Which Low-Abundance Proteins are Present in the Human Milieu of Gamete/Embryo Maternal Interaction?
Abstract
1. Introduction
2. Results
2.1. Sample Collection and Immunoaffinity Depletion
2.2. Data Analysis and Quantification
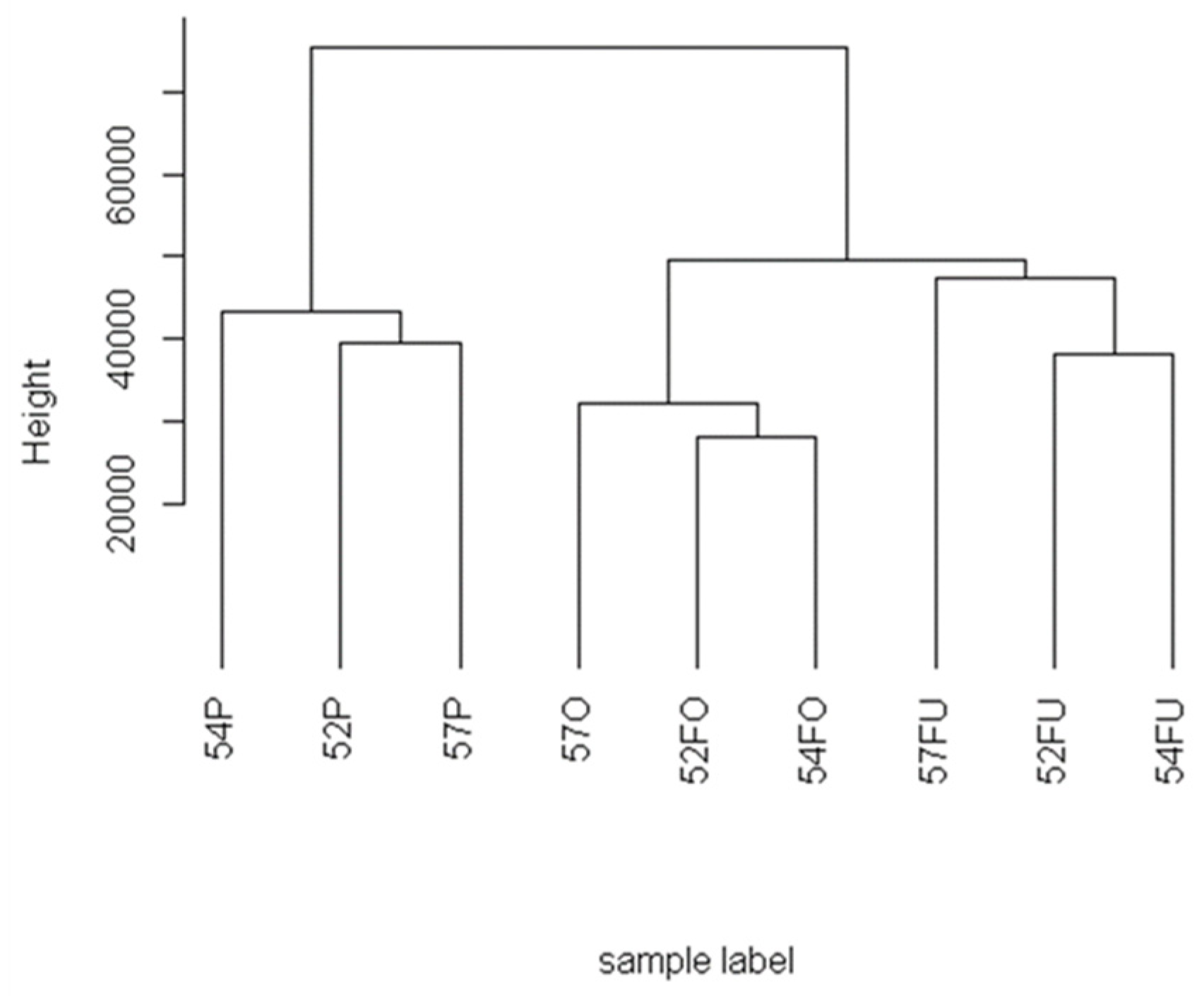
2.2.1. Hierarchical Clustering
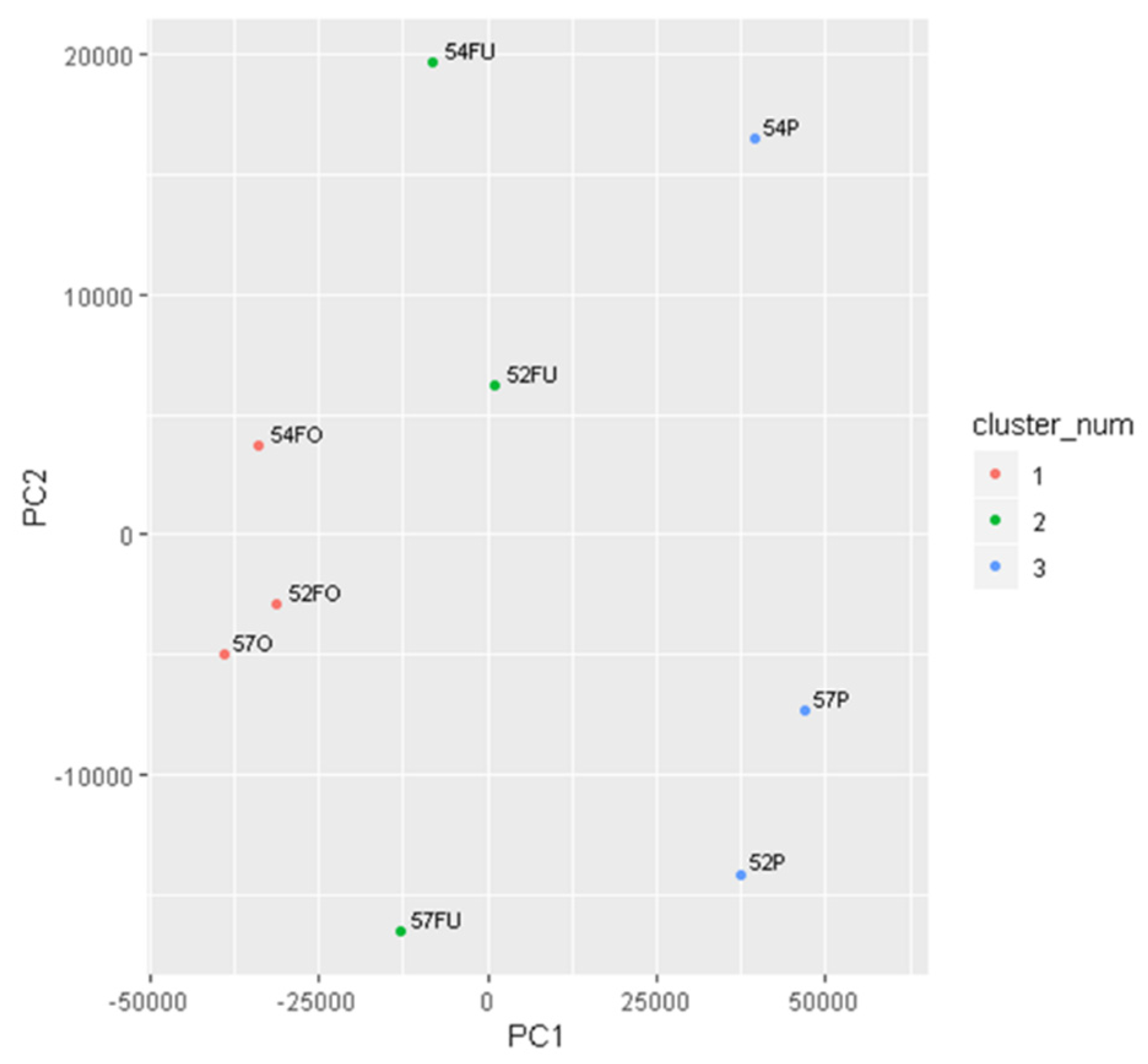
2.2.2. Principal Component Analysis (PCA)
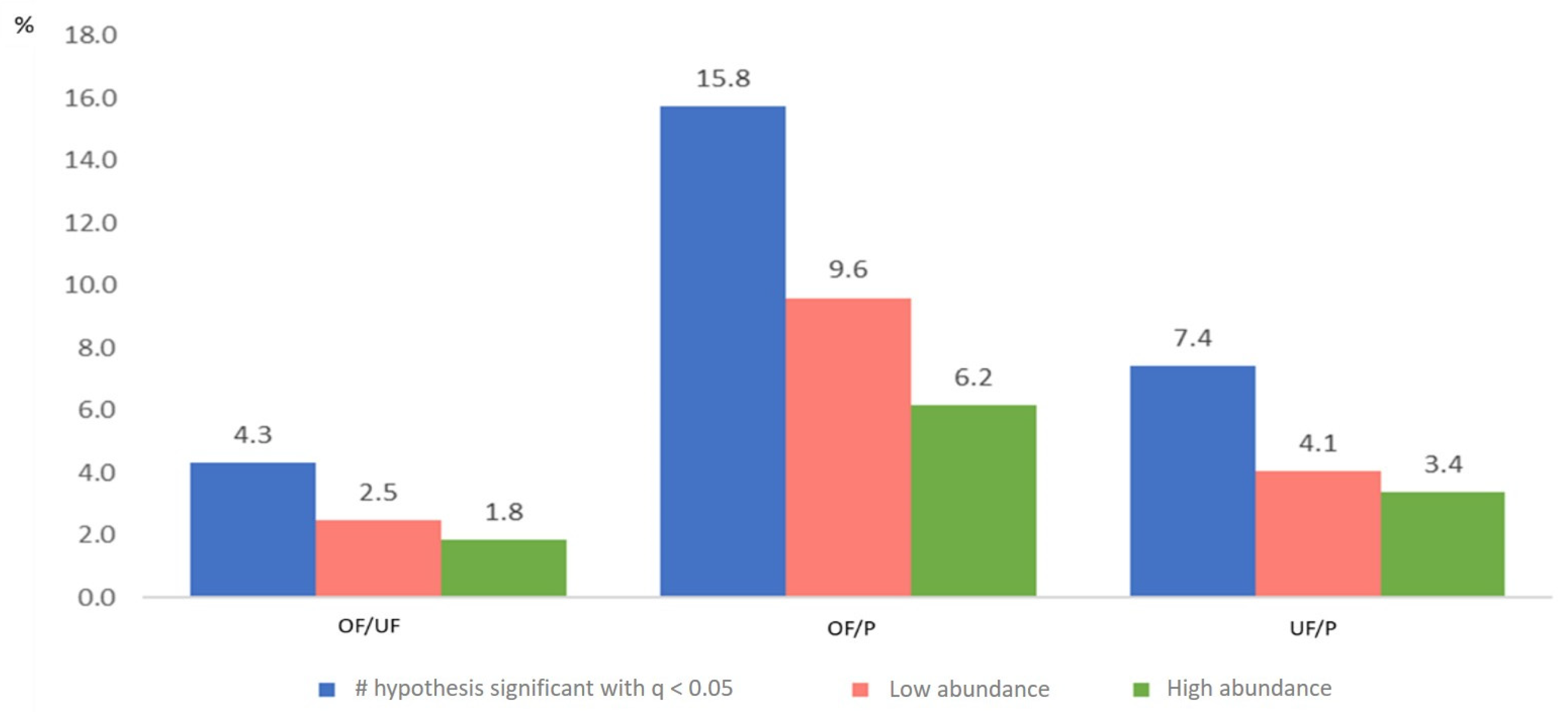
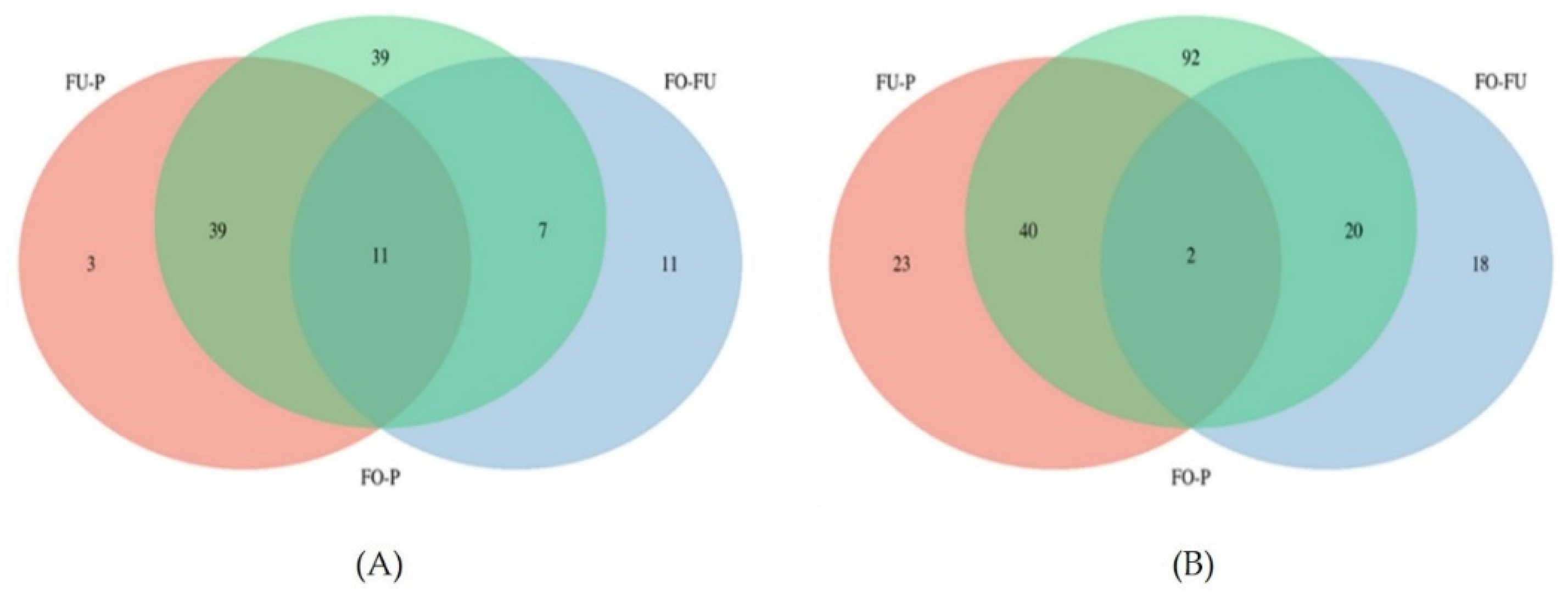
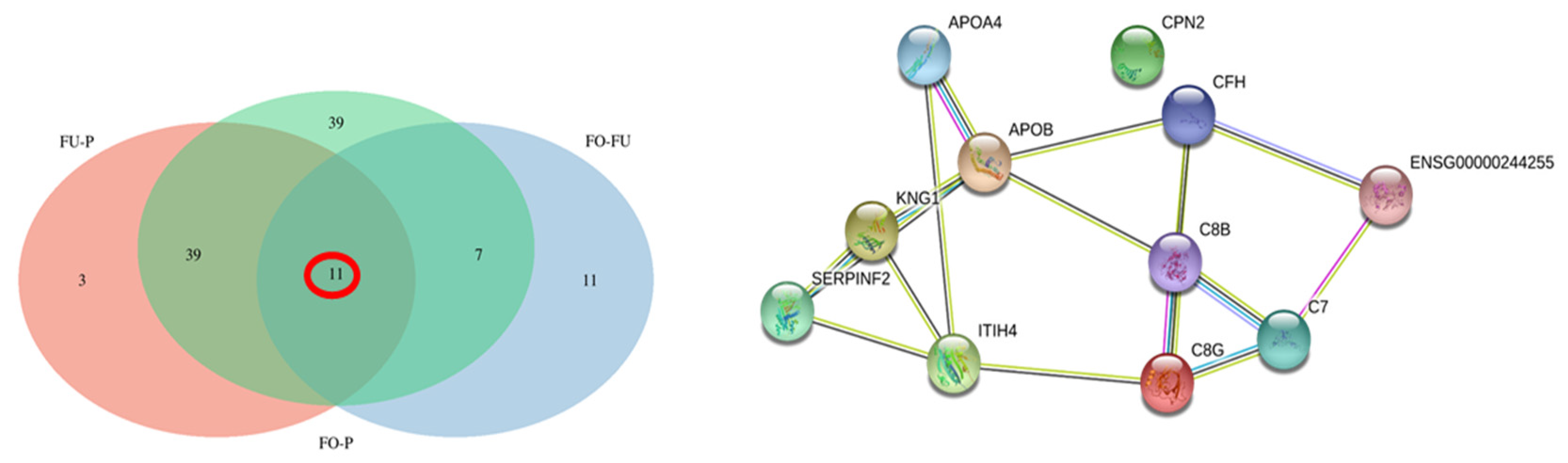
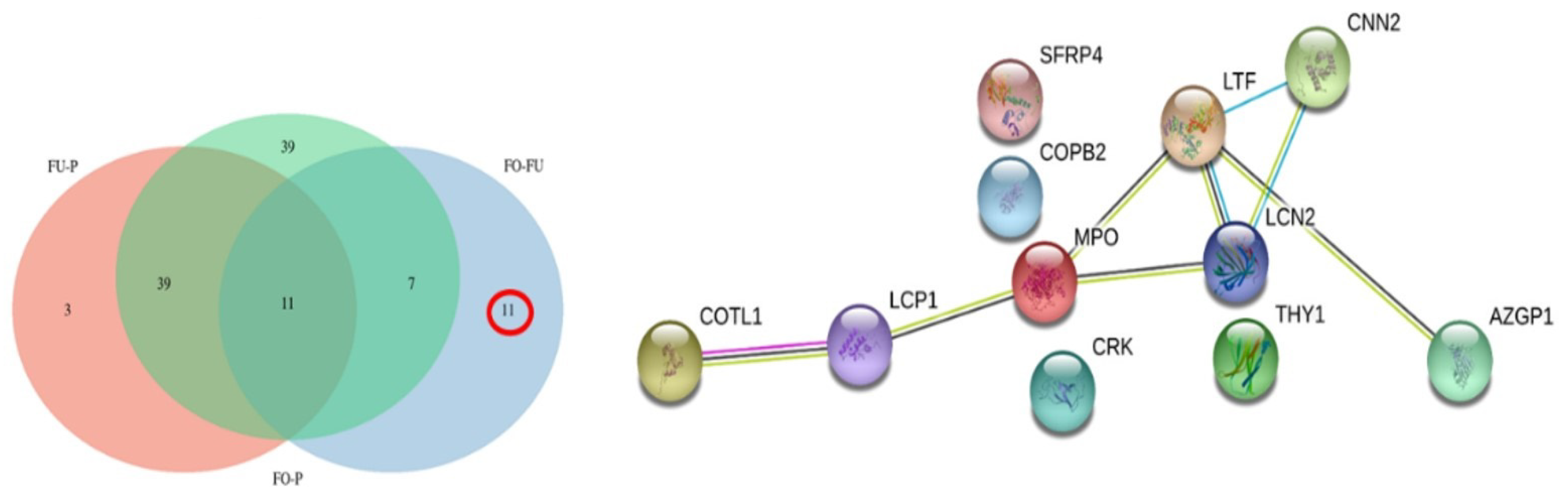
2.2.3. Comparative Analysis
2.2.4. Targeted Mass Spectrometry (MRM/SRM)
3. Discussion
3.1. How to Analyze the Reproductive Fluids?
3.1.1. The Most Adequate Method to Collect and Perform A Proteomic Characterization
3.1.2. The Most Suitable Study Population
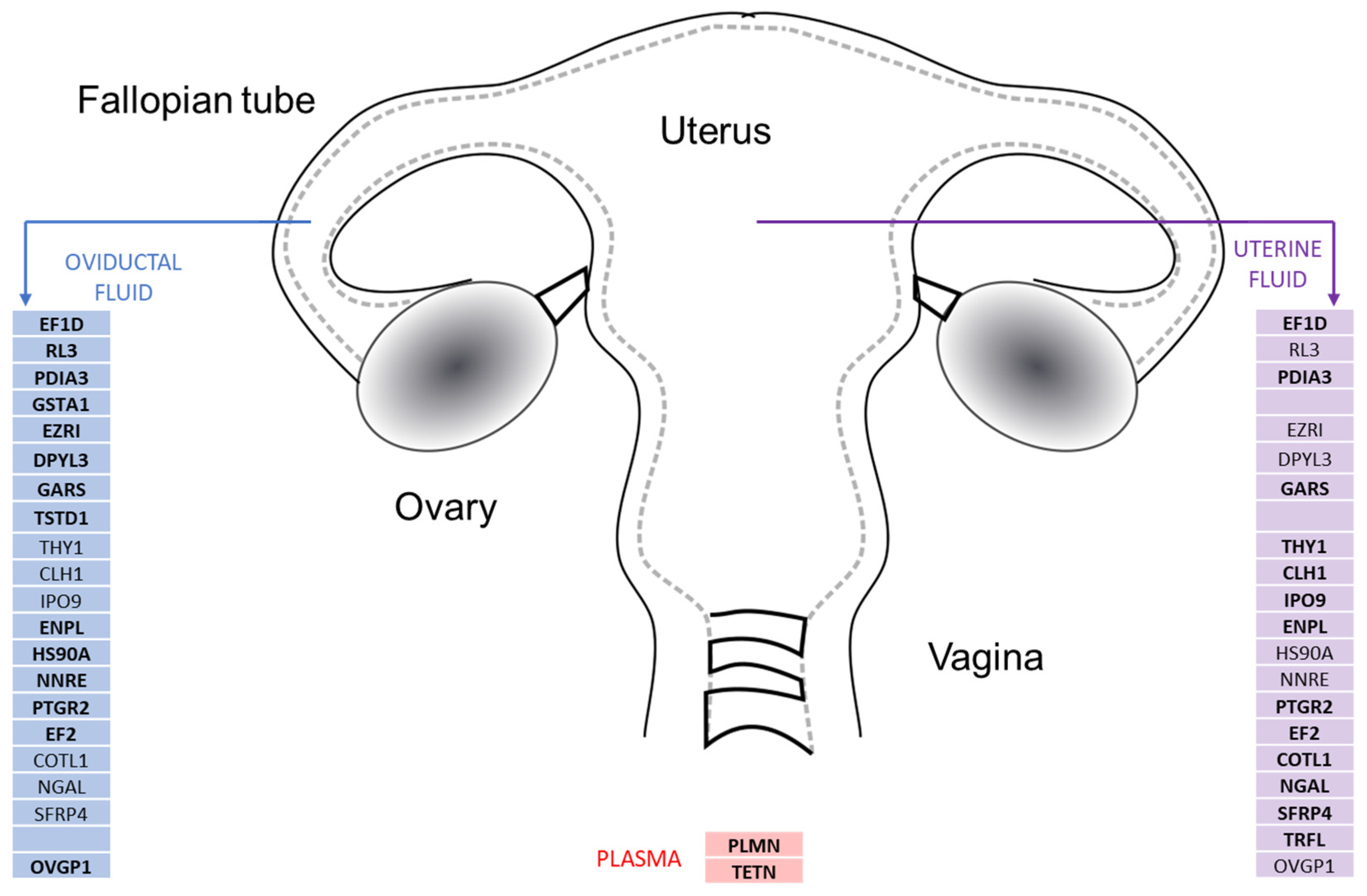
3.2. Common Differences in Protein abundance of Reproductive Fluids Versus Plasma
3.3. Differentially abundance Proteins in Oviductal Fluid
3.4. Differentially abundant Proteins in Uterine Fluid
4. Materials and Methods
4.1. Study Population
4.2. Sample Collection
4.3. Immunoaffinity Depletion
4.4. Tryptic Digestion
4.5. Liquid Chromatography and Mass Spectrometer Analysis
4.6. Targeted Mass Spectrometry (MRM/SRM)
4.7. Data Analysis and Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OF | Oviductal fluids |
| UF | Uterine fluids |
| P | Plasma |
References
- Adamson, G.D.; de Mouzon, J.; Chambers, G.M.; Zegers-Hochschild, F.; Mansour, R.; Ishihara, O.; Banker, M.; Dyer, S. International Committee for Monitoring Assisted Reproductive Technology: World report on assisted reproductive technology, 2011. Fertil. Steril. 2011, 110, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Chronopoulou, E.; Harper, J.C. IVF culture media: Past, present and future. Hum. Reprod. Update 2015, 21, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Sunde, A.; Brison, D.; Dumoulin, J.; Harper, J.; Lundin, K.; Magli, M.C.; Van Den Abbeel, E.; Veiga, A. Time to take human embryo culture seriously. Hum. Reprod. 2016, 31, 2174–2182. [Google Scholar] [CrossRef] [PubMed]
- Morbeck, D.E.; Paczkowski, M.; Fredrickson, J.R.; Krisher, R.L.; Hoff, H.S.; Baumann, N.A.; Moyer, T.; Matern, D. Composition of protein supplements used for human embryo culture. J. Assist. Reprod. Genet. 2014, 31, 1703–1711. [Google Scholar] [CrossRef]
- Blake, D.; Svalander, P.; Jin, M.; Silversand, C.; Hamberger, L. Protein supplementation of human IVF culture media. J. Assist. Reprod. Genet. 2002, 19, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Bavister, B.D. Culture of preimplantation embryos: Facts and artifacts *. Hum. Reprod. Update 1995, 1, 91–148. [Google Scholar] [CrossRef]
- Bungum, M.; Humaidan, P.; Bungum, L. Recombinant human albumin as protein source in culture media used for IVF: A prospective randomized study. Reprod. Biomed. Online 2002, 4, 233–236. [Google Scholar] [CrossRef]
- Youssef, M.M.; Mantikou, E.; van Wely, M.; Van der Veen, F.; Al-Inany, H.G.; Repping, S.; Mastenbroek, S. Culture media for human pre-implantation embryos in assisted reproductive technology cycles. Cochrane Database Syst. Rev. 2015, 8e, 291–295. [Google Scholar] [CrossRef]
- Sinclair, K.D.; Rutherford, K.M.D.; Wallace, J.M.; Brameld, J.M.; Stöger, R.; Alberio, R.; Sweetman, D.; Gardner, D.S.; Perry, V.E.A.; Adam, C.L.; et al. Epigenetics and developmental programming of welfare and production traits in farm animals. Reprod. Fertil. Dev. 2016, 28, 1443–1478. [Google Scholar] [CrossRef]
- Fernández-Gonzalez, R.; Moreira, P.; Bilbao, A.; Jiménez, A.; Pérez-Crespo, M.; Ramírez, M.A.; De Fonseca, F.R.; Pintado, B.; Gutiérrez-Adán, A. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc. Natl. Acad. Sci. USA 2004, 101, 5880–5885. [Google Scholar] [CrossRef]
- El Hajj, N.; Haaf, T. Epigenetic disturbances in in vitro cultured gametes and embryos: Implications for human assisted reproduction. Fertil. Steril. 2013, 99, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Canovas, S.; Campos, R.; Aguilar, E.; Cibelli, J.B. Progress towards human primordial germ cell specification in vitro. Mol. Hum. Reprod. 2017, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Mantikou, E.; Youssef, M.A.F.M.; van Wely, M.; van der Veen, F.; Al-Inany, H.G.; Repping, S.; Mastenbroek, S. Embryo culture media and IVF/ICSI success rates: A systematic review. Hum. Reprod. Update 2013, 19, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, C.; Léandri, R.; Desch, L.; Ernst, A.; Bruno, C.; Cerf, C.; Chiron, A.; Souchay, C.; Burguet, A.; Jimenez, C.; et al. Does Embryo Culture Medium Influence the Health and Development of Children Born after In Vitro Fertilization? PLoS ONE 2016, 11, e0150857. [Google Scholar] [CrossRef]
- Kleijkers, S.H.M.; Mantikou, E.; Slappendel, E.; Consten, D.; Van Echten-Arends, J.; Wetzels, A.M.; Van Wely, M.; Smits, L.J.M.; Van Montfoort, A.P.A.; Repping, S.; et al. Influence of embryo culture medium (G5 and HTF) on pregnancy and perinatal outcome after IVF: A multicenter RCT. Hum. Reprod. 2016, 31, 2219–2230. [Google Scholar] [CrossRef] [PubMed]
- Avilés, M.; Gutiérrez-Adán, A.; Coy, P. Oviductal secretions: Will they be key factors for the future ARTs? MHR Basic Sci. Reprod. Med. 2010, 16, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Galileo, D.S.; Bathala, P.; Fereshteh, Z.; Martin-DeLeon, P.A.; Li, K.; Al-Dossary, A.A. Oviductal extracellular vesicles (oviductosomes, OVS) are conserved in humans: Murine OVS play a pivotal role in sperm capacitation and fertility. MHR Basic Sci. Reprod. Med. 2018, 24, 143–157. [Google Scholar]
- Schramm, R.D.; Al-Sharhan, M. Fertilization and early embryology: Development of in-vitro-fertilized primate embryos into blastocysts in a chemically defined, protein-free culture medium. Hum. Reprod. 1996, 11, 1690–1697. [Google Scholar] [CrossRef]
- Tarahomi, M.; Vaz, F.; Zafardoust, S.; Fatemi, F.; VanWely, M.; Mohammadzadeh, A.; Repping, S.; Hamer, G.; Mastenbroek, S. Human uterine fluid composition is distinct from clinically used preimplantation embryo culture media. Fertil. Steril. 2018, 110, e364. [Google Scholar] [CrossRef]
- Leese, H.J. The formation and function of oviduct fluid. J. Reprod. Fert 1988, 82, 843–856. [Google Scholar] [CrossRef]
- Coy, P.; Yanagimachi, R. The Common and Species-Specific Roles of Oviductal Proteins in Mammalian Fertilization and Embryo Development. Bioscience 2015, 65, 973–984. [Google Scholar] [CrossRef]
- Hannan, N.J.; Stoikos, C.J.; Stephens, A.N.; Salamonsen, L.A. Depletion of high-abundance serum proteins from human uterine lavages enhances detection of lower-abundance proteins. J. Proteome Res. 2009, 8, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Mondéjar, I.; Grullón, L.A.; García-Vázquez, F.A.; Romar, R.; Coy, P. Fertilization outcome could be regulated by binding of oviductal plasminogen to oocytes and by releasing of plasminogen activators during interplay between gametes. Fertil. Steril. 2012, 97, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Coy, P.; Cánovas, S.; Mondéjar, I.; Saavedra, M.D.; Romar, R.; Grullón, L.; Matás, C.; Avilés, M. Oviduct-specific glycoprotein and heparin modulate sperm-zona pellucida interaction during fertilization and contribute to the control of polyspermy. Proc. Natl. Acad. Sci. USA 2008, 105, 15809–15814. [Google Scholar] [CrossRef]
- Coy, P.; García-Vázquez, F.A.; Visconti, P.E.; Avilés, M. Roles of the oviduct in mammalian fertilization. Reproduction 2012, 144, 649–660. [Google Scholar] [CrossRef]
- Mondéjar, I.; Martínez-Martínez, I.; Avilés, M.; Coy, P. Identification of potential oviductal factors responsible for zona pellucida hardening and monospermy during fertilization in mammals. Biol. Reprod. 2013, 89, 67. [Google Scholar] [CrossRef]
- Canovas, S.; Ivanova, E.; Romar, R.; García-Martínez, S.; Soriano-Úbeda, C.; García-Vázquez, F.A.; Saadeh, H.; Andrews, S.; Kelsey, G.; Coy, P. DNA methylation and gene expression changes derived from assisted reproductive technologies can be decreased by reproductive fluids. Elife 2017, 6, e23670. [Google Scholar] [CrossRef]
- Berlanga, O.; Bradshaw, H.B.; Vilella-Mitjana, F.; Garrido-Gómez, T.; Simón, C. How endometrial secretomics can help in predicting implantation. Placenta 2011, 32, S271–S275. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Q.; Wang, H.; Duan, E. Uterine Fluid in Pregnancy: A Biological and Clinical Outlook. Trends Mol. Med. 2017, 23, 604–614. [Google Scholar] [CrossRef]
- Salamonsen, L.A.; Edgell, T.; Rombauts, L.J.F.; Stephens, A.N.; Robertson, D.M.; Rainczuk, A.; Nie, G.; Hannan, N.J. Proteomics of the human endometrium and uterine fluid: A pathway to biomarker discovery. Fertil. Steril. 2013, 99, 1086–1092. [Google Scholar] [CrossRef]
- Di Quinzio, M.K.W.; Oliva, K.; Holdsworth, S.J.; Ayhan, M.; Walker, S.P.; Rice, G.E.; Georgiou, H.M.; Permezel, M. Proteomic analysis and characterisation of human cervico-vaginal fluid proteins. Aust. New Zeal. J. Obstet. Gynaecol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Ametzazurra, A.; Matorras, R.; García-Velasco, J.A.; Prieto, B.; Simón, L.; Martínez, A.; Nagore, D. Endometrial fluid is a specific and non-invasive biological sample for protein biomarker identification in endometriosis. Hum. Reprod. 2009, 24, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Hannan, N.J.; Nie, G.; Rainzcuk, A.; Rombauts, L.J.F.; Salamonsen, L.A. Uterine Lavage or Aspirate: Which View of the Intrauterine Environment? Reprod. Sci. 2012, 19, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.; Carneiro, I.; Corrales, F.; Elortza, F.; Paradela, A.; del Pino, M.S.; Iloro, I.; Marcilla, M.; Mora, M.I.; Valero, L.; et al. Multicentric study of the effect of pre-analytical variables in the quality of plasma samples stored in biobanks using different complementary proteomic methods. J. Proteom. 2017, 150, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Casado-Vela, J.; Rodriguez-Suarez, E.; Iloro, I.; Ametzazurra, A.; Alkorta, N.; García-Velasco, J.A.; Matorras, R.; Prieto, B.; González, S.; Nagore, D.; et al. Comprehensive proteomic analysis of human endometrial fluid aspirate. J. Proteome Res. 2009, 10, 4622–4632. [Google Scholar] [CrossRef] [PubMed]
- Tjokronegoro, A.; Sirisinha, S. Quantitative Analysis of Immunoglobulins and Albumin in Secretion of Female Reproductive Tract**Supported by Research Grants GA BMS 7209 and GA BMS 7309 from The Rockefeller Foundation and by the National Research Council of Thailand. Fertil. Steril. 1975, 26, 413–417. [Google Scholar] [CrossRef]
- Ramos-Fernández, A.; Paradela, A.; Navajas, R.; Albar, J.P. Generalized method for probability-based peptide and protein identification from tandem mass spectrometry data and sequence database searching. Mol. Cell. Proteom. 2008, 7, 1748–1754. [Google Scholar] [CrossRef]
- Xu, H.; Zhu, Q.; Ji, X.; Wang, Y.; He, Y.; Chen, Z.; Sun, Y.; Hu, S.; Yao, G. Transcriptomic Changes During the Pre-Receptive to Receptive Transition in Human Endometrium Detected by RNA-Seq. J. Clin. Endocrinol. Metab. 2014, 99, E2744–E2753. [Google Scholar]
- Woo, M.M.M.; Alkushi, A.; Verhage, H.G.; Magliocco, A.M.; Leung, P.C.K.; Gilks, C.B.; Auersperg, N. Gain of OGP, an estrogen-regulated oviduct-specific glycoprotein, is associated with the development of endometrial hyperplasia and endometrial cancer. Clin. Cancer Res. 2004, 10, 7958–7964. [Google Scholar] [CrossRef]
- Brosens, J.J.; Gellersen, B. Cyclic Decidualization of the Human Endometrium in Reproductive Health and Failure. Endocr. Rev. 2014, 35, 851–905. [Google Scholar]
- Bazer, F.W.; Spencer, T.E.; Johnson, G.A.; Burghardt, R.C.; Wu, G. Comparative aspects of implantation. Reproduction 2009, 138, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Strandell, A.; Lindhard, A. Why does hydrosalpinx reduce fertility? The importance of hydrosalpinx fluid. Hum. Reprod. 2002, 17, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.H.; Ajonuma, L.C.; Lau, E.Y.; Yeung, W.S.; Ho, P.C. Adverse effects of hydrosalpinx fluid on sperm motility and survival. Hum. Reprod. 2000, 15, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Meyer, W.R.; Castelbaum, A.J.; Somkuti, S.; Sagoskin, A.W.; Doyle, M.; Harris, J.E.; Lessey, B.A. Hydrosalpinges adversely affect markers of endometrial receptivity. Hum. Reprod. 1997, 12, 1393–1398. [Google Scholar] [CrossRef] [PubMed]
- Kodithuwakku, S.P.; Miyamoto, A.; Wijayagunawardane, M.P.B. Spermatozoa stimulate prostaglandin synthesis and secretion in bovine oviductal epithelial cells. Reproduction 2007, 133, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; He, Y.; Wang, X.-L.; Zhang, Y.-X.; Wu, Y.-M. Differentially expressed proteins among normal cervix, cervical intraepithelial neoplasia and cervical squamous cell carcinoma. Clin. Transl. Oncol. 2015, 17, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Manohar, M.; Khan, H.; Sirohi, V.K.; Das, V.; Agarwal, A.; Pandey, A.; Siddiqui, W.A.; Dwivedi, A. Alteration in endometrial proteins during early- and mid-secretory phases of the cycle in women with unexplained infertility. PLoS ONE 2014, 9, e111687. [Google Scholar] [CrossRef]
- Papp, S.M.; Fröhlich, T.; Radefeld, K.; Havlicek, V.; Kösters, M.; Yu, H.; Mayrhofer, C.; Brem, G.; Arnold, G.J.; Besenfelder, U. A novel approach to study the bovine oviductal fluid proteome using transvaginal endoscopy. Theriogenology 2019, 132, 53–61. [Google Scholar] [CrossRef]
- Coy, P.; Jimenez-Movilla, M.; Garcia-Vazquez, F.A.; Mondejar, I.; Grullon, L.; Romar, R. Oocytes use the plasminogen-plasmin system to remove supernumerary spermatozoa. Hum. Reprod. 2012, 7, 1985–1993. [Google Scholar] [CrossRef]
- Grullón, L.A.; Gadea, J.; Mondéjar, I.; Matás, C.; Romar, R.; Coy, P. How Is Plasminogen/Plasmin System Contributing to Regulate Sperm Entry into the Oocyte? Reprod. Sci. 2013, 20, 1075–1082. [Google Scholar]
- Roldan-Olarte, M.; Garcia, D.C.; Jimenez-Diaz, M.; Valdecantos, P.A.; Miceli, D.C. In vivo and in vitro expression of the plasminogen activators and urokinase type plasminogen activator receptor (u-PAR) in the pig oviduct. Anim. Reprod. Sci. 2012, 136, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Holtet, T.L.; Graversen, J.H.; Clemmensen, I.; Thogersen, H.C.; Etzerodt, M. Tetranectin, a trimeric plasminogen-binding C-type lectin. Protein Sci. 1997, 6, 1511–1515. [Google Scholar] [CrossRef] [PubMed]
- Bhusane, K.; Bhutada, S.; Chaudhari, U.; Savardekar, L.; Katkam, R.; Sachdeva, G. Secrets of Endometrial Receptivity: Some Are Hidden in Uterine Secretome. Am. J. Reprod. Immunol. 2016, 75, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Revel, A.; Achache, H. Endometrial receptivity markers, the journey to successful embryo implantation. Hum. Reprod. Update 2006, 12, 731–746. [Google Scholar]
- Aghajanova, L.; Hamilton, A.E.; Giudice, L.C. Uterine receptivity to human embryonic implantation: Histology, biomarkers, and transcriptomics. Semin. Cell Dev. Biol. 2008, 19, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Redgrove, K.A.; Anderson, A.L.; Dun, M.D.; McLaughlin, E.A.; O’Bryan, M.K.; Aitken, R.J.; Nixon, B. Involvement of multimeric protein complexes in mediating the capacitation-dependent binding of human spermatozoa to homologous zonae pellucidae. Dev. Biol. 2011, 356, 460–474. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Unruh, J.R.; Deng, M.; Slaughter, B.D.; Rubinstein, B.; Li, R. Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat. Cell Biol. 2011, 13, 1252–1258. [Google Scholar] [CrossRef]
- Kumar, A.; Dumasia, K.; Gaonkar, R.; Sonawane, S.; Kadam, L.; Balasinor, N.H. Estrogen and androgen regulate actin-remodeling and endocytosis-related genes during rat spermiation. Mol. Cell. Endocrinol. 2015, 404, 91–101. [Google Scholar] [CrossRef]
- Melideo, S.L.; Jackson, M.R.; Jorns, M.S. Biosynthesis of a central intermediate in hydrogen sulfide metabolism by a novel human sulfurtransferase and its yeast ortholog. Biochemistry 2014, 53, 4739–4753. [Google Scholar] [CrossRef]
- Haverinen, M.; Passinen, S.; Syvälä, H.; Pasanen, S.; Manninen, T.; Tuohimaa, P.; Ylikomi, T. Heat shock protein 90 and the nuclear transport of progesterone receptor. Cell Stress Chaperones 2001, 6, 256–262. [Google Scholar] [CrossRef]
- Wetendorf, M.; DeMayo, F.J. The progesterone receptor regulates implantation, decidualization, and glandular development via a complex paracrine signaling network. Mol. Cell. Endocrinol. 2012, 357, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Reed, C.A.; Osborne, E.; Kirk, E.A.; Bailey, J.A.; Fan, M.; Nephew, K.P.; Long, X.; Gize, E.A.; Bigsby, R.M. The Activating Enzyme of NEDD8 Inhibits Steroid Receptor Function. Mol. Endocrinol. 2002, 16, 315–330. [Google Scholar]
- Boeddeker, S.J.; Hess, A.P. The role of apoptosis in human embryo implantation. J. Reprod. Immunol. 2015, 108, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, C.; Greco, E.; Ubaldi, F.; Martinez, F.; Tesarik, J.; Rienzi, L.; Iacobelli, M. Caspase activity in preimplantation human embryos is not associated with apoptosis. Hum. Reprod. 2002, 17, 1584–1590. [Google Scholar]
- Bukovský, A.; Presl, J.; Zidovský, J. Association of some cell surface antigens of lymphoid cells and cell surface differentiation antigens with early rat pregnancy. Immunology 1984, 52, 631–640. [Google Scholar]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Jefferson, W.N.; Padilla–Banks, E.; Newbold, R.R. Lactoferrin is an estrogen responsive protein in the uterus of mice and rats. Reprod. Toxicol. 2000, 14, 103–110. [Google Scholar] [CrossRef]
- Wu, K.; Hoy, M.A. Clathrin heavy chain is important for viability, oviposition, embryogenesis and, possibly, systemic RNAi response in the predatory mite Metaseiulus occidentalis. PLoS ONE 2014, 9, e110874. [Google Scholar] [CrossRef]
- Biedka, S.; Micic, J.; Wilson, D.; Brown, H.; Diorio-Toth, L.; Woolford, J.L.J. Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre-60S ribosomes. J. Cell Biol. 2018, 217, 2503–2518. [Google Scholar] [CrossRef]
- Matsumiya, T.; Xing, F.; Ebina, M.; Hayakari, R.; Imaizumi, T.; Yoshida, H.; Kikuchi, H.; Topham, M.K.; Satoh, K.; Stafforini, D.M. Novel role for molecular transporter importin 9 in posttranscriptional regulation of IFN-epsilon expression. J. Immunol. 2013, 191, 1907–1915. [Google Scholar] [CrossRef]
- Hermant, P.; Francius, C.; Clotman, F.; Michiels, T. IFN-epsilon is constitutively expressed by cells of the reproductive tract and is inefficiently secreted by fibroblasts and cell lines. PLoS ONE 2013, 8, e71320. [Google Scholar] [CrossRef] [PubMed]
- López-Ferrer, D.; Martínez-Bartolomé, S.; Villar, M.; Campillos, M.; Martín-Maroto, F.; Vázquez, J. Statistical Model for Large-Scale Peptide Identification in Databases from Tandem Mass Spectra Using SEQUEST. Anal. Chem. 2004, 76, 6853–6860. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Serra, P.; Marcilla, M.; Villanueva, A.; Ramos-Fernandez, A.; Palau, A.; Leal, L.; Wahi, J.E.; Setien-Baranda, F.; Szczesna, K.; Moutinho, C.; et al. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat. Commun. 2014, 5, 3608. [Google Scholar] [CrossRef] [PubMed]
- Algarra, B.; Maillo, V.; Avilés, M.; Gutiérrez-Adán, A.; Rizos, D.; Jiménez-Movilla, M. Effects of recombinant OVGP1 protein on in vitro bovine embryo development. J. Reprod. Dev. 2018, 64, 433–443. [Google Scholar] [CrossRef] [PubMed]
| Collected Fluids | BRA-52 | BRA-54 | BRA-57 |
|---|---|---|---|
| OF—oviductal fluid (µL) | 57 | 70.9 | 49 |
| UF—uterine fluid (µL) | 260 | 100 | 59 |
| OF/UF | OF/P | UF/P | Abundance Color-Code According to the Statistical Confidence |
|---|---|---|---|
| 77.90% | 60.40% | 77.90% | truly null fraction (π0, proportion of contrasts under the null hypothesis) |
| 15 | 49 | 13 | Confident high abundant (q-value < 0.01, positive log fold change) |
| 25 | 107 | 52 | Likely high abundant (q-value < 0.05, positive log fold change) |
| 308 | 601 | 398 | Putative high abundant (q-value > 0.05 but p-value < 1−π0, positive log fold change) |
| 1016 | 552 | 958 | no differentially abundant |
| 232 | 216 | 127 | Putative low abundant (q-value > 0.05 but p-value < 1−π0, negative log fold change) |
| 16 | 33 | 27 | Likely low abundant (q-value < 0.05, negative log fold change) |
| 14 | 67 | 27 | Confident low abundant (q-value < 0.01, negative log fold change) |
| 1626 | 1625 | 1602 | # hypotheses tested |
| 70 | 256 | 119 | # hypotheses significant with q < 0.05 |
| Demographic Data | BRA-52 | BRA-54 | BRA-57 |
|---|---|---|---|
| Age (years) | 33 | 39 | 31 |
| Menarche (years) | 11 | 14 | 10 |
| Parity | G4C4 | G2P2 | G2C1P1 |
| Menstrual cycle duration | 30 days | 30 days | 29/30 days |
| Sample collection day (Menstrual Cycle phase) | Day 18: early secretory phase | Day 17: early secretory phase | Day 22: secretory phase |
| Gene Name | Gene Description | Protein Class |
|---|---|---|
| EF1D | Elongation factor 1-delta | Plasma proteins Predicted intracellular proteins |
| RL3 | 60S ribosomal protein L3 | FDA approved drug targets Plasma proteins Predicted intracellular proteins Predicted secreted proteins Ribosomal proteins |
| PDIA3 | Protein disulfide-isomerase A3 | Enzymes Plasma proteins Predicted secreted proteins |
| GSTA1 | Glutathione S-transferase A1 | Enzymes Plasma proteins Predicted intracellular proteins |
| EZRI | Ezrin | Cancer-related genes Plasma proteins Predicted intracellular proteins |
| DPYL3 | Isoform LCRMP-4 of Dihydropyrimidinase-related protein 3 | Predicted intracellular proteins |
| GARS | Glycine--tRNA ligase | Disease-related genes Plasma proteins Potential drug targets Predicted secreted proteins |
| TSTD1 | Thiosulfate:glutathione sulfurtransferase | Predicted intracellular proteins |
| THY1 | Thy-1 membrane glycoprotein | CD markers Plasma proteins Predicted membrane proteins Predicted secreted proteins |
| CLH1 | Clathrin heavy chain 1 | Cancer-related genes Plasma proteins Predicted intracellular proteins |
| IPO9 | Importin-9 | Predicted intracellular proteins Predicted secreted proteins Transporters Predicted localization Intracellular, Secreted |
| ENPL | Endoplasmin | Cancer-related genes Plasma proteins Predicted intracellular proteins Predicted secreted proteins |
| HS90A | Heat shock protein HSP 90-alpha | Cancer-related genes Plasma proteins Predicted intracellular proteins |
| NNRE | NAD(P)H-hydrate epimerase | Disease related genes Enzymes Potential drug targets Predicted secreted proteins |
| PTGR2 | Prostaglandin reductase 2 | Enzymes Predicted intracellular proteins |
| EF2 | Elongation factor 2 | Cancer-related genes Disease-related genes Plasma proteins Predicted intracellular proteins |
| COTL1 | Coactosin-like protein | Plasma proteins Predicted intracellular proteins |
| NGAL | Neutrophil gelatinase-associated lipocalin | Candidate cardiovascular disease genes Plasma proteins Predicted secreted proteins |
| SFRP4 | Secreted frizzled-related protein 4 | Candidate cardiovascular disease genes Plasma proteins Predicted secreted proteins |
| TRFL | Lactotransferrin | Cancer-related genes Plasma proteins Predicted intracellular proteins Predicted secreted proteins |
| PLMN | Plasminogen | Cancer-related genes Candidate cardiovascular disease genes Disease-related genes Enzymes FDA approved drug targets Plasma proteins Predicted secreted proteins |
| TETN | Tetranectin | Cancer-related genes Plasma proteins Predicted intracellular proteins Predicted secreted proteins |
| OVGP1 | Oviduct-specific glycoprotein | Plasma proteins Predicted secreted proteins |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canha-Gouveia, A.; Paradela, A.; Ramos-Fernández, A.; Prieto-Sánchez, M.T.; Sánchez-Ferrer, M.L.; Corrales, F.; Coy, P. Which Low-Abundance Proteins are Present in the Human Milieu of Gamete/Embryo Maternal Interaction? Int. J. Mol. Sci. 2019, 20, 5305. https://doi.org/10.3390/ijms20215305
Canha-Gouveia A, Paradela A, Ramos-Fernández A, Prieto-Sánchez MT, Sánchez-Ferrer ML, Corrales F, Coy P. Which Low-Abundance Proteins are Present in the Human Milieu of Gamete/Embryo Maternal Interaction? International Journal of Molecular Sciences. 2019; 20(21):5305. https://doi.org/10.3390/ijms20215305
Chicago/Turabian StyleCanha-Gouveia, Analuce, A. Paradela, António Ramos-Fernández, Maria Teresa Prieto-Sánchez, Maria Luisa Sánchez-Ferrer, Fernando Corrales, and Pilar Coy. 2019. "Which Low-Abundance Proteins are Present in the Human Milieu of Gamete/Embryo Maternal Interaction?" International Journal of Molecular Sciences 20, no. 21: 5305. https://doi.org/10.3390/ijms20215305
APA StyleCanha-Gouveia, A., Paradela, A., Ramos-Fernández, A., Prieto-Sánchez, M. T., Sánchez-Ferrer, M. L., Corrales, F., & Coy, P. (2019). Which Low-Abundance Proteins are Present in the Human Milieu of Gamete/Embryo Maternal Interaction? International Journal of Molecular Sciences, 20(21), 5305. https://doi.org/10.3390/ijms20215305






