Downregulated Expression of Virulence Factors Induced by Benzyl Isothiocyanate in Staphylococcus Aureus: A Transcriptomic Analysis
Abstract
1. Introduction
2. Results
2.1. Antibacterial Assay
2.2. Transcriptome Library Construction and Sequencing Data Quality Control
2.3. Reads and Reference Genome Comparison
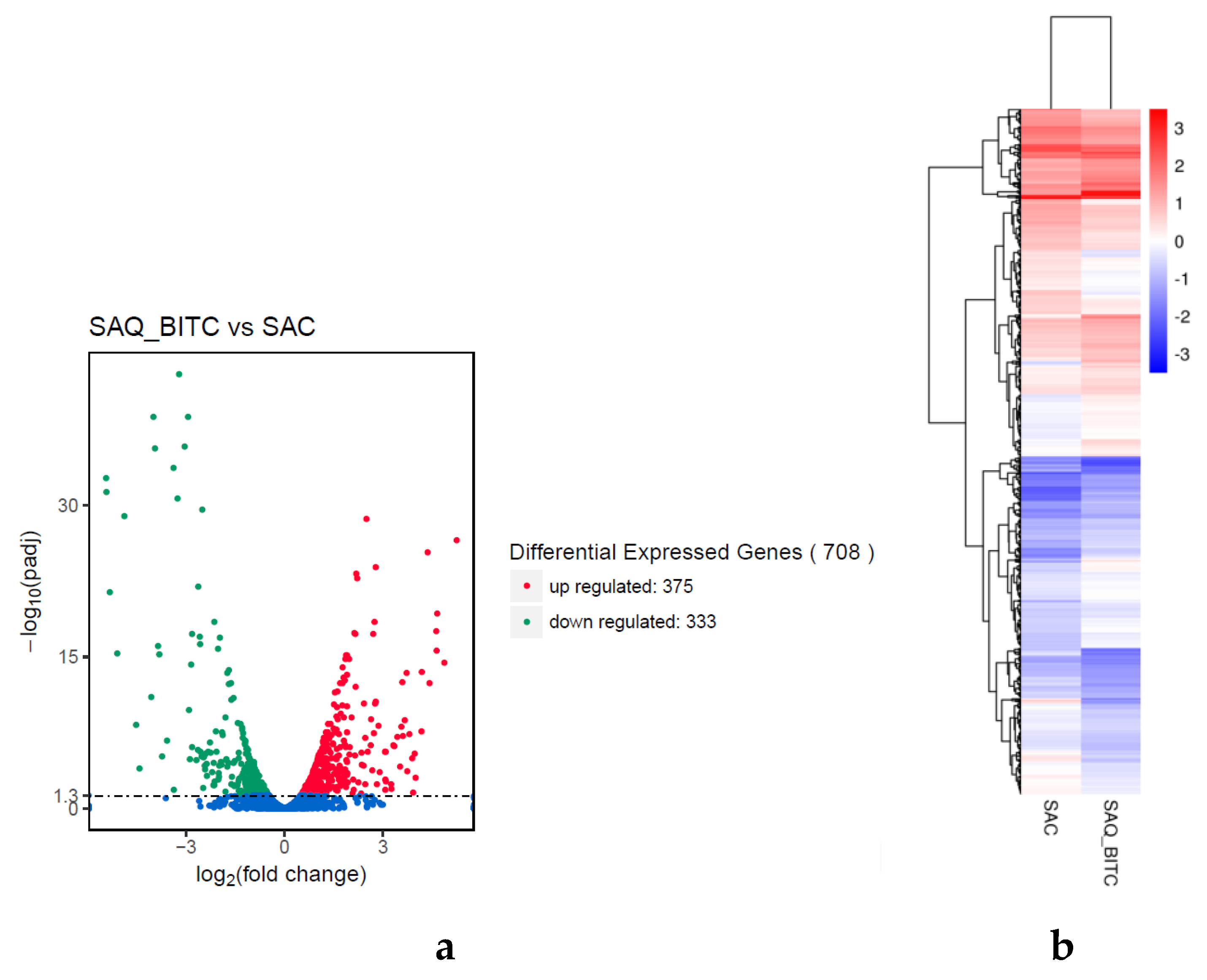
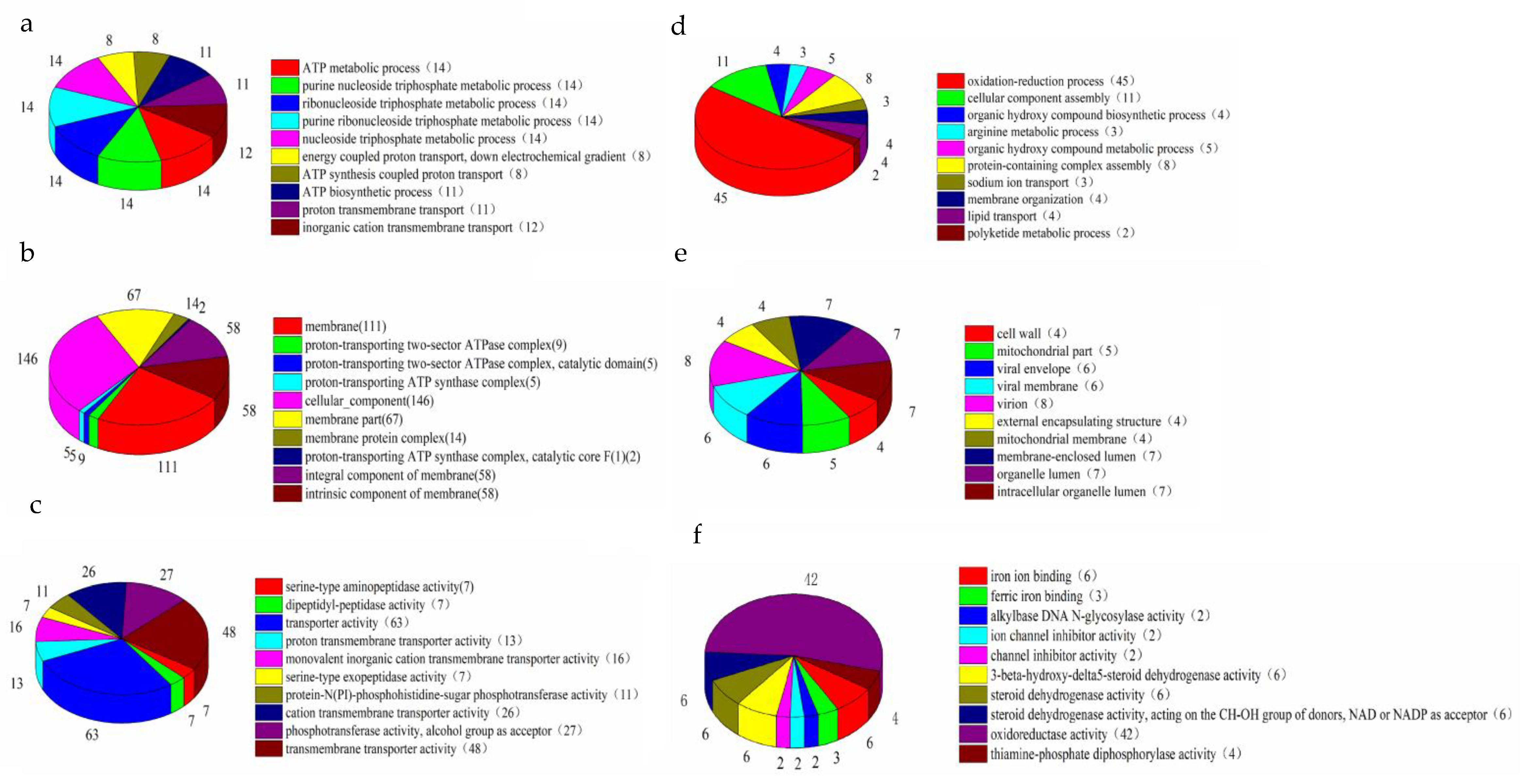
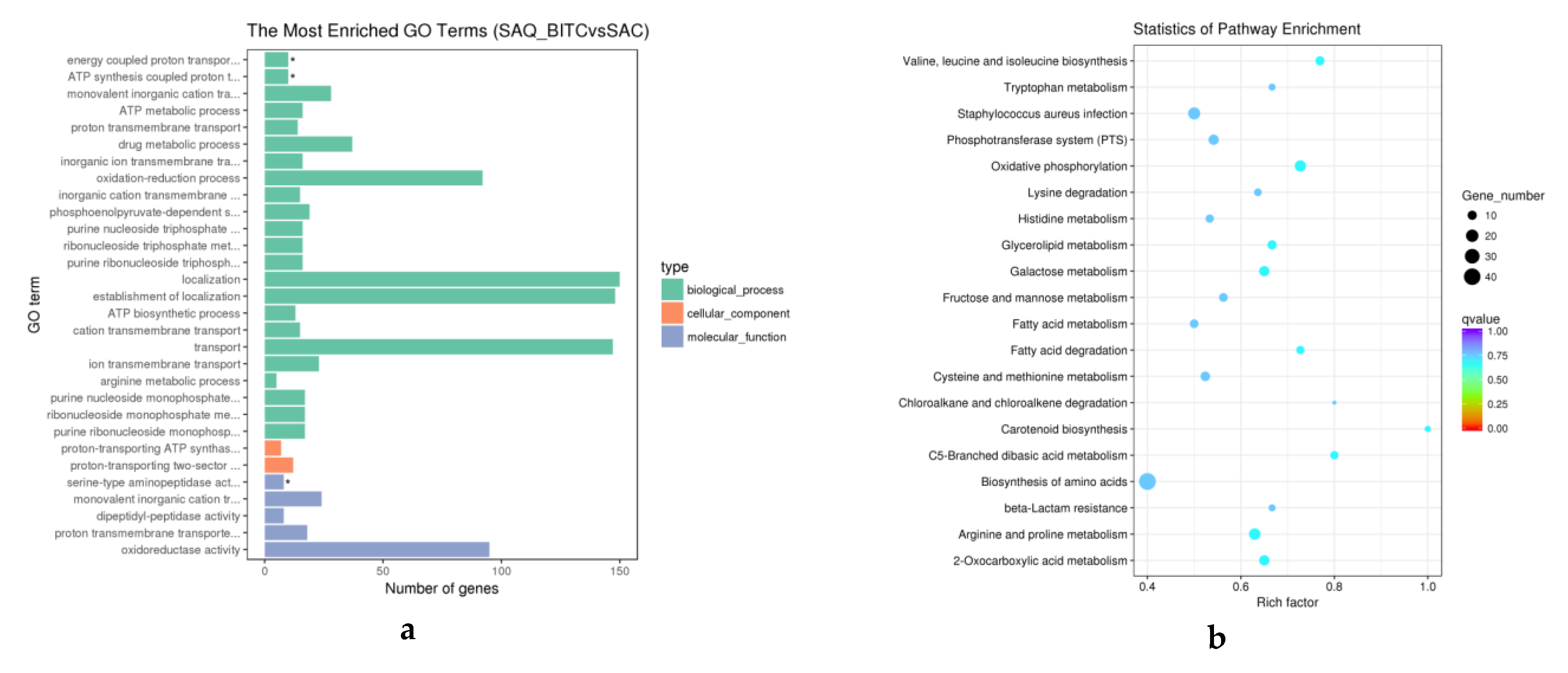
2.4. Analysis of Differentially Expressed Genes (DEGs)
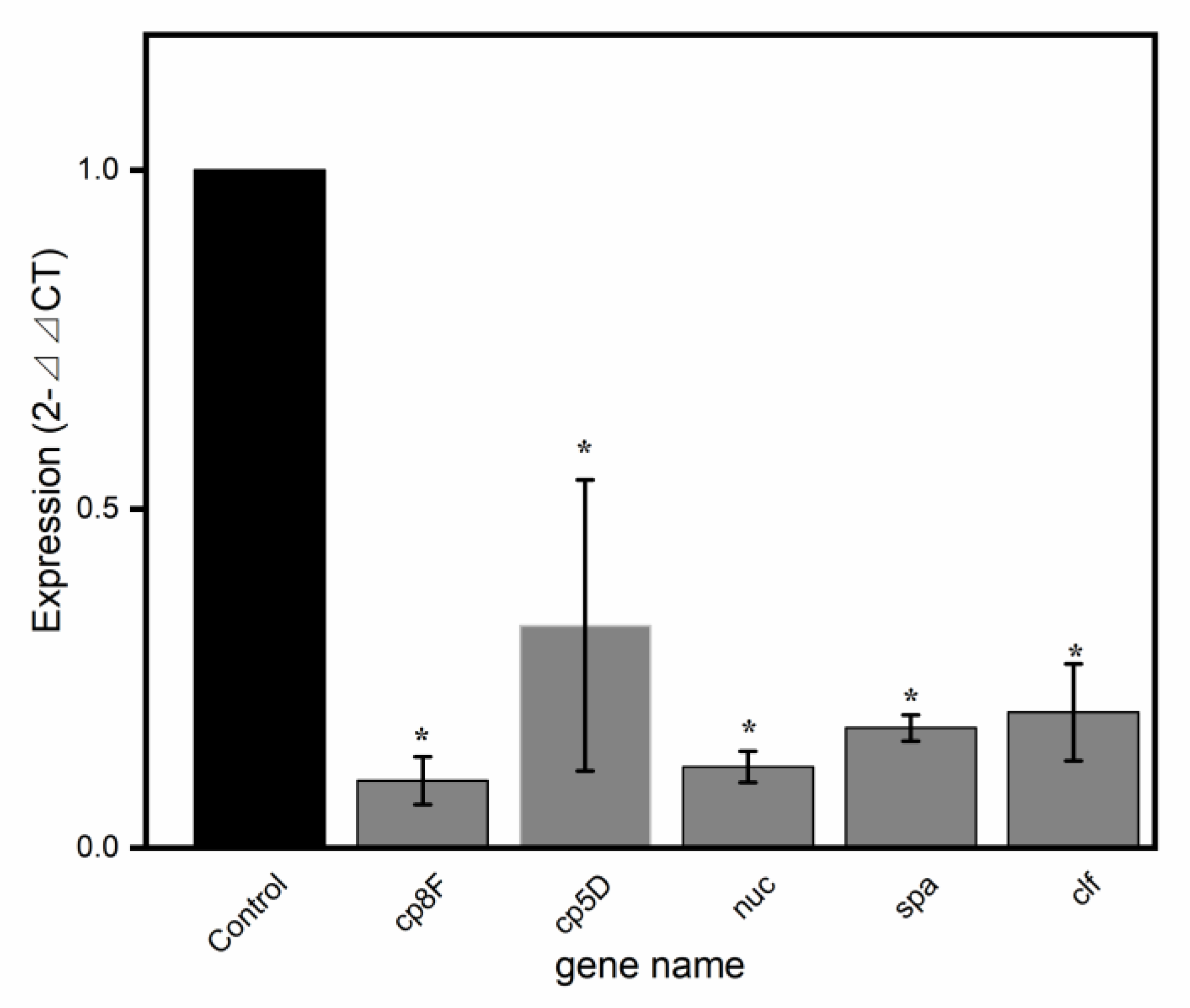
2.5. Validation of Virulence-Related Gene Results by qRT-PCR
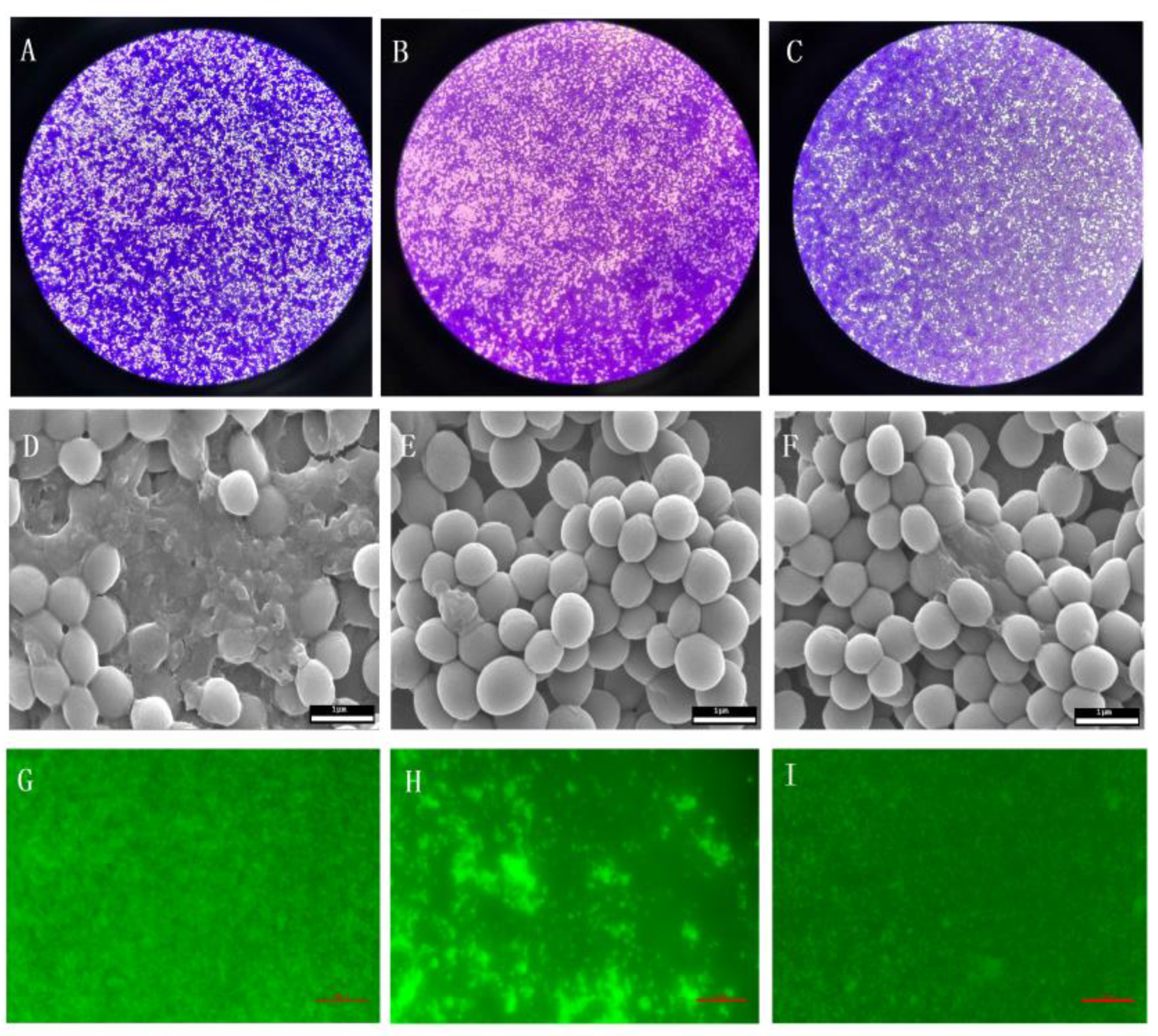
2.6. Effect of BITC on the S. Aureus Biofilm
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture
4.2. Antibacterial Assays
4.3. Extraction and Detection of RNA Samples
4.4. Library Construction and Sequencing
4.5. Biological Information Analysis
4.6. qRT-PCR Validation of Differential Genes
4.7. Effects of BITC on Formation of S. Aureus Biofilm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ITCs | isothiocyanates |
| BITC | benzyl isothiocyanate |
| BP | biological process |
| CC | cellular component |
| MF | molecular function |
| DEGs | differential expression genes |
| FPKMs | Fragments Per Kilobase of exon model per Million mapped reads |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | quantitative Real-time Polymerase Chain Reaction |
References
- Vanderhaeghen, W.; Hermans, K.; Haesebrouck, F.; Butaye, P. Methicillin-resistant staphylococcus aureus (mrsa) in food production animals. Epidemiol. Infect. 2010, 138, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Graveland, H.; Duim, B.; Duijkeren, E.V.; Heederik, D.; Wagenaar, J.A. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int. J. Med. Microbiol. 2011, 301, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Rasooly, A. Analytical chromatography for recovery of small amounts of staphylococcal enterotoxins from food. Int. J. Food Microbiol. 2001, 64, 33–40. [Google Scholar] [CrossRef]
- Krishnamoorthy, K.; Moon, J.Y.; Hyun, H.B.; Cho, S.K.; Kim, S.J. Mechanistic investigation on the toxicity of mgo nanoparticles toward cancer cells. J. Mater. Chem. 2012, 22, 24610–24617. [Google Scholar] [CrossRef]
- Loir, Y.L.; Baron, F.; Gautier, M. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2003, 2, 63–76. [Google Scholar] [PubMed]
- Shi, C.; Che, M.Y.; Zhang, X.W.; Liu, Z.J.; Meng, R.Z.; Bu, X.J.; Ye, H.Q.; Guo, N. Antibacterial activity and mode of action of totarol against Staphylococcus aureus in carrot juice. J. Food Sci. Technol. 2018, 55, 924–934. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Liu, X.Y.; Wang, Y.F.; Jiang, P.P.; Siew, Y.Q. Antibacterial activity and mechanism of cinnamon essential oil against Escherichia coli and Staphylococcus aureus. Food Control. 2016, 59, 282–289. [Google Scholar] [CrossRef]
- Kang, J.; Jin, W.; Wang, J.; Sun, Y.; Wu, X.; Liu, L. Antibacterial and anti-biofilm activities of peppermint essential oil against Staphylococcus aureus. LWT—Food Sci. Technol. 2019, 101, 639–645. [Google Scholar] [CrossRef]
- Molinavargas, L.F. Mechanism of action of isothiocyanates. a review. Agron. Colomb. 2013, 31, 68–75. [Google Scholar]
- Khamis, I.; Heikkila, J.J. Effect of isothiocyanates, bitc and peitc, on stress protein accumulation, protein aggregation and aggresome-like structure formation in xenopus a6 kidney epithelial cells. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2018, 204, 1–13. [Google Scholar] [CrossRef]
- Higdon, J.V.; Delage, B.; Williams, D.E.; Dashwood, R.H. Cruciferous vegetables and human cancer risk: Epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007, 55, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, P.; Kim, J.S. Anti-Carcinogenic Glucosinolates in Cruciferous Vegetables and Their Antagonistic Effects on Prevention of Cancers. Molecules 2018, 23, 2983. [Google Scholar] [CrossRef] [PubMed]
- Dufour, V.; Stahl, M.; Baysse, C. The antibacterial properties of isothiocyanates. Microbiology 2015, 161, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Hou, H.M.; Wu, H.Y.; Li, K.L.; Wang, Y.; Zhou, Q.Q.; Gong-Liang Zhang, G.L. Transcriptomic Analysis of Vibrio parahaemolyticus Reveals Different Virulence Gene Expression in Response to Benzyl Isothiocyanate. Molecules 2019, 24, 761. [Google Scholar] [CrossRef]
- Slany, M.; Oppelt, J.; Cincarova, L. Formation of Staphylococcus aureus biofilm in the presence of sublethal concentrations of disinfectants studied via a transcriptomic analysis using transcriptome sequencing (rna-seq). Appl. Environ. Microbiol. 2017, 83, AEM.01643-17. [Google Scholar] [CrossRef]
- Duan, J.; Li, M.; Hao, Z.; Shen, X.; Liu, L.; Jin, Y.; Wang, S.; Guo, Y.; Yang, L.; Wang, L.; et al. Subinhibitory concentrations of resveratrol reduce alpha-hemolysin production in Staphylococcus aureus isolates by downregulating saers. Emerg. Microbes Infect. 2018, 7, 136. [Google Scholar] [CrossRef]
- Lowy, F.D. Medical progress: Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Miyafusa, T.; Tanaka, Y.; Kuroda, M.; Ohta, T.; Tsumoto, K. Expression, purification, crystallization and preliminary diffraction analysis of capf, a capsular polysaccharide-synthesis enzyme from Staphylococcus aureus. Acta. Crystallogr Sect. F. Struct. Biol. Cryst. Commun. 2008, 64, 512–516. [Google Scholar] [CrossRef]
- Sandel, M.K.; Mckillip, J.L. Virulence and recovery of Staphylococcus aureus relevant to the food industry using improvements on traditional approaches. Food Control. 2004, 15, 5–10. [Google Scholar] [CrossRef]
- Alarcón, B.; Vicedo, B.; Aznar, R. Pcr-based procedures for detection and quantification of Staphylococcus aureus and their application in food. J. Appl. Microbil. 2006, 100, 352–364. [Google Scholar] [CrossRef]
- Hu, Y.; Meng, J.; Shi, C.; Hervin, K.; Fratamico, P.M.; Shi, X. Characterization and comparative analysis of a second thermonuclease from Staphylococcus aureus. Microbil. Res. 2013, 168, 174–182. [Google Scholar] [CrossRef]
- Thakker, M.; Park, J.S.; Carey, V.; Lee, J.C. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect. Immun. 1998, 66, 5183–5189. [Google Scholar]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Salimena, A.P.S.; Lange, C.C.; Camussone, C.; Signorini, M.; Calvinho, L.F.; Brito, M.A.; Borges, C.A.; Guimarães, A.S.; Ribeiro, J.B.; Mendonça, L.C.; et al. Genotypic and phenotypic detection of capsular polysaccharide and biofilm formation instaphylococcus aureusisolated from bovine milk collected from brazilian dairy farms. Vet. Re. Commun. 2016, 40, 97–106. [Google Scholar] [CrossRef]
- Hyldgaard, M.; Sutherland, D.S.; Sundh, M.; Mygind, T.; Meyer, R.L. Antimicrobial mechanism of monocaprylate. Appl. Environ. Microbiol. 2012, 78, 2957–2965. [Google Scholar] [CrossRef]
- Zhang, S.; Xiong, J.; Lou, W.; Ning, Z.; Zhang, D.; Yang, J. Antimicrobial activity and action mechanism of triglycerol monolaurate on common foodborne pathogens. Food Control. 2018, 98, 113–119. [Google Scholar] [CrossRef]
- Hennekinne, J.A.; De Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef]
- Suo, B.; Wang, X.; Pan, Z.; Wang, N.; Ai, Z.; Yu, S.; Salazar, J.K. Inactivation and sublethal injury kinetics of Staphylococcus aureus in broth at low temperature storage. J. Food Prot. 2014, 77, 1689–1695. [Google Scholar] [CrossRef]
- Iqbal, Z.; Seleem, M.N.; Hussain, H.I.; Huang, L.; Hao, H.; Yuan, Z. Comparative virulence studies and transcriptome analysis of Staphylococcus aureus strains isolated from animals. Sci. Rep. 2016, 6, 35442. [Google Scholar] [CrossRef]
- Murphy, E.; Lin, S.L.; Nunez, L.; Andrew, L.; Fink, P.S.; Dilts, D.A.; Hoiseth, S.K.; Jansen, K.U.; Anderson, A.S. Challenges for the evaluation of Staphylococcus aureus protein based vaccines: Monitoring antigenic diversity. Hum. Vaccin. 2011, 7, 51–59. [Google Scholar] [CrossRef][Green Version]
- Nanra, J.S.; Buitrago, S.M.; Crawford, S.; Ng, J.; Fink, P.S.; Hawkins, J.; Scully, I.L.; McNeil, L.K.; Aste-Amézaga, J.M.; Cooop, D. Capsular polysaccharides are an important immune evasion mechanism for Staphylococcus aureus. Hum. Vaccin. Immunother. 2013, 9, 480–487. [Google Scholar]
- Rodrigues, M.V.; Fortaleza, C.M.; Riboli, D.F.; Rocha, R.S.; Rocha, C.; da Cunha Mde, L. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in a burn unit from Brazil. Burns 2013, 39, 1242–12499. [Google Scholar] [CrossRef]
- Costa, L.; Faustino, M.A.; Tomé, J.P.; Neves, M.G.; Tomé, A.C.; Cavaleiro, J.A. Involvement of typeI and typeII mechanisms on the photo in activation of non-enveloped DNA and RNA bacteriophages. J. Photochem. Photobiol. B. 2013, 120, 10–16. [Google Scholar]
- Cassat, J.E.; Hammer, N.D.; Campbell, J.P.; Benson, M.A.; Perrien, D.S.; Mrak, L.N.; Smeltzer, M.S.; Torres, V.J.; Skaar, E.P. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe. 2013, 13, 759–772. [Google Scholar] [CrossRef]
- Mendoza Bertelli, A.; Delpino, M.V.; Lattar, S.; Giai, C.; Llana, M.N.; Sanjuan, N.; Cassat, J.E.; Sordelli, D.; Gómez, M.I. Staphylococcus aureus protein A enhances osteoclastogenesis via TNFR1 and EGFR signaling. Biochim. Biophys. Acta 2016, 10, 1975–1983. [Google Scholar] [CrossRef]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker, K.G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sara loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Andes, D.; Diekema, D.J.; Espinel, I.A.; Sheehan, D. Wild-type mic distributions, epidemiological cutoff values and species-specific clinical breakpoints for fluconazole and candida: Time for harmonization of clsi and eucast broth microdilution methods. Drug Resist. Updat. 2010, 13, 180–195. [Google Scholar] [CrossRef]
- Miladi, H.; Zmantar, T.; Kouidhi, B.; Chaabouni, Y.; Mahdouani, K.; Bakhrouf, A.; Chaieb, K. Use of carvacrol, thymol, and eugenol for biofilm eradication and resistance modifying susceptibility of salmonella enterica serovar typhimurium strains to nalidixic acid. Microb. Pathog. 2017, 104, 56–63. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jinwei, Z.; Shiyuan, B.; Hongjuan, C.; Tongtong, C.; Rui, Y.; Minghui, L.; Fu, Y.L.; Jia, A.Q. Anti-biofilm and antivirulence activities of metabolites from plectosphaerella cucumerina against pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 769. [Google Scholar]
| Sample Name | Total Reads | Total Mapped | Multiple Mapped | Uniquely Mapped |
|---|---|---|---|---|
| SAC1 | 17,425,772 | 16,699,263 (95.83%) | 912,234 (5.23%) | 15,787,029 (90.6%) |
| SAC2 | 19,641,306 | 18,949,431 (96.48%) | 995,830 (5.07%) | 17,953,601 (91.41%) |
| SAC3 | 19,752,162 | 18,917,775 (95.78%) | 1,151,497 (5.83%) | 17,766,278 (89.95%) |
| SAQ_BITC1 | 17,015,102 | 16,523,451 (97.11%) | 782,977 (4.6%) | 15,740,474 (92.51%) |
| SAQ_BITC2 | 19,245,460 | 18,643,857 (96.87%) | 928,383 (4.82%) | 17,715,474 (92.05%) |
| SAQ_BITC3 | 13,327,608 | 12,947,341 (97.15%) | 602,910 (4.52%) | 12,344,431 (92.62%) |
| Gene_Id | Gene Name | log2FoldChange (SAQ_BITC vs SAC) | Pval (SAQ_BITC vs SAC) | Padj (SAQ_BITC vs SAC) | Significant (SAQ_BITC vs SAC) |
|---|---|---|---|---|---|
| SAOUHSC_00119 | Cp8F | −3.9945 | 2.33 × 10−42 | 2.02 × 10−39 | DOWN |
| SAOUHSC_00117 | CP5D | −5.4282 | 1.73 × 10−34 | 5.63 × 10−32 | DOWN |
| SAOUHSC_00818 | nuc | −2.1358 | 2.85 × 10−21 | 3.53 × 10−19 | DOWN |
| SAOUHSC_00069 | spa | −1.55 | 2.54 × 10−13 | 1.10 × 10−11 | DOWN |
| SAOUHSC_00812 | clf | −1.1057 | 2.92 × 10−7 | 4.77 × 10−6 | DOWN |
| Gene | Primer | Sequence (5’–3’) |
|---|---|---|
| 16S rRNA | 16S rRNA-F | CGTGCTACAATGGACAATACA |
| 16S rRNA-R | ACAATCCGAACTGAGAACAAC | |
| Cp8F | Cp8F-F | ACAGACTTTAGTTATCCCTTAC |
| Cp8F-R | TGATGCCAGTGATTACCTTTA | |
| CP5D | CP5D-F | CTTTAGTTGTTGGTGCTGGTC |
| CP5D-R | CGGTTCAAGTTTCATTTCGTC | |
| nuc | nuc-F | GAAAGGGCAATACGCAAAG |
| nuc-R | ACGCCATTATCTGTTTG | |
| spa | spa-F | ATAAGAAGCAACCAGCAAAC |
| spa-R | GGCTAATGATAATCCACCAA | |
| clf | clf-F | ACGAATGGCGATGTTGTAGC |
| clf-R | CTCGGTCTGTAAATAAAGGTAATG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wu, H.; Niu, T.; Bi, J.; Hou, H.; Hao, H.; Zhang, G. Downregulated Expression of Virulence Factors Induced by Benzyl Isothiocyanate in Staphylococcus Aureus: A Transcriptomic Analysis. Int. J. Mol. Sci. 2019, 20, 5441. https://doi.org/10.3390/ijms20215441
Wang X, Wu H, Niu T, Bi J, Hou H, Hao H, Zhang G. Downregulated Expression of Virulence Factors Induced by Benzyl Isothiocyanate in Staphylococcus Aureus: A Transcriptomic Analysis. International Journal of Molecular Sciences. 2019; 20(21):5441. https://doi.org/10.3390/ijms20215441
Chicago/Turabian StyleWang, Xiaoning, Hongyan Wu, Tongxin Niu, Jingran Bi, Hongman Hou, Hongshun Hao, and Gongliang Zhang. 2019. "Downregulated Expression of Virulence Factors Induced by Benzyl Isothiocyanate in Staphylococcus Aureus: A Transcriptomic Analysis" International Journal of Molecular Sciences 20, no. 21: 5441. https://doi.org/10.3390/ijms20215441
APA StyleWang, X., Wu, H., Niu, T., Bi, J., Hou, H., Hao, H., & Zhang, G. (2019). Downregulated Expression of Virulence Factors Induced by Benzyl Isothiocyanate in Staphylococcus Aureus: A Transcriptomic Analysis. International Journal of Molecular Sciences, 20(21), 5441. https://doi.org/10.3390/ijms20215441







