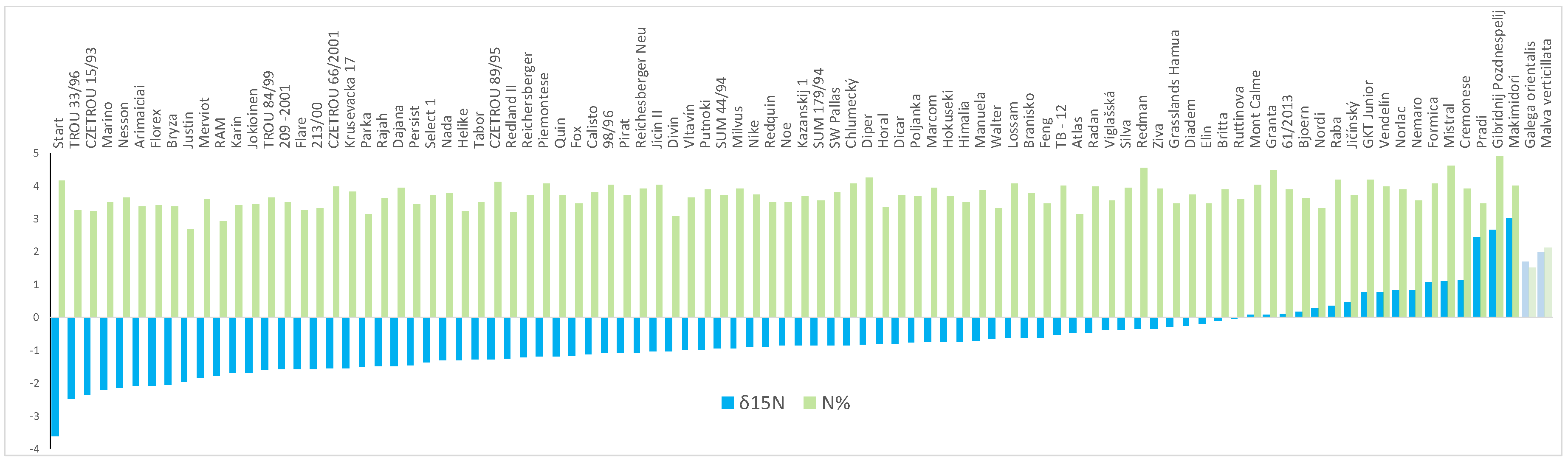
2.1. Nitrogen Fixation Assays
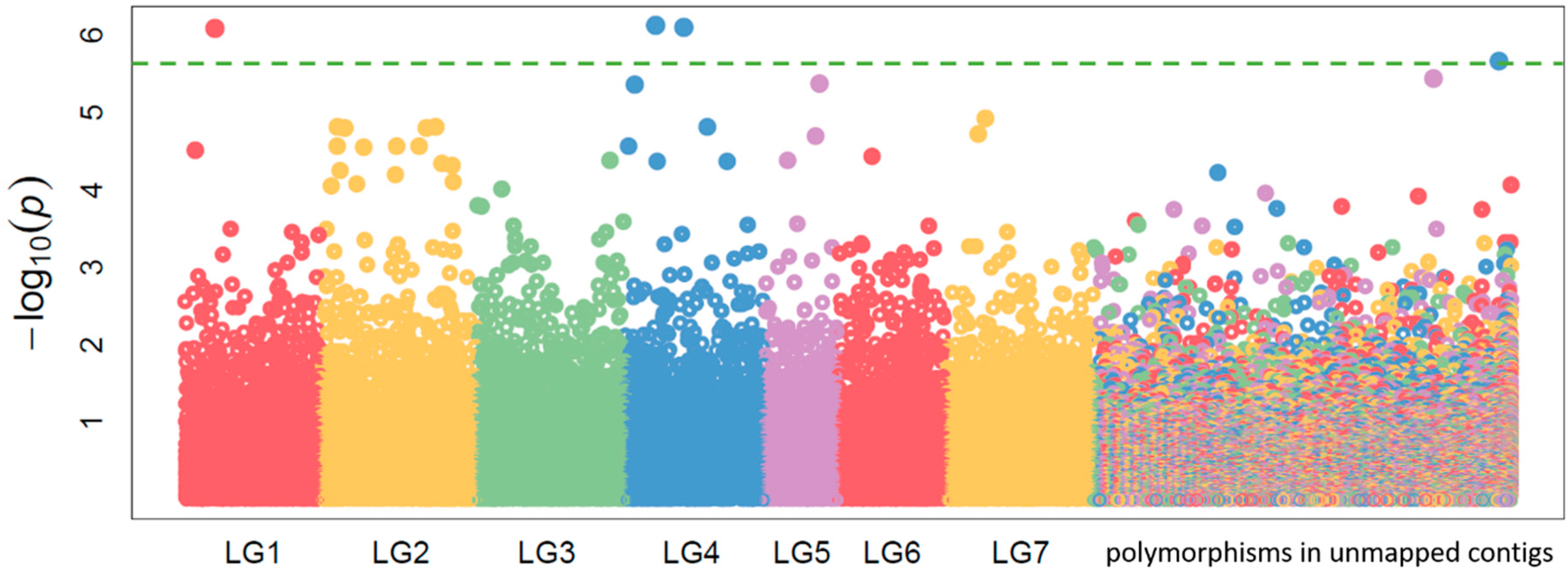
2.2. Candidate Gene Target Sequencing
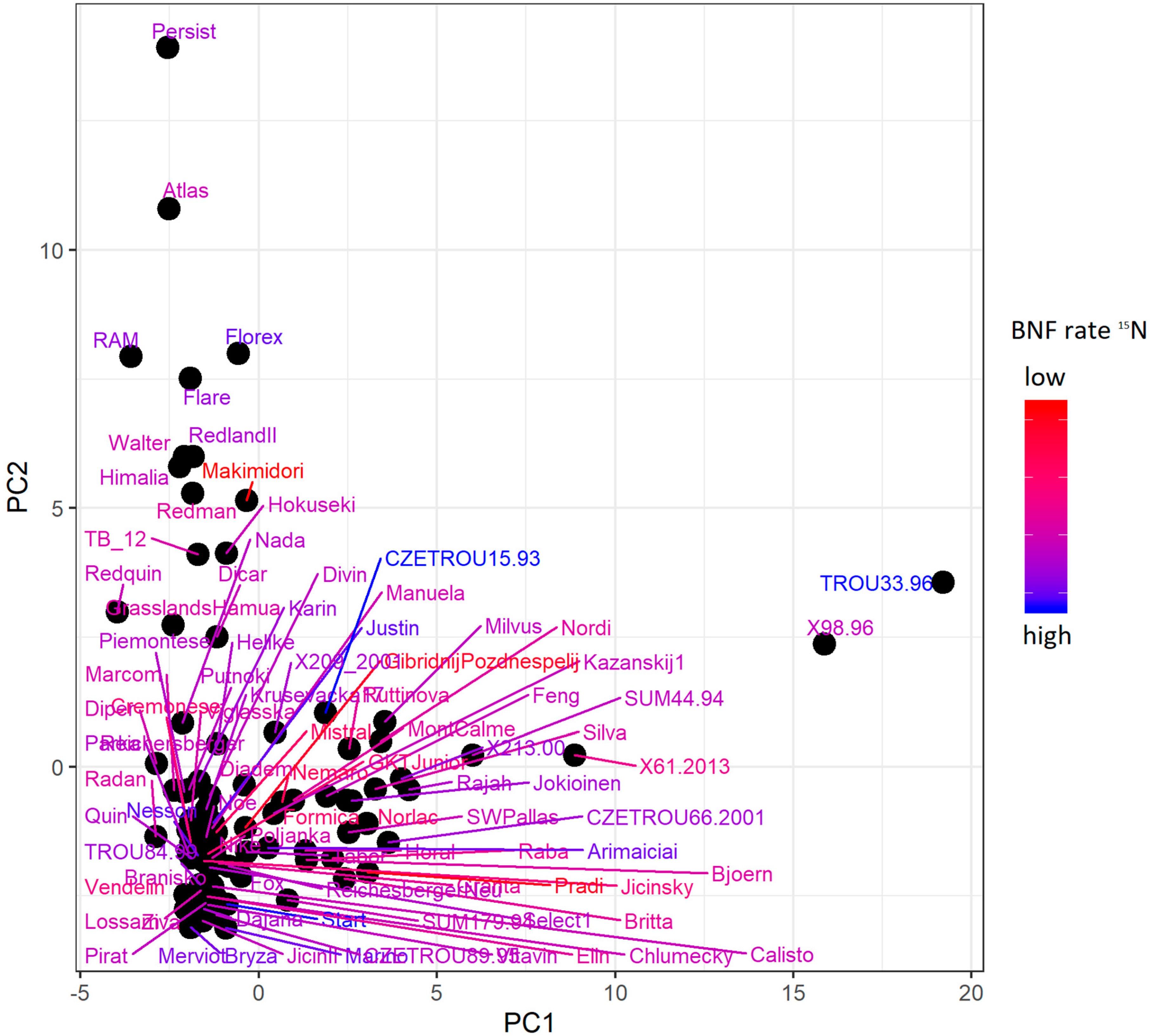
2.3. ddRADseq and N Isotopic Composition
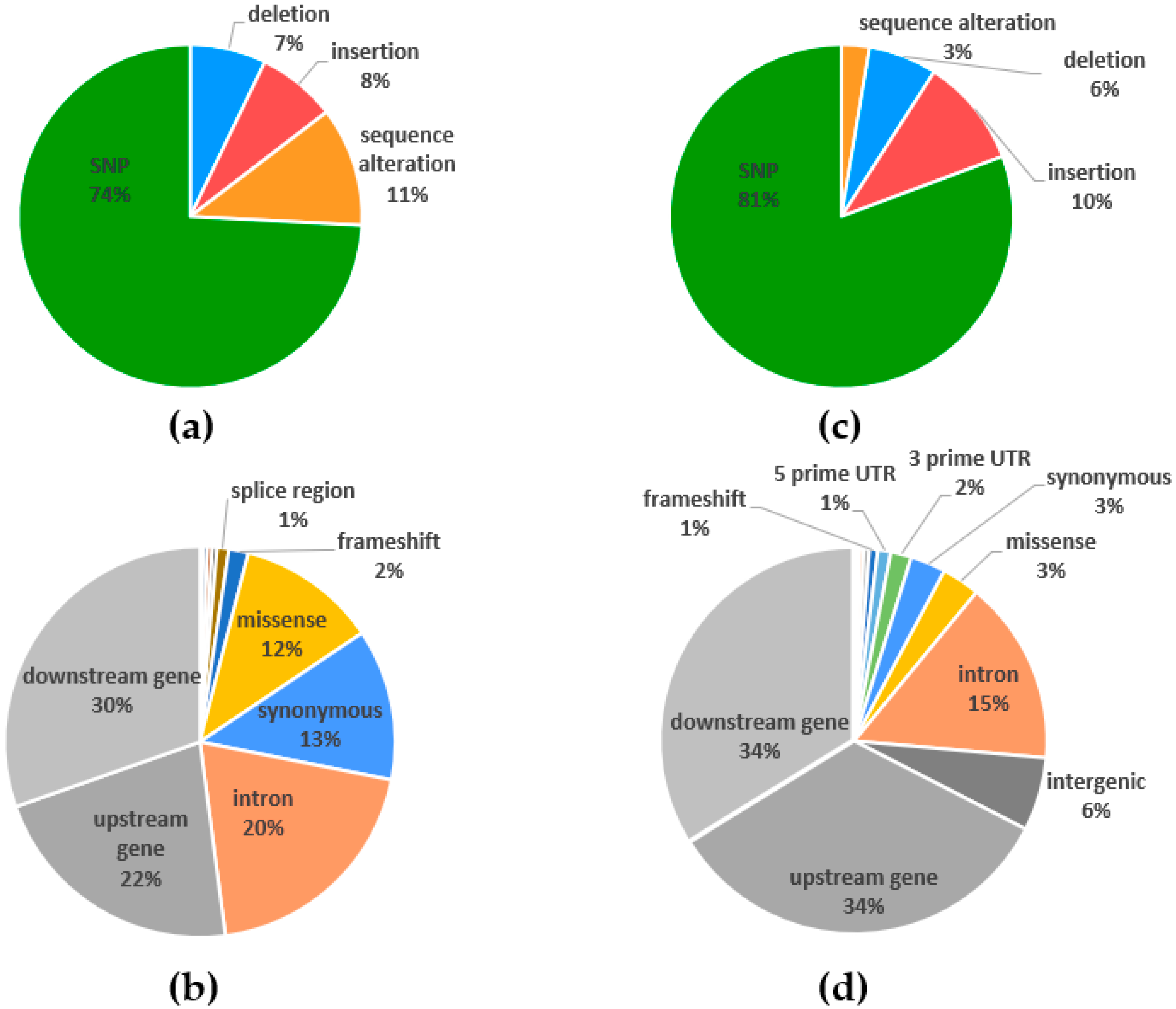
2.4. Polymorphism Annotations
2.5. Validation of Selected InDel Polymorphisms
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Growth Conditions and Evaluation of Nitrogen Fixation by Acetylene Reduction Assay
4.3. Evaluation of Nitrogen Fixation by Natural 15N-Abundance Method
4.4. Selection of Candidate Genes and Procedure of Targeted Sequencing
4.5. ddRADseq Library Preparation and Sequence Processing of T. pratense Population Set
4.6. Bioinformatic Analysis
4.7. Statistical Analysis
4.8. Validation of Selected InDel Polymorphisms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AP2 | APETALA2 |
| ARA | Acetylene reduction assay |
| ATP | Adenosine triphosphate |
| BNF | Biological fixation of atmospheric nitrogen |
| CE | Ethylene molar concentration |
| CRE | Cytokinin response 1 |
| CTAB | Cetyl trimethylammonium bromide |
| ddRADseq | Double-digest RAD sequencing |
| DNF2 | Defective in nitrogen fixation 2 |
| EFD | Ethylene response factor required for nodule differentiation |
| ERF | Ethylene response factor |
| ERF3 | Ethylene-responsive transcription factor 3 |
| FLOT | Flotillin |
| GATK | Genome Analysis Toolkit |
| Hs | Expected heterozygosity |
| InDel | Insertion/deletions |
| LG | Linkage group |
| MOT1 | Molybdate transporter 1 |
| ModABC | ATP-binding cassette transporter involved in molybdate transport |
| Mt | Medicago truncatula |
| NFP | Nod factor perception |
| NF-YC2 | Nuclear transcription factor Y subunit C2 |
| NGS | Next-generation sequencing |
| Nod factors | Nodulation factors |
| PC | Principal component |
| PCA | Principal component analysis |
| PNO1 | partner of NOB1-like |
| RLK | Receptor-like kinases |
| RR2 | Cytokinin responsive gene |
| SeqCap | Hybridization-based sequence capture |
| SNP | Single-nucleotide polymorphism |
| SST | Symbiotic sulfate transporter |
| Tp | Trifolium pratense |
| VEP | Variant effect predictor |
| WOX11 | WUSCHEL-related homeobox gene |
References
- Zohary, M.; Heller, D. The genus Trifolium; Israel Academy of Sciences and Humanities: Jerusalem, Israel, 1984. [Google Scholar]
- Gillett, J.M.; Taylor, N.L. The World of Clovers; Iowa State University Press: Ames, Iowa, IA, USA, 2001. [Google Scholar]
- Ellison, N.W.; Liston, A.; Steiner, J.J.; Williams, W.M.; Taylor, N.L. Molecular phylogenetics of the clover genus (Trifolium-Leguminosae). Mol. Phylogenet. Evol. 2006, 39, 688–705. [Google Scholar] [CrossRef] [PubMed]
- Kintl, A.; Elbl, J.; Lošák, T.; Vaverková, M.D.; Nedělník, J. Mixed intercropping of wheat and white clover to enhance the sustainability of the conventional cropping system: Effects on biomass production and leaching of mineral nitrogen. Sustain 2018, 10, 3367. [Google Scholar] [CrossRef]
- Hauer, R.F.; Lamberti, G.A. Methods in Stream Ecology: Volume 1: Ecosystem Structure; Academic Press: Cambridge, MA, USA, 2017; ISBN 9780124165786. [Google Scholar]
- Lerouge, P. Symbiotic host specificity between leguminous plants and rhizobia is determined by substituted and acylated glucosamine oligosaccharide signals. Glycobiology 1994, 4, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Luna, R.; Planchon, C. Genotype x Bradyrhizobium japonicum strain interactions in dinitrogen fixation and agronomic traits of soybean (Glycine max L. Merr.). Euphytica 1995, 86, 127–134. [Google Scholar] [CrossRef]
- Kouchi, H.; Imaizumi-Anraku, H.; Hayashi, M.; Hakoyama, T.; Nakagawa, T.; Umehara, Y.; Suganuma, N.; Kawaguchi, M. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010, 51, 1381–1397. [Google Scholar] [CrossRef]
- Carlsson, G.; Huss-Danell, K. Nitrogen fixation in perennial forage legumes in the field. Plant Soil 2003, 253, 353–372. [Google Scholar] [CrossRef]
- Provorov, N.A.; Tikhonovich, I.A. Genetic resources for improving nitrogen fixation in legume-rhizobia symbiosis. Genet. Resour. Crop. Evol. 2003, 50, 89–99. [Google Scholar] [CrossRef]
- Freiberg, C.; Fellay, R.; Bairoch, A.; Broughton, W.J.; Rosenthal, A.; Perret, X. Molecular basis of symbiosis between Rhizobium and legumes. Nature 1997, 387, 394–401. [Google Scholar] [CrossRef]
- Catoira, R.; Galera, C.; De Billy, F.; Penmetsa, R.V.; Journet, E.P.; Maillet, F.; Rosenberg, C.; Cook, D.; Gough, C.; Denarie, J. Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 2000, 12, 1647–1665. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Long, S.R. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in nod factor signaling. Plant Physiol. 2003, 131, 1027–1032. [Google Scholar] [CrossRef]
- Gleason, C.; Yang, T. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition Article W342F Mutation in CCaMK Enhances Its Affinity to Calmodulin But Compromises Its Role in Supporting Root Nodule Symbiosis in Medicago truncatula View pr. Nature 2006, 441, 1149–1152. [Google Scholar] [CrossRef] [PubMed]
- Delves, A.C.; Mathews2, A.; Day, D.A.; Carter, A.S.; Carroll, B.J.; Gresshoff, P.M. Regulation of the Soybean-Rhizobium Nodule Symbiosis by Shoot and Root Factors’. Plant Physiol. 1986, 82, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anolles, G.; Gresshoff, P.M. Plant Genetic Control of Nodulation. Annu. Rev. Microbiol. 1991, 45, 345–382. [Google Scholar] [CrossRef] [PubMed]
- Gage, D.J. 2004 Infection and Invasion of Roots by Symbiotic, Nitrogen-Fixing Rhizobia during Nodulation of Temperate Legumes. Microbiol. Mol. Biol. Rev. 2017, 68, 203. [Google Scholar]
- Marchal, K.; Vanderleyden, J. The “oxygen paradox” of dinitrogen-fixing bacteria. Biol. Fertil. Soils 2000, 30, 363–373. [Google Scholar] [CrossRef]
- Kundu, S.; Trent, J.T.; Hargrove, M.S. Plants, humans and hemoglobins. Trends Plant Sci. 2003, 8, 387–393. [Google Scholar] [CrossRef]
- Penmetsa, R.V.; Cook, D.R. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 1997, 275, 527–530. [Google Scholar] [CrossRef]
- Mitra, R.M.; Gleason, C.A.; Edwards, A.; Hadfield, J.; Downie, J.A.; Oldroyd, G.E.D.; Long, S.R. A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 2004, 101, 4701–4705. [Google Scholar] [CrossRef]
- Domonkos, A.; Horvath, B.; Marsh, J.F.; Halasz, G.; Ayaydin, F.; Oldroyd, G.E.D.; Kalo, P. The identification of novel loci required for appropriate nodule development in Medicago truncatula. BMC Plant Biol. 2013, 13, 157. [Google Scholar] [CrossRef]
- Kang, Y.; Li, M.; Sinharoy, S.; Verdier, J. A snapshot of functional genetic studies in Medicago truncatula. Front. Plant Sci. 2016, 7, 1175. [Google Scholar] [CrossRef][Green Version]
- Veerappan, V.; Jani, M.; Kadel, K.; Troiani, T.; Gale, R.; Mayes, T.; Shulaev, E.; Wen, J.; Mysore, K.S.; Azad, R.K.; et al. Rapid identification of causative insertions underlying Medicago truncatula Tnt1 mutants defective in symbiotic nitrogen fixation from a forward genetic screen by whole genome sequencing. BMC Genom. 2016, 17, 141. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Aoki, S.; Liu, M.; Umehara, Y.; Suganuma, N.; Iwasaki, W.; Sato, S.; Soyano, T.; Kouchi, H.; Kawaguchi, M. Function and evolution of a Lotus japonicus AP2/ERF family transcription factor that is required for development of infection threads. DNA Res. 2017, 24, 193–203. [Google Scholar] [PubMed]
- Cregan, P.B.; Jarvik, T.; Bush, A.L.; Shoemaker, R.C.; Lark, K.G.; Kahler, A.L.; Kaya, N.; VanToai, T.T.; Lohnes, D.G.; Chung, J. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999, 39, 1464–1490. [Google Scholar] [CrossRef]
- Santos, M.A.D.; Nicolás, M.F.; Hungria, M. Identificação de QTL associados à simbiose entre Bradyrhizobium japonicum, B. elkanii e soja. Pesqui. Agropecuária Bras. 2006, 41, 67–75. [Google Scholar] [CrossRef]
- Santos, M.A.; Geraldi, I.O.; Garcia, A.A.F.; Bortolatto, N.; Schiavon, A.; Hungria, M. Mapping of QTLs associated with biological nitrogen fixation traits in soybean. Hereditas 2013, 150, 17–25. [Google Scholar] [CrossRef]
- Nicolás, M.F.; Hungria, M.; Arias, C.A.A. Identification of quantitative trait loci controlling nodulation and shoot mass in progenies from two Brazilian soybean cultivars. Field Crop. Res. 2006, 95, 355–366. [Google Scholar] [CrossRef]
- Kim, D.H.; Parupalli, S.; Azam, S.; Lee, S.H.; Varshney, R.K. Comparative sequence analysis of nitrogen fixation-related genes in six legumes. Front. Plant Sci. 2013, 4, 300. [Google Scholar] [CrossRef]
- Alves-Carvalho, S.; Aubert, G.; Carrère, S.; Cruaud, C.; Brochot, A.L.; Jacquin, F.; Klein, A.; Martin, C.; Boucherot, K.; Kreplak, J.; et al. Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J. 2015, 84, 1–19. [Google Scholar] [CrossRef]
- Qiao, Z.; Pingault, L.; Nourbakhsh-Rey, M.; Libault, M. Comprehensive comparative genomic and transcriptomic analyses of the legume genes controlling the nodulation process. Front. Plant Sci. 2016, 7, 34. [Google Scholar] [CrossRef]
- Vižintin, L.; Javornik, B.; Bohanec, B. Genetic characterization of selected Trifolium species as revealed by nuclear DNA content and ITS rDNA region analysis. Plant Sci. 2006, 170, 859–866. [Google Scholar] [CrossRef]
- Ištvánek, J.; Jaroš, M.; Krenek, A.; Řepková, J. Genome assembly and annotation for red clover (Trifolium pratense; Fabaceae). Am. J. Bot. 2014, 101, 327–337. [Google Scholar] [CrossRef] [PubMed]
- De Vega, J.J.; Ayling, S.; Hegarty, M.; Kudrna, D.; Goicoechea, J.L.; Ergon, Å.; Rognli, O.A.; Jones, C.; Swain, M.; Geurts, R.; et al. Red clover (Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.D.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.X.; Gouzy, J.; Schoof, H.; et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Stanton-Geddes, J.; Paape, T.; Epstein, B.; Briskine, R.; Yoder, J.; Mudge, J.; Bharti, A.K.; Farmer, A.D.; Zhou, P.; Denny, R.; et al. Candidate Genes and Genetic Architecture of Symbiotic and Agronomic Traits Revealed by Whole-Genome, Sequence-Based Association Genetics in Medicago truncatula. PLoS ONE 2013, 8, e65688. [Google Scholar] [CrossRef] [PubMed]
- Curtin, S.J.; Tiffin, P.; Guhlin, J.; Trujillo, D.; Burghart, L.; Atkins, P.; Baltes, N.J.; Denny, R.; Voytas, D.F.; Stupar, R.M.; et al. Validating genome-wide association candidates controlling quantitative variation in nodulation. Plant Physiol. 2017, 173, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Grunvald, A.K.; Torres, A.R.; Luiz de Lima Passianotto, A.; Santos, M.A.; Jean, M.; Belzile, F.; Hungria, M. Identification of QTLs associated with biological nitrogen fixation traits in soybean using a genotyping-by-sequencing approach. Crop. Sci. 2018, 58, 2523–2532. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Byrne, S.; Czaban, A.; Studer, B.; Panitz, F.; Bendixen, C.; Asp, T. Genome Wide Allele Frequency Fingerprints (GWAFFs) of Populations via Genotyping by Sequencing. PLoS ONE 2013, 8, e57438. [Google Scholar] [CrossRef]
- Kozarewa, I.; Armisen, J.; Gardner, A.F.; Slatko, B.E.; Hendrickson, C.L. Overview of target enrichment strategies. Curr. Protoc. Mol. Biol. 2015, 112, 7–21. [Google Scholar]
- Roux, B.; Rodde, N.; ßoise Jardinaud, M.-F.; Timmers, T.; Sauviac, L.; Cottret, L.; Ebastien Carr Ere, S.; Sallet, E.; Courcelle, E.; Moreau, S.; et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014, 77, 817–837. [Google Scholar] [CrossRef]
- Tejada-Jiménez, M.; Gil-Díez, P.; León-Mediavilla, J.; Wen, J.; Mysore, K.S.; Imperial, J.; González-Guerrero, M. Medicago truncatula Molybdate Transporter type 1 (MtMOT1.3) is a plasma membrane molybdenum transporter required for nitrogenase activity in root nodules under molybdenum deficiency. New Phytol. 2017, 216, 1223–1235. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, Q.; Li, X.; Ai, W.; Liu, D.; Qi, W.; Zhang, M.; Yang, C.; Liao, H. Characterization of genetic basis on synergistic interactions between root architecture and biological nitrogen fixation in soybean. Front. Plant Sci. 2017, 8, 1466. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P. Legume Symbiotic Nitrogen Fixation: Agronomic Aspects. In The Rhizobiaceae; Springer Netherlands: Heidelberg, Germany, 1998; pp. 509–530. [Google Scholar]
- Yang, Q.; Yang, Y.; Xu, R.; Lv, H.; Liao, H. Genetic analysis and mapping of QTLs for soybean biological nitrogen fixation traits under varied field conditions. Front. Plant Sci. 2019, 10, 75. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.R.; Knight, W.E.; Peterson, H.H. Variation among Inbred Lines of Crimson Clover for N2 Fixation (C2H2) Efficiency. Crop Sci. 1982, 22, 716–719. [Google Scholar] [CrossRef]
- Nutman, P.S. Improving nitrogen fixation in legumes by plant breeding; the relevance of host selection experiments in red clover (Trifolium pratense L.) and subterranean clover (T. subterraneum L.). Plant Soil 1984, 82, 285–301. [Google Scholar] [CrossRef]
- Unkovich, M.; Herridge, D.; Peoples, M.; Cadisch, G.; Boddey, B.; Giller, K.; Alves, B.; Chalk, P. Measuring plant-associated nitrogen fixation in agricultural systems. Aust. Cent. Int. Agric. Res. 2008, 136, 132–188. [Google Scholar]
- Sulieman, S.; Schulze, J. The efficiency of nitrogen fixation of the model legume Medicago truncatula (Jemalong A17) is low compared to Medicago sativa. J. Plant Physiol. 2010, 167, 683–692. [Google Scholar] [CrossRef]
- Thilakarathna, M.S.; Papadopoulos, Y.A.; Rodd, A.V.; Grimmett, M.; Fillmore, S.A.E.; Crouse, M.; Prithiviraj, B. Nitrogen fixation and transfer of red clover genotypes under legume–grass forage based production systems. Nutr. Cycl. Agroecosystems 2016, 106, 233–247. [Google Scholar] [CrossRef]
- Thilakarathna, M.S.; Papadopoulos, Y.A.; Grimmett, M.; Fillmore, S.A.E.; Crouse, M.; Prithiviraj, B. Red Clover Varieties with Nitrogen Fixing Advantage during the Early Stages of Seedling Development. Can. J. Plant Sci. 2018, 98, 517–526. [Google Scholar] [CrossRef]
- Hardy, R.W.F.; Burns, R.C.; Holsten, R.D. Applications of the acetylene-ethylene assay for measurement of nitrogen fixation. Soil Biol. Biochem. 1973, 5, 47–81. [Google Scholar] [CrossRef]
- Mckenna, P.; Cannon, N.; Dooley, J.; Conway, J. The use of red clover (Trifolium pratense) in soil fertility-building: A Review. Field Crop. Res. 2018, 221, 38–49. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Mens, C.; Hastwell, A.H.; Zhang, M.; Su, H.; Jones, C.H.; Chu, X.; Gresshoff, P.M. Legume nodulation: The host controls the party. Plant Cell Environ. 2018, 42, 41–51. [Google Scholar] [CrossRef]
- Roughley, R.J.; Dart, P.J. Reduction of acetylene by nodules of Trifolium subterraneum as affected by root temperature, Rhizobium strain and host cultivar. Arch. Mikrobiol. 1969, 69, 171–179. [Google Scholar] [CrossRef]
- Bergersen, F.J. The Quantitative Relationship Between Nitrogen Fixation And The Acetylene-Reduction assay. Aust. J. Biol. Sci. 1970, 23, 1015–1026. [Google Scholar] [CrossRef]
- Vessey, J.K. Measurement of nitrogenase activity in legume root nodules: In defense of the acetylene reduction assay. Plant Soil 1994, 158, 151–162. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Vernié, T.; Moreau, S.; De Billy, F.; Plet, J.; Combier, J.P.; Rogers, C.; Oldroyd, G.; Frugier, F.; Niebel, A.; Gamas, P. EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 2008, 20, 2696–2713. [Google Scholar] [CrossRef]
- Tejada-Jiménez, M.; Chamizo-Ampudia, A.; Galván, A.; Fernández, E.; Llamas, Á. Molybdenum metabolism in plants. Metallomics 2013, 5, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Krusell, L.; Krause, K.; Ott, T.; Desbrosses, G.; Krämer, U.; Sato, S.; Nakamura, Y.; Tabata, S.; James, E.K.; Sandal, N.; et al. The Sulfate Transporter SST1 Is Crucial for Symbiotic Nitrogen Fixation in Lotus japonicus Root Nodules. Plant Cell 2005, 17, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Karunakaran, R.; East, A.K.; Poole, P.S. Multiplicity of Sulfate and Molybdate Transporters and Their Role in Nitrogen Fixation in Rhizobium leguminosarum bv. viciae Rlv3841. Mol. Plant Microbe. Interact. 2016, 29, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Delgado, M.J.; Tresierra-Ayala, A.; Talbi, C.; Bedmar, E.J. Functional characterization of the Bradyrhizobium japonicum modA and modB genes involved in molybdenum transport. Microbiology 2006, 152, 199–207. [Google Scholar] [CrossRef]
- Avenhaus, U.; Cabeza, R.A.; Liese, R.; Lingner, A.; Dittert, K.; Salinas-Riester, G.; Pommerenke, C.; Schulze, J. Short-term molecular acclimation processes of legume nodules to increased external oxygen concentration. Front. Plant Sci. 2016, 6. [Google Scholar] [CrossRef]
- Minchin, F.R. Regulation of oxygen diffusion in legume nodules. Soil Biol. Biochem. 1997, 29, 881–888. [Google Scholar] [CrossRef]
- Mylona, P.; Pawlowski, K.; Bisseling, T. Symbiotic Nitrogen Fixation. Plant Cell 1995, 7, 869–885. [Google Scholar] [CrossRef]
- Cabeza, R.; Koester, B.; Liese, R.; Lingner, A.; Baumgarten, V.; Dirks, J.; Salinas-Riester, G.; Pommerenke, C.; Dittert, K.; Schulze, J. An RNA Sequencing Transcriptome Analysis Reveals Novel Insights into Molecular Aspects of the Nitrate Impact on the Nodule Activity of Medicago truncatula. Plant Physiol. 2014, 164, 400–411. [Google Scholar] [CrossRef]
- Bogusz, D.; Appleby, C.A.; Landsmann, J.; Dennis, E.S.; Trinick, M.J.; Peacock, W.J. Functioning haemoglobin genes in non-nodulating plants. Nature 1988, 331, 178–180. [Google Scholar] [CrossRef]
- Arredondo-Peter, R.; Hargrove, S.; Sarath, C.; Moran, J.F.; Lohrman, J.; Olson, J.S.; Klucas, R. V Rice Hemoglobins. Plant Physiol. 1997, 115, 1259–1266. [Google Scholar] [CrossRef]
- Trevaskis, B.; Watts, R.A.; Andersson, C.R.; Llewellyn, D.J.; Hargrove, M.S.; Olson, J.S.; Dennis, E.S.; Peacock, W.J. Two hemoglobin genes in Arabidopsis thaliana: The evolutionary origins of leghemoglobins. Proc. Natl. Acad. Sci. USA 1997, 94, 12230–12234. [Google Scholar] [CrossRef] [PubMed]
- Andersson, C.R.; Ostergaard Jensen, E.; Llewellyn, D.J.; Dennis, E.S.; Peacock, W.J. A new hemoglobin gene from soybean: A role for hemoglobin in all plants (nonsymbiotic/leghemoglobin/evolution). Plant Biol. 1996, 93, 5682–5687. [Google Scholar]
- Bustos-Sanmamed, P.; Tovar-Méndez, A.; Crespi, M.; Sato, S.; Tabata, S.; Becana, M. Regulation of nonsymbiotic and truncated hemoglobin genes of Lotus japonicus in plant organs and in response to nitric oxide and hormones. New Phytol. 2011, 189, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Begueria, L.; Cuypers, B.; Van Doorslaer, S.; Abbruzzetti, S.; Bruno, S.; Berghmans, H.; Dewilde, S.; Ramos, J.; Viappiani, C.O.; Becana, M. Characterization of the heme pocket structure and ligand binding kinetics of non-symbiotic hemoglobins from the model legume lotus japonicus. Front. Plant Sci. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Igamberdiev, A.U. Nitrate, NO and haemoglobin in plant adaptation to hypoxia: An alternative to classic fermentation pathways. J. Exp. Bot. 2004, 55, 2473–2482. [Google Scholar] [CrossRef]
- Gupta, K.J.; Mur, L.A.J.; Wany, A.; Kumari, A.; Fernie, A.R.; Ratcliffe, R.G. The role of nitrite and nitric oxide under low oxygen conditions in plants. New Phytol. 2019. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Breakspear, A.; Liu, C.; Roy, S.; Stacey, N.; Rogers, C.; Trick, M.; Morieri, G.; Mysore, K.S.; Wen, J.; Oldroyd, G.E.D.; et al. The root hair “infectome” of medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infectionw. Plant Cell 2014, 26, 4680–4701. [Google Scholar] [CrossRef]
- Becana, M.; Wienkoop, S.; Matamoros, M.A. Sulfur Transport and Metabolism in Legume Root Nodules. Front. Plant Sci. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Ehrhardt, D.W.; Atkinson, M.E.; Faull, K.F.; Freedberg, D.I.; Sutherlin, D.P.; Armstrong, R.; Long, S.R. In vitro sulfotransferase activity of NodH, a nodulation protein of Rhizobium meliloti required for host-specific nodulation. J. Bacteriol. 1995, 177, 6237–6245. [Google Scholar] [CrossRef]
- Zhao, Y.; Cheng, S.; Song, Y.; Huang, Y.; Zhou, S.; Liu, X.; Zhou, D.-X. The Interaction between Rice ERF3 and WOX11 Promotes Crown Root Development by Regulating Gene Expression Involved in Cytokinin Signaling. Plant Cell 2015, 27, 2469–2483. [Google Scholar] [CrossRef] [PubMed]
- Dündar, E.; Bush, D.R. BAT1, a bidirectional amino acid transporter in Arabidopsis. Planta 2009, 229, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from plant tissues. In Plant Molecular Biology Manual; Gelvin, S.B., Schilperoort, R.A., Verma, D.P.S., Eds.; Springer Netherlands: Heidelberg, Germany, 1989; pp. 73–83. ISBN 978-94-009-0951-9. [Google Scholar]
- Andrews, S. Babraham Bioinformatics-FastQC A Quality Control tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 9 September 2019).
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H. Stacks: Building and Genotyping Loci De Novo From Short-Read Sequences. G3 Genes Genomes Genet. 2011, 1, 171–182. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Depristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–501. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Meirmans, P.G.; Liu, S.; Van Tienderen, P.H. The Analysis of Polyploid Genetic Data. J. Hered. 2018, 109, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pcaMethods-A bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
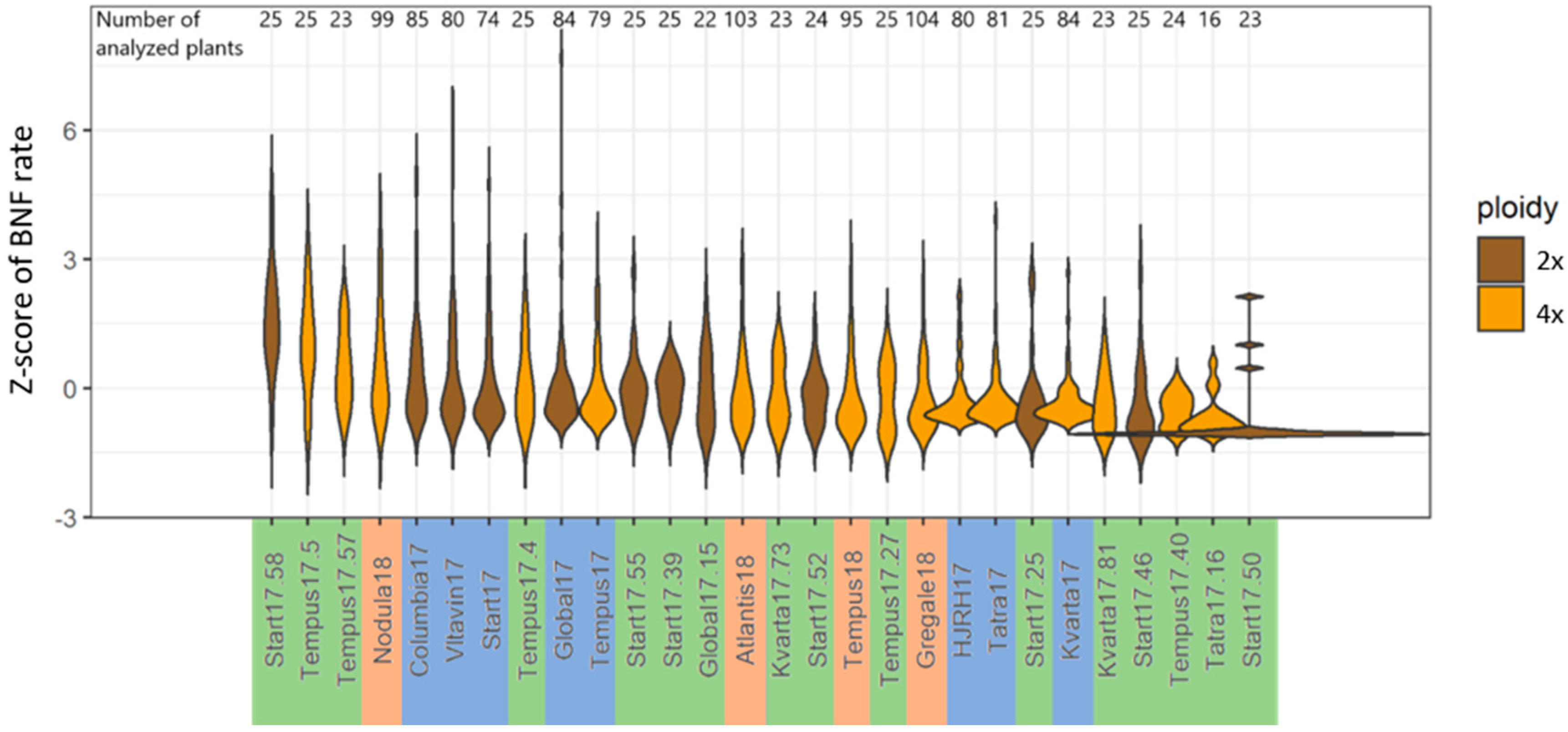
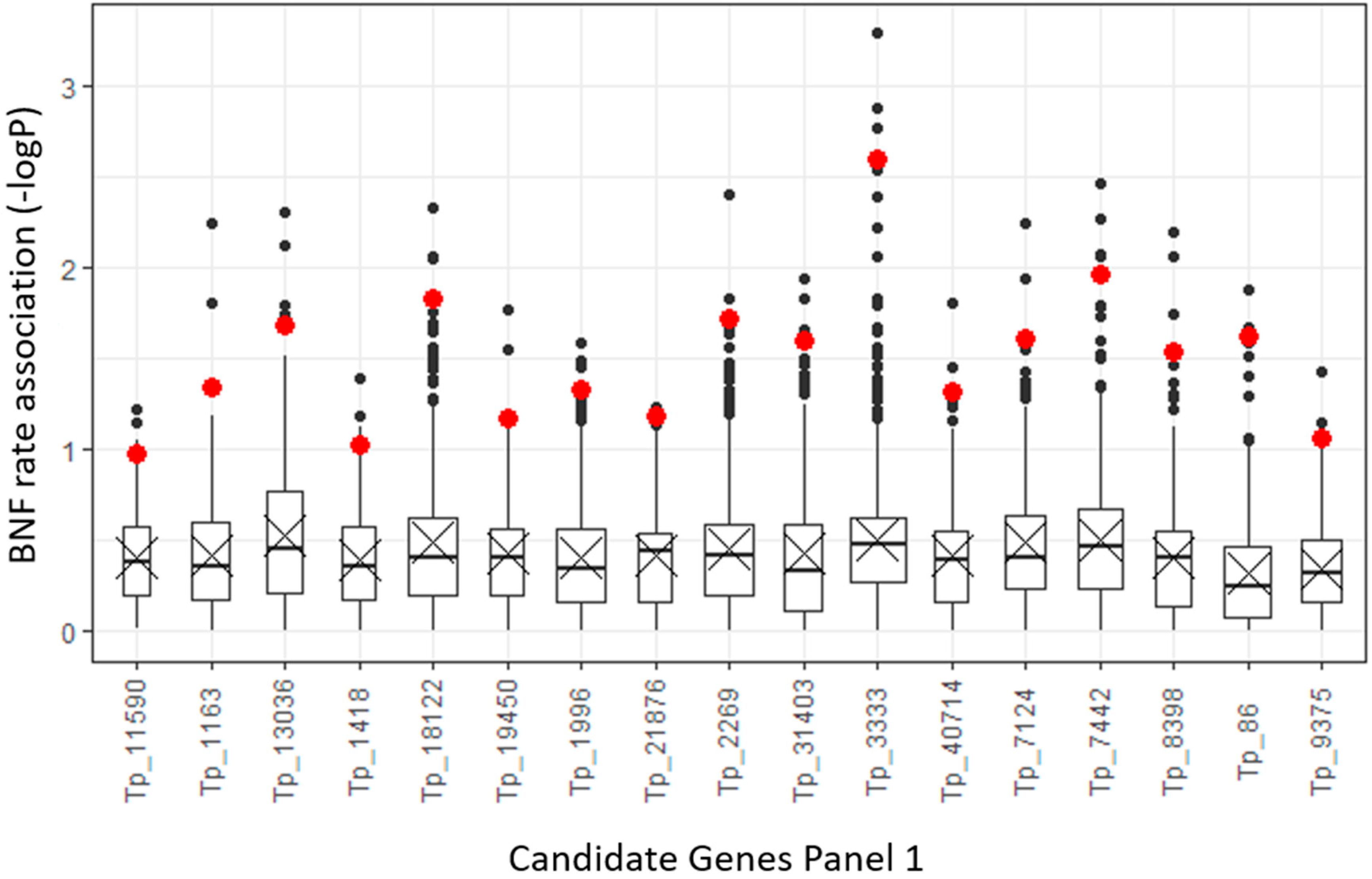
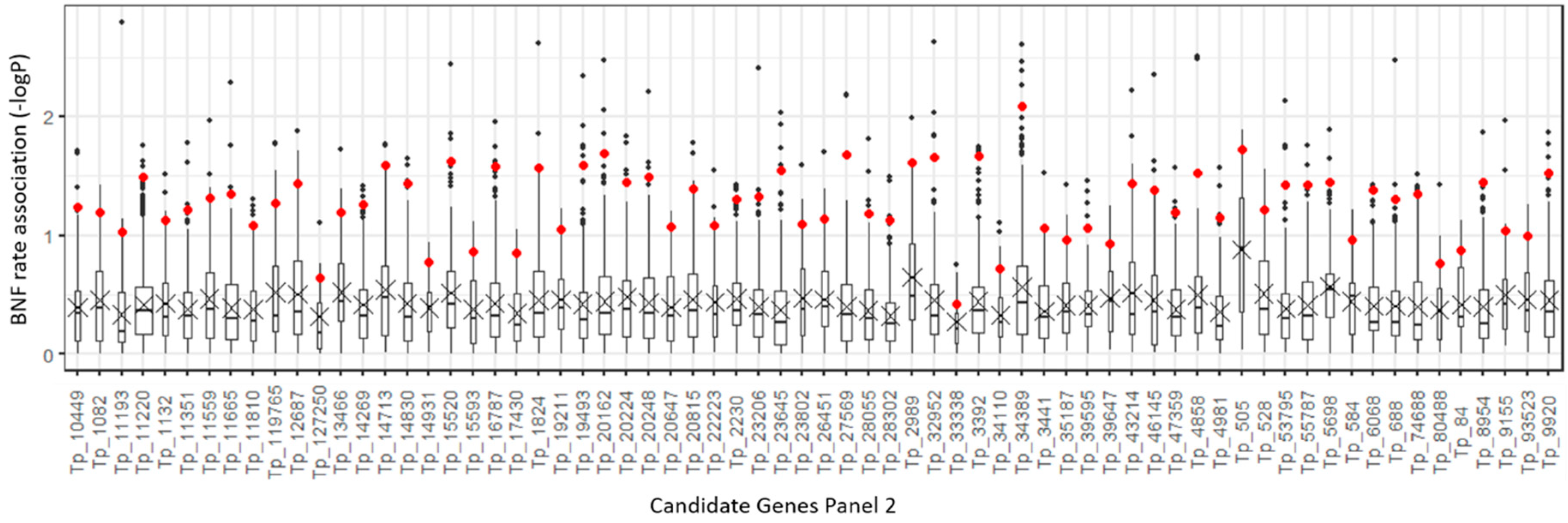
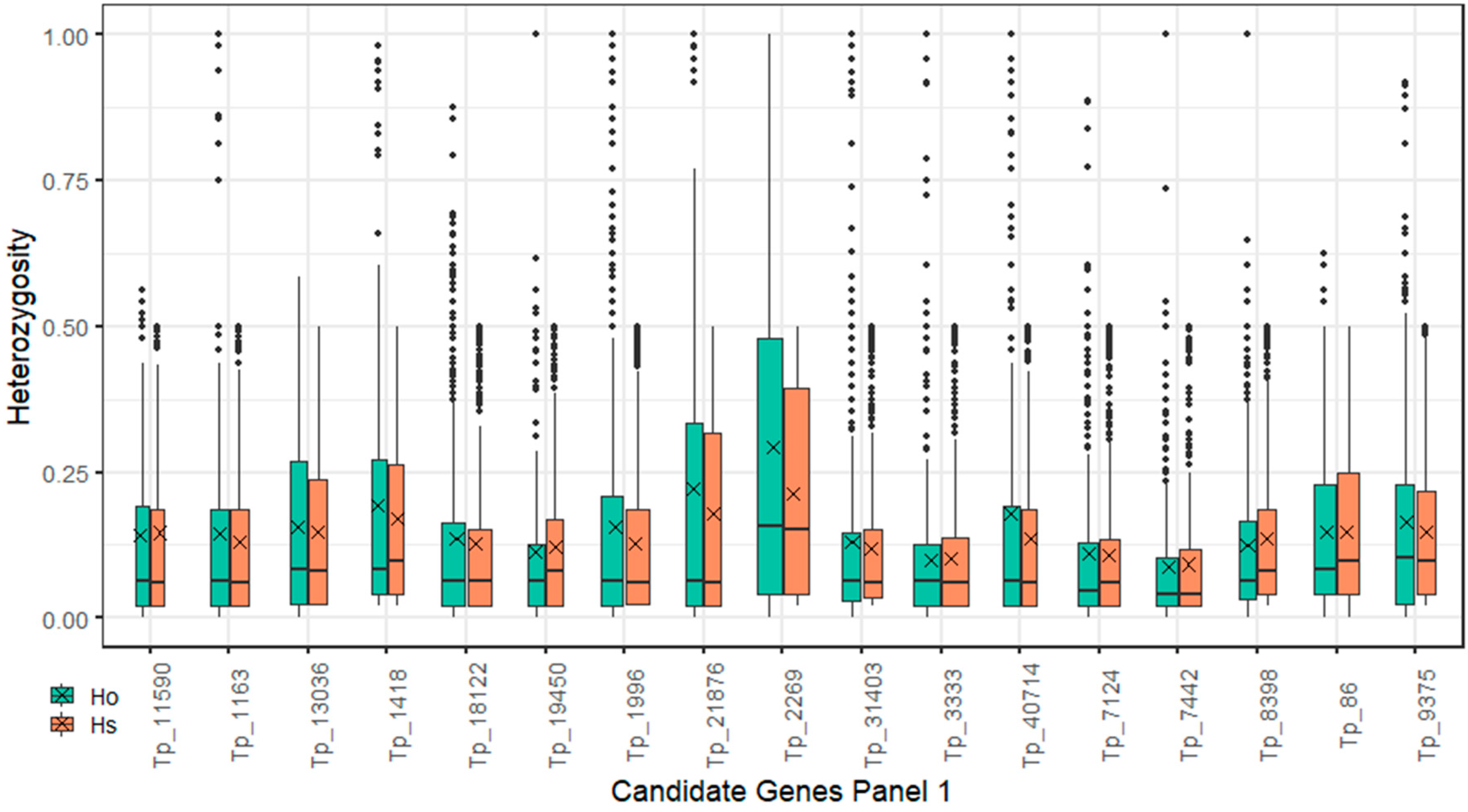

| Plant Set | p-Value 1 | Different Pairs of Accessions 2 |
|---|---|---|
| 1 | 3.413 × 10−6 | Columbia17-HJRH17, Columbia17-Kvarta17 |
| 2 | 1.151 × 10−6 | Nodula18-Gregale18, Nodula18-Tempus18 |
| 3 | 2.2 × 10−16 | Kvarta17.73-Start17.58, Kvarta17.81-Start17.58, Start17.25-Start17.58, Start17.39-Start17.50, Start17.46-Start17.58, Start17.46-Tempus17.5, Start17.50-Start17.55, Start17.50-Start17.58, Start17.50-Tempus17.4, Start17.50-Tempus17.5, Start17.50-Tempus17.57, Start17.52-Start17.58, Start17.58-Tatra17.16, Start17.58-Tempus17.27, Start17.58-Tempus17.40, Tatra17.16-Tempus17.5, Tempus17.40-Tempus17.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trněný, O.; Vlk, D.; Macková, E.; Matoušková, M.; Řepková, J.; Nedělník, J.; Hofbauer, J.; Vejražka, K.; Jakešová, H.; Jansa, J.; et al. Allelic Variants for Candidate Nitrogen Fixation Genes Revealed by Sequencing in Red Clover (Trifolium pratense L.). Int. J. Mol. Sci. 2019, 20, 5470. https://doi.org/10.3390/ijms20215470
Trněný O, Vlk D, Macková E, Matoušková M, Řepková J, Nedělník J, Hofbauer J, Vejražka K, Jakešová H, Jansa J, et al. Allelic Variants for Candidate Nitrogen Fixation Genes Revealed by Sequencing in Red Clover (Trifolium pratense L.). International Journal of Molecular Sciences. 2019; 20(21):5470. https://doi.org/10.3390/ijms20215470
Chicago/Turabian StyleTrněný, Oldřich, David Vlk, Eliška Macková, Michaela Matoušková, Jana Řepková, Jan Nedělník, Jan Hofbauer, Karel Vejražka, Hana Jakešová, Jan Jansa, and et al. 2019. "Allelic Variants for Candidate Nitrogen Fixation Genes Revealed by Sequencing in Red Clover (Trifolium pratense L.)" International Journal of Molecular Sciences 20, no. 21: 5470. https://doi.org/10.3390/ijms20215470
APA StyleTrněný, O., Vlk, D., Macková, E., Matoušková, M., Řepková, J., Nedělník, J., Hofbauer, J., Vejražka, K., Jakešová, H., Jansa, J., Piálek, L., & Knotová, D. (2019). Allelic Variants for Candidate Nitrogen Fixation Genes Revealed by Sequencing in Red Clover (Trifolium pratense L.). International Journal of Molecular Sciences, 20(21), 5470. https://doi.org/10.3390/ijms20215470





