Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development
Abstract
1. Introduction
1.1. Role of BAF Complex in Chromatin Remodeling
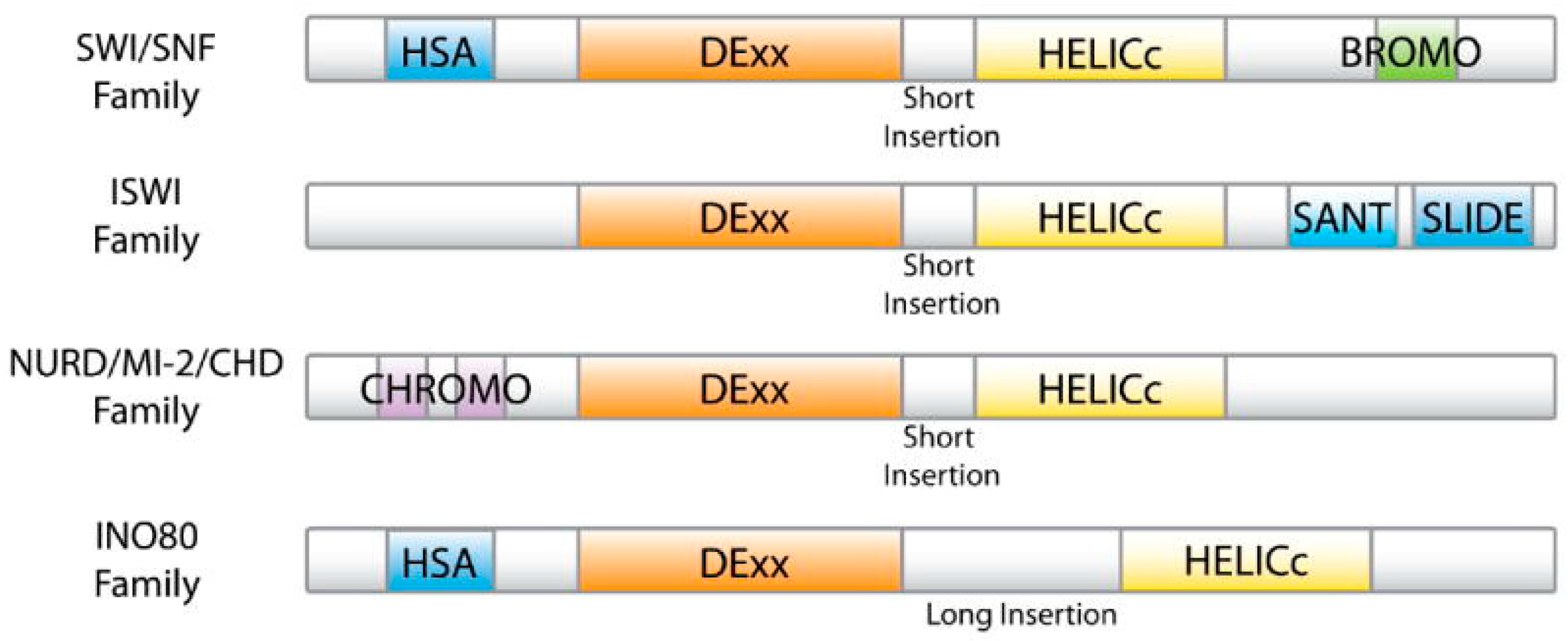
1.2. Structure of BAF Complex
1.3. Deregulation of BAF Complex Associated with Diseases
1.4. Intrinsically Disordered Proteins (IDPs)
2. Results and Discussion
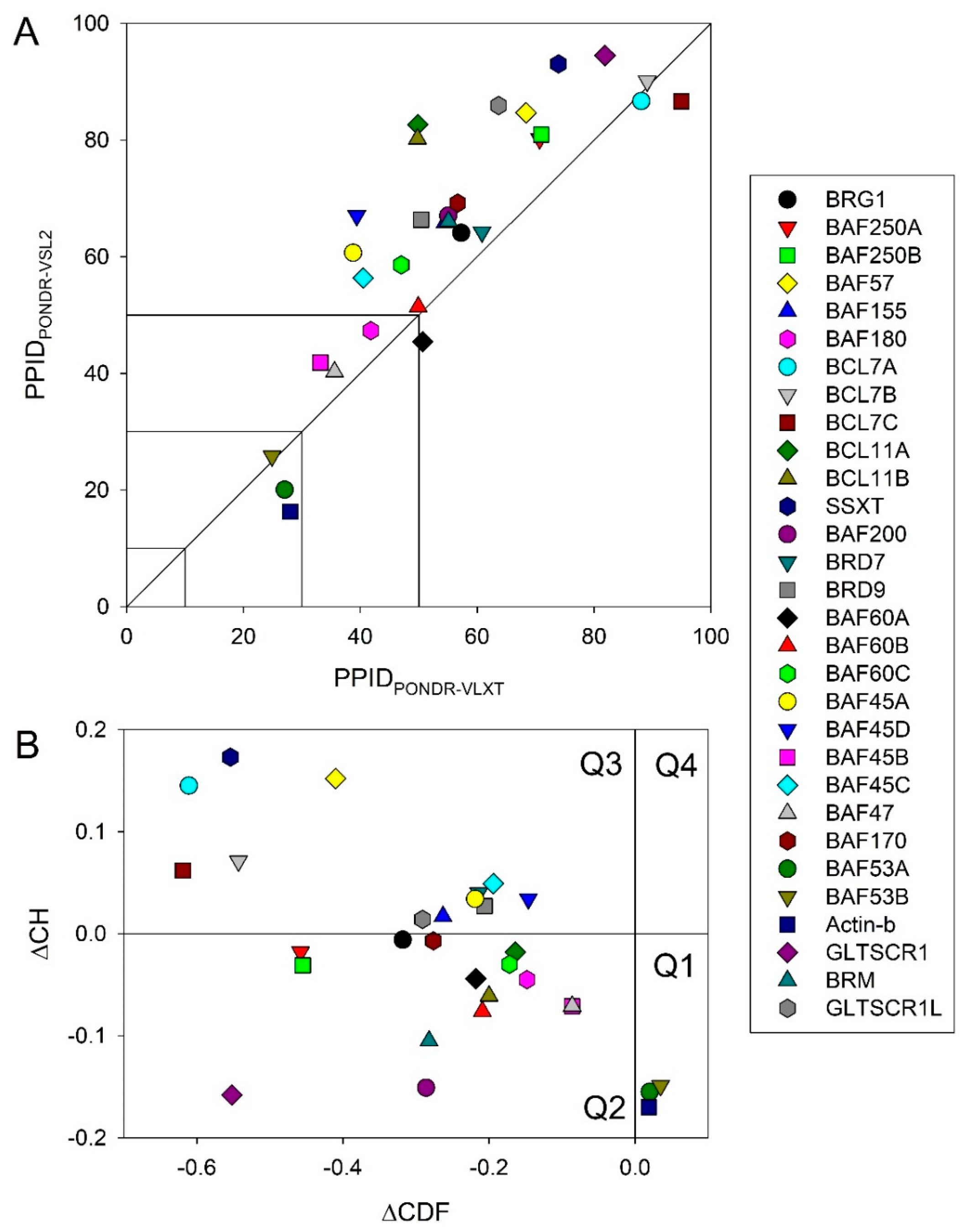
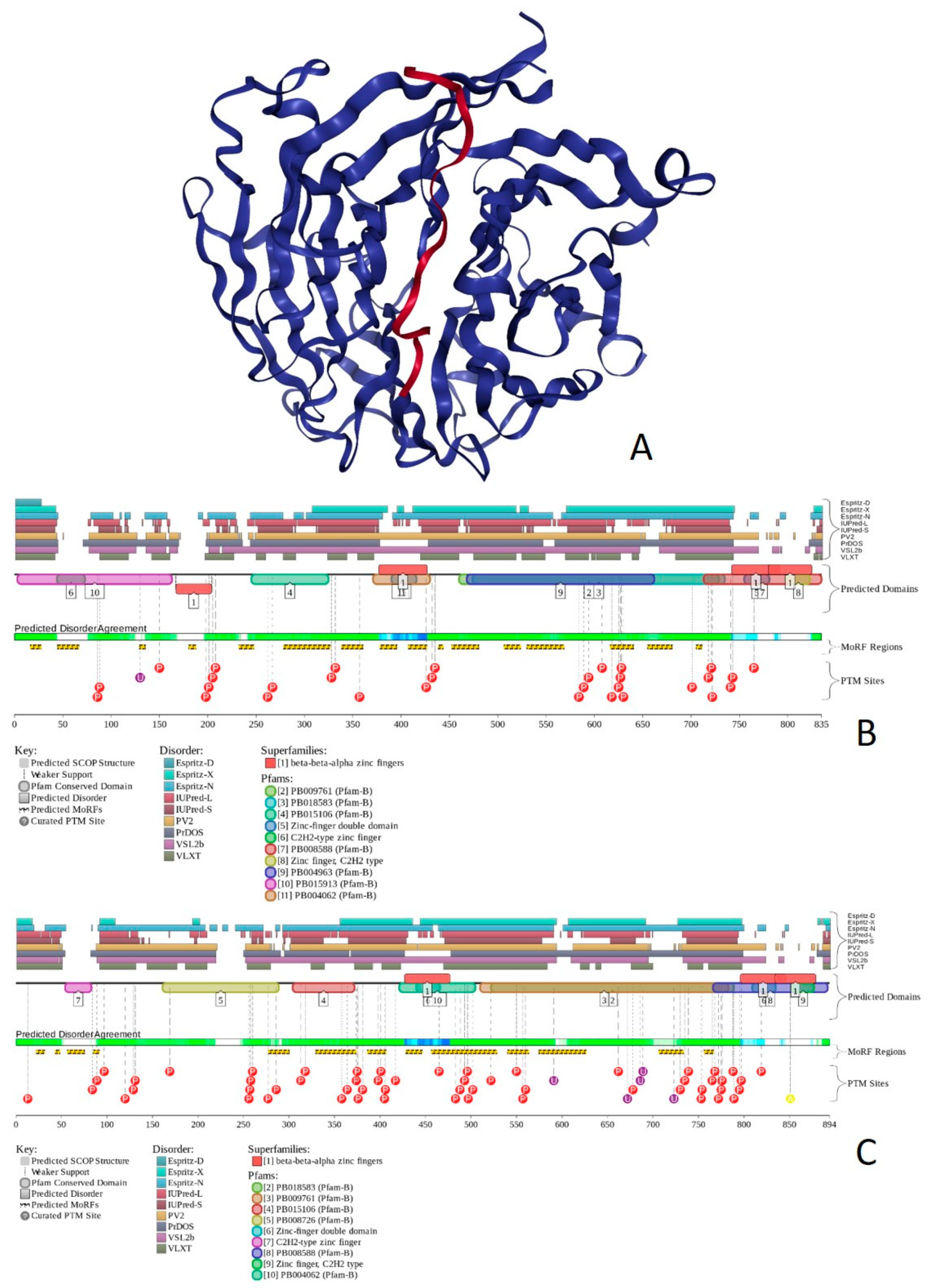
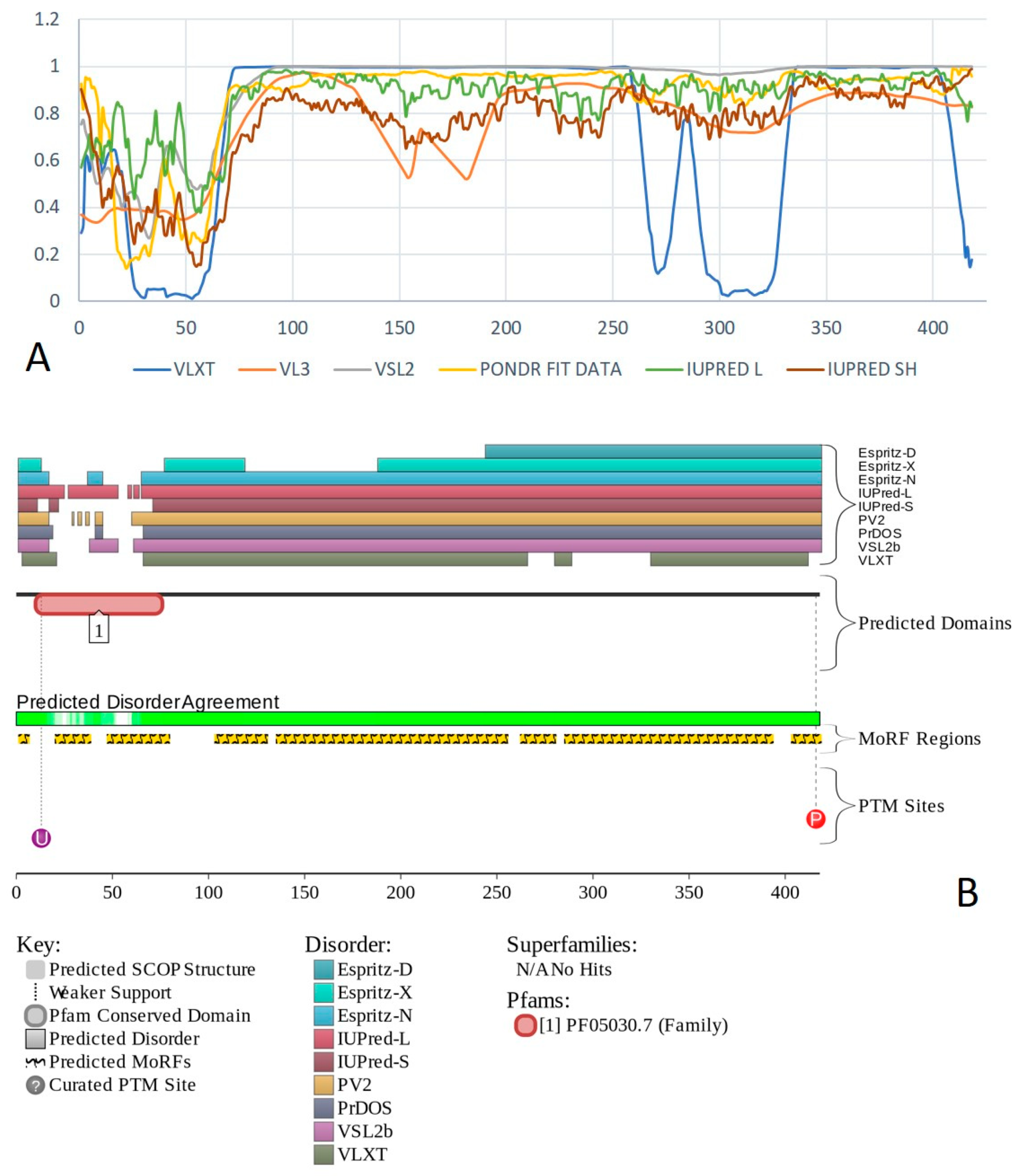
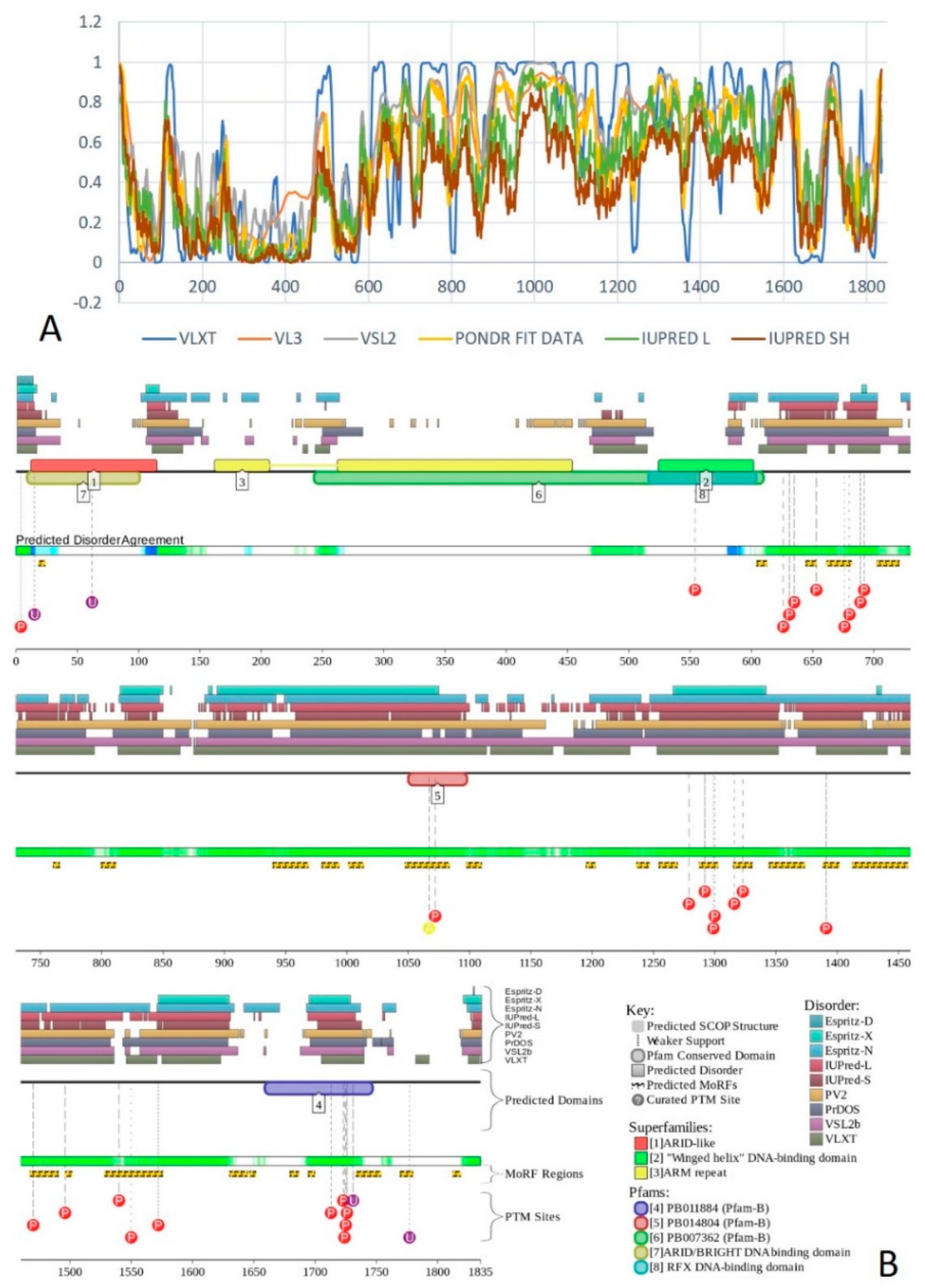
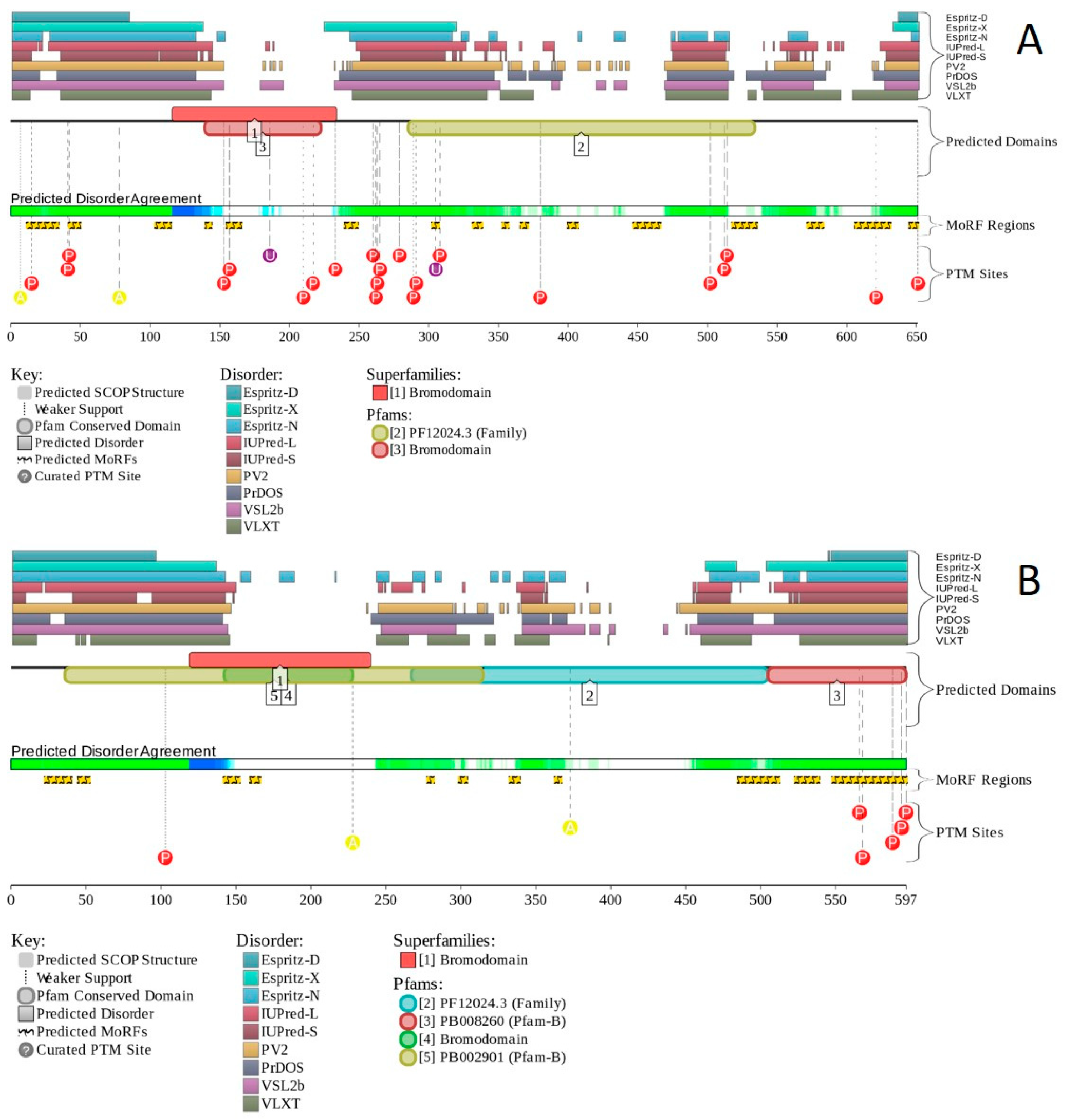
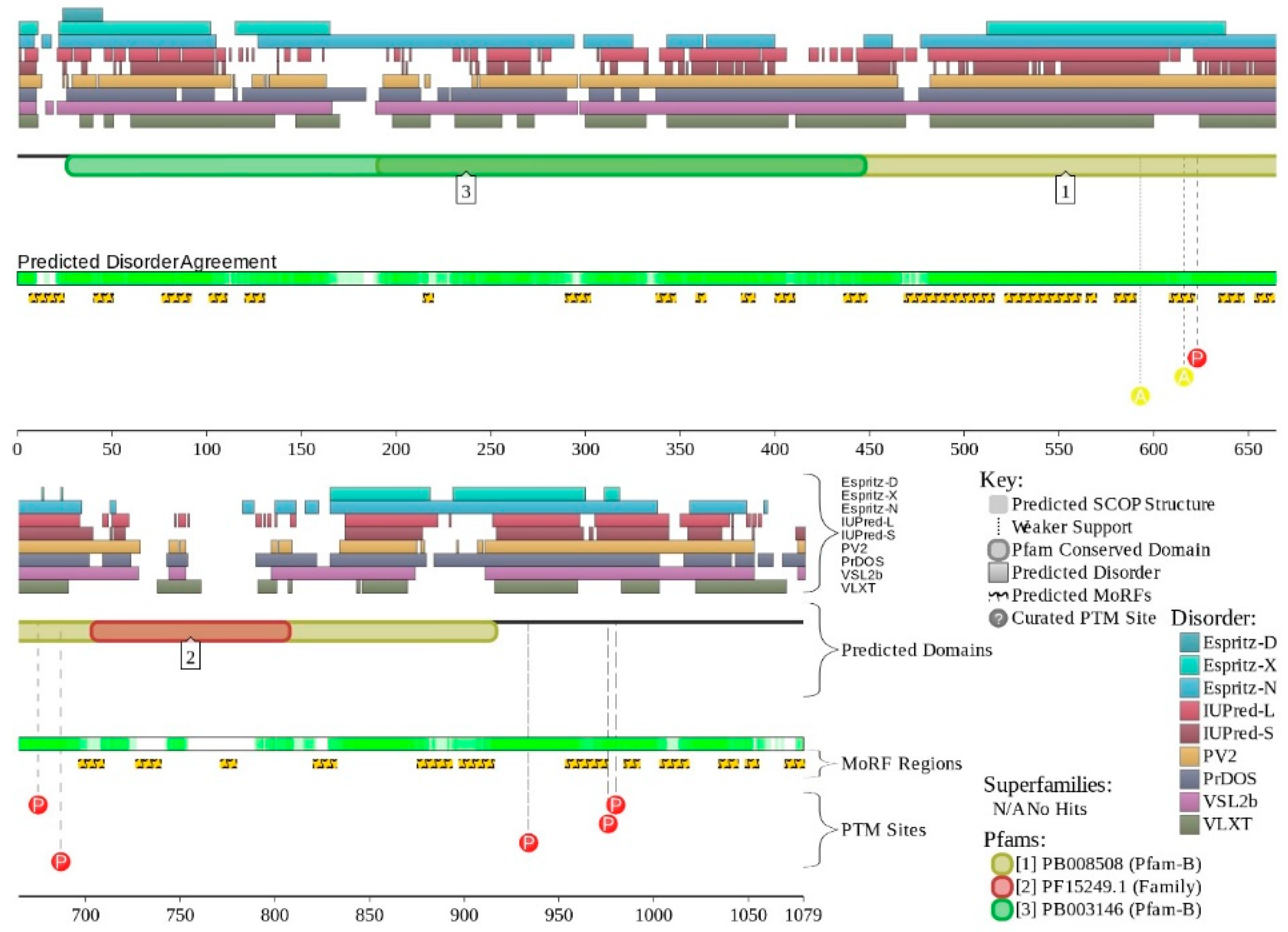
2.1. Global Disorder Analysis of the BAF Subunits
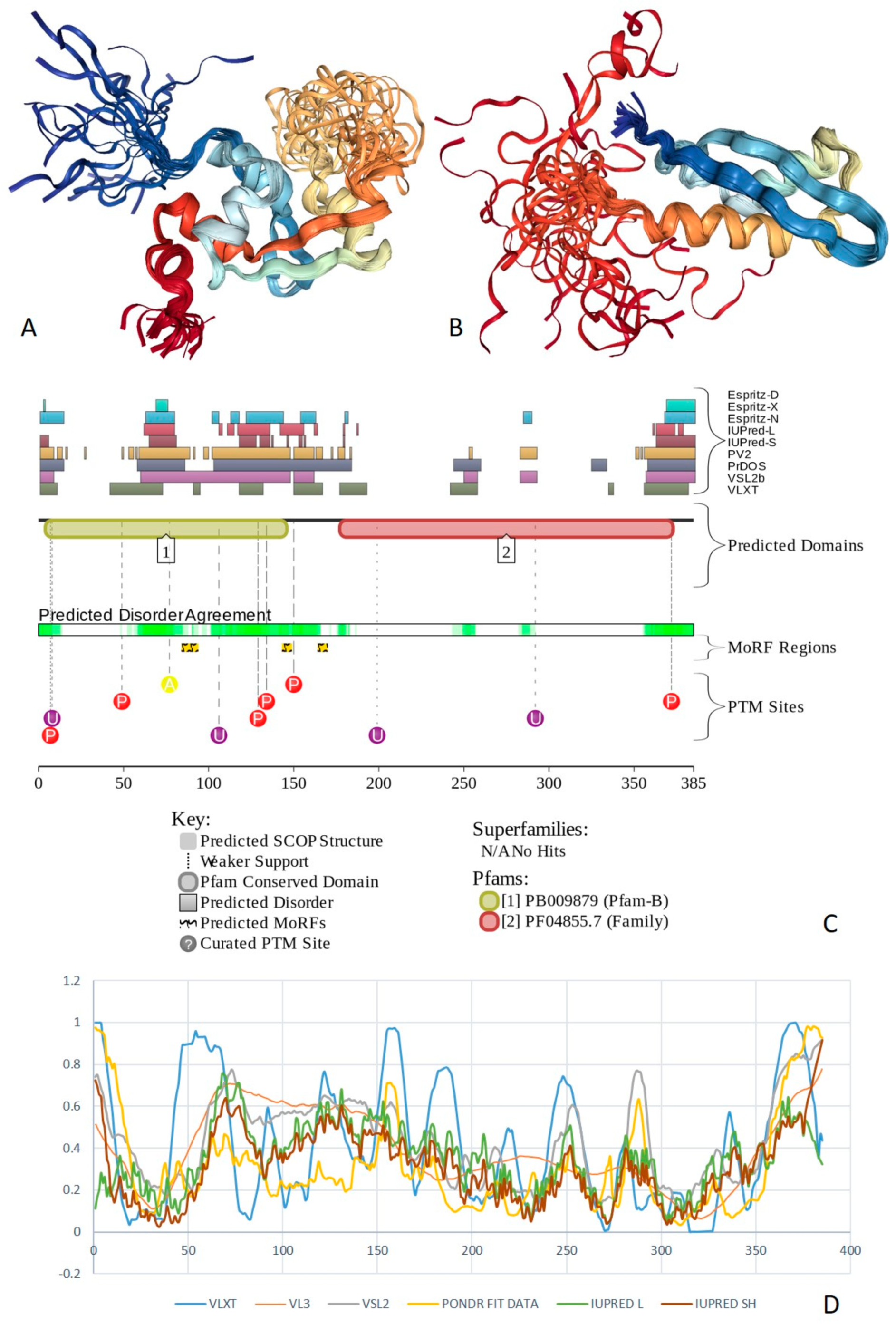
2.2. Transcriptional Activator BRG1 or ATP-Dependent Helicase SMARCA4 (PPID = 61.3%)
2.3. Protein Brahma Homolog BRM or Probable Global Transcription Activator SNF2L2 (PPID = 60.6%)
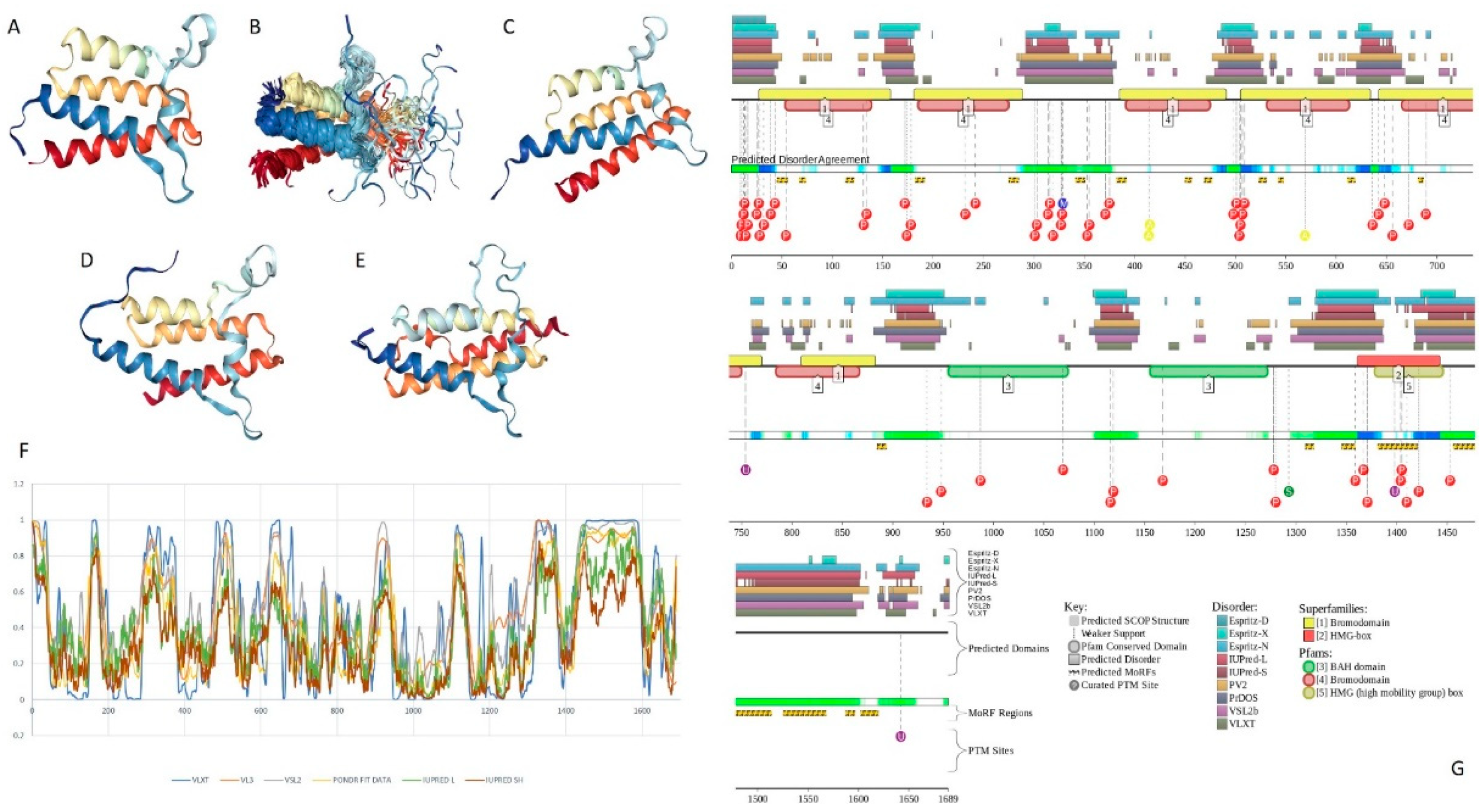
2.4. BAF155 (PPID = 59.2%)
2.5. BAF170 (PPID = 63.1%)
2.6. BAF47 (PPID = 35.8%)
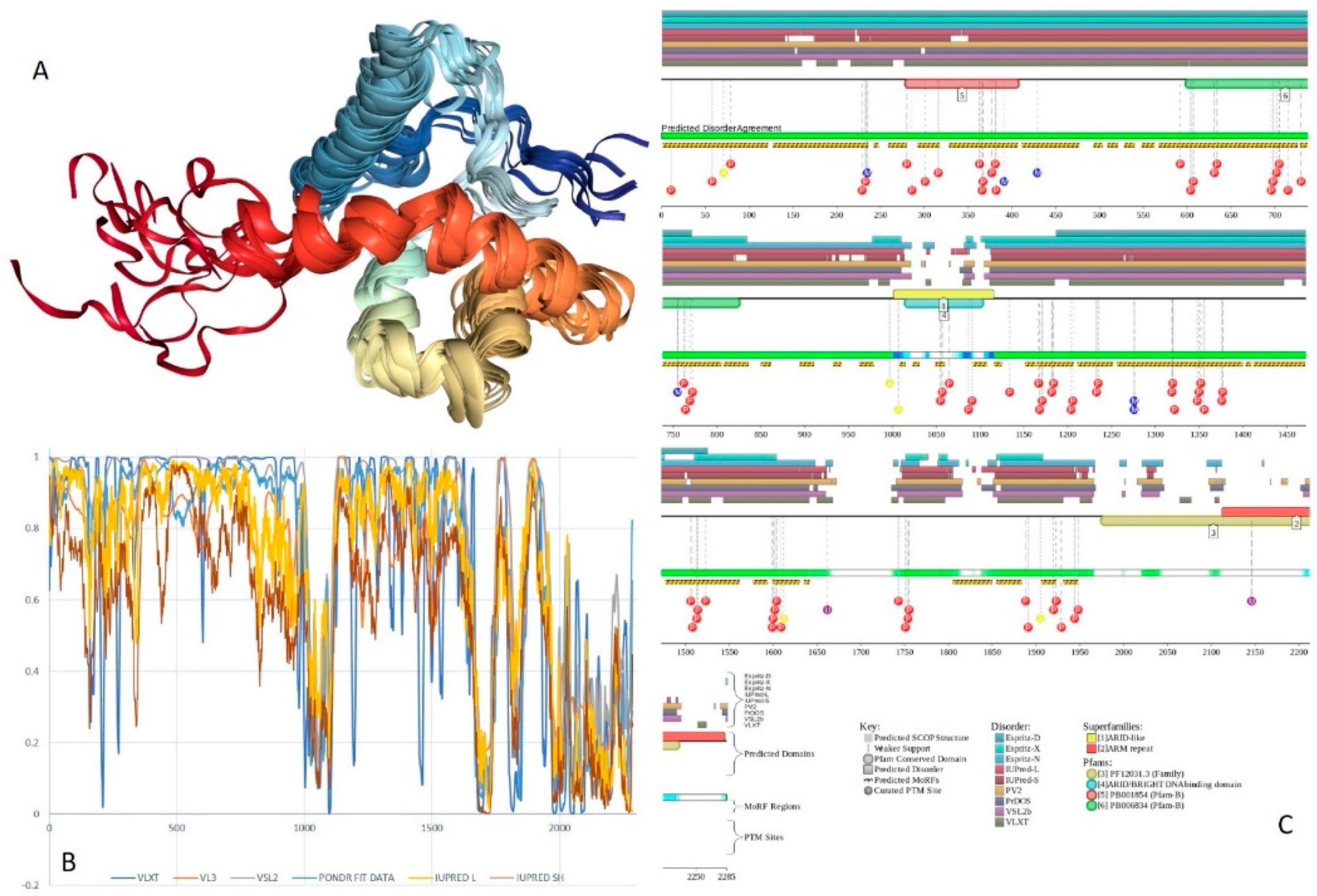
2.7. BAF250A (PPID = 75.8%)
2.8. BAF250B (PPID = 76.9%)
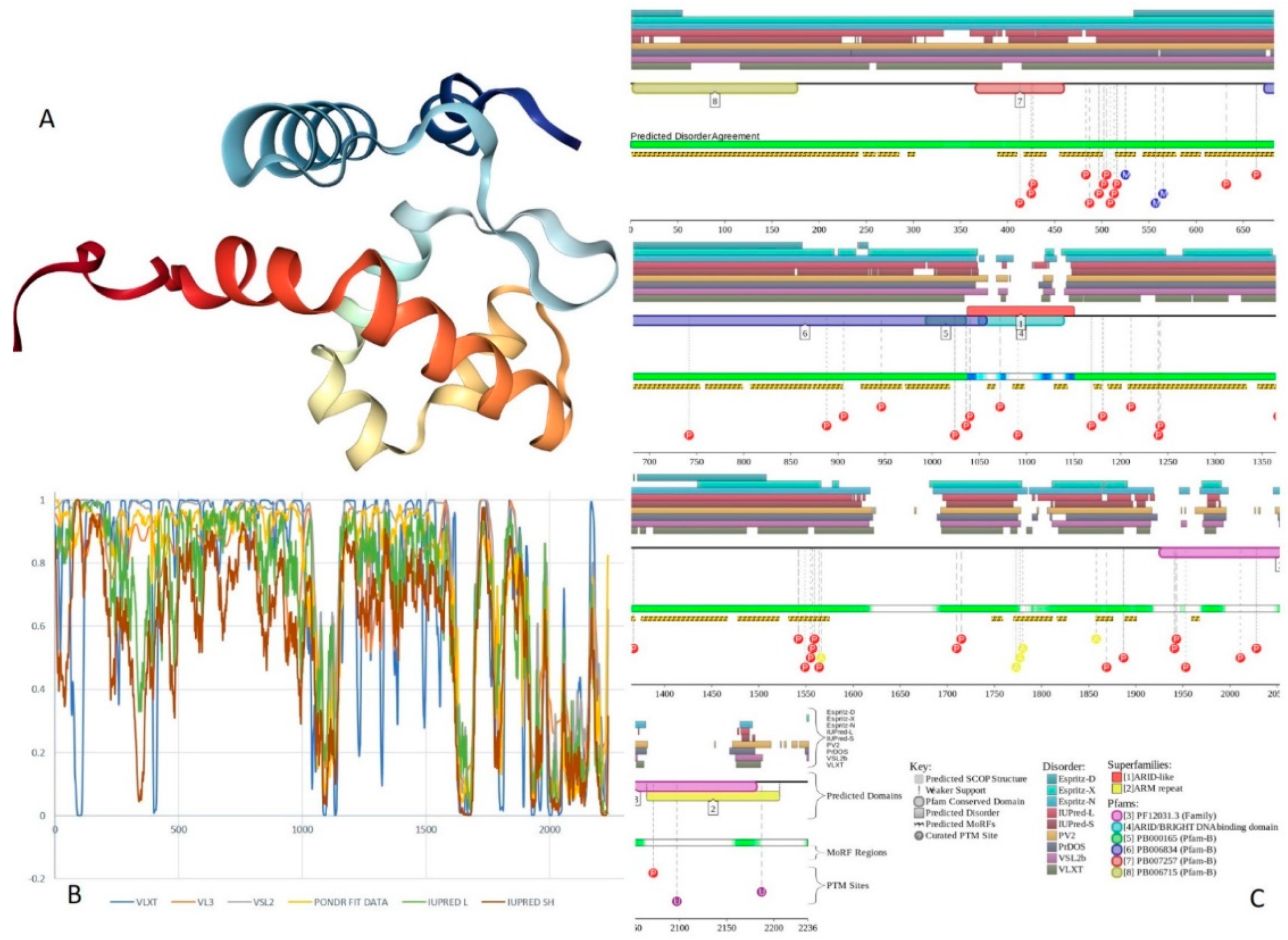
2.9. BAF57 (PPID = 77.6%)
2.10. BAF180 (PPID = 43.2%)
2.11. BCL7A (PPID = 87.6%), BCL7B (PPID = 89.6%), and BCL7C (PPID = 90.0%)
2.12. BCL11A (PPID = 69.0%) and BCL11B (PPID = 69.8%)
2.13. SSXT (PPID = 84.1%)
2.14. BAF200/ARID2 (PPID = 61.3%)
2.15. BRD7 (PPID = 61.2%) and BRD9 (PPID = 60.1%)
2.16. BAF60A/SMARCD1 (PPID = 43.8%), BAF60B/SMARCD2 (PPID = 49.3%), and BAF60C/SMARCD3 (PPID = 50.5%)
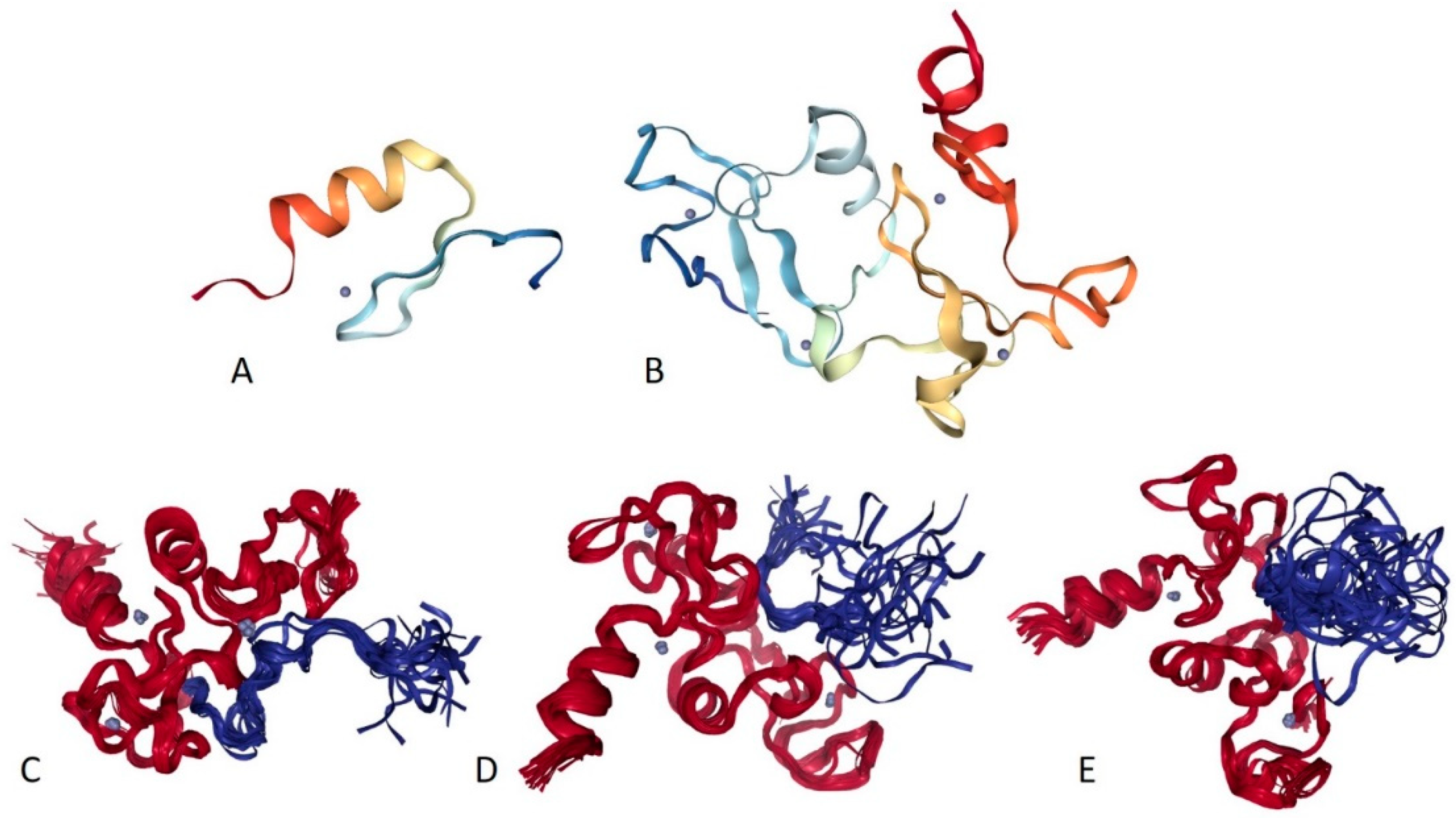
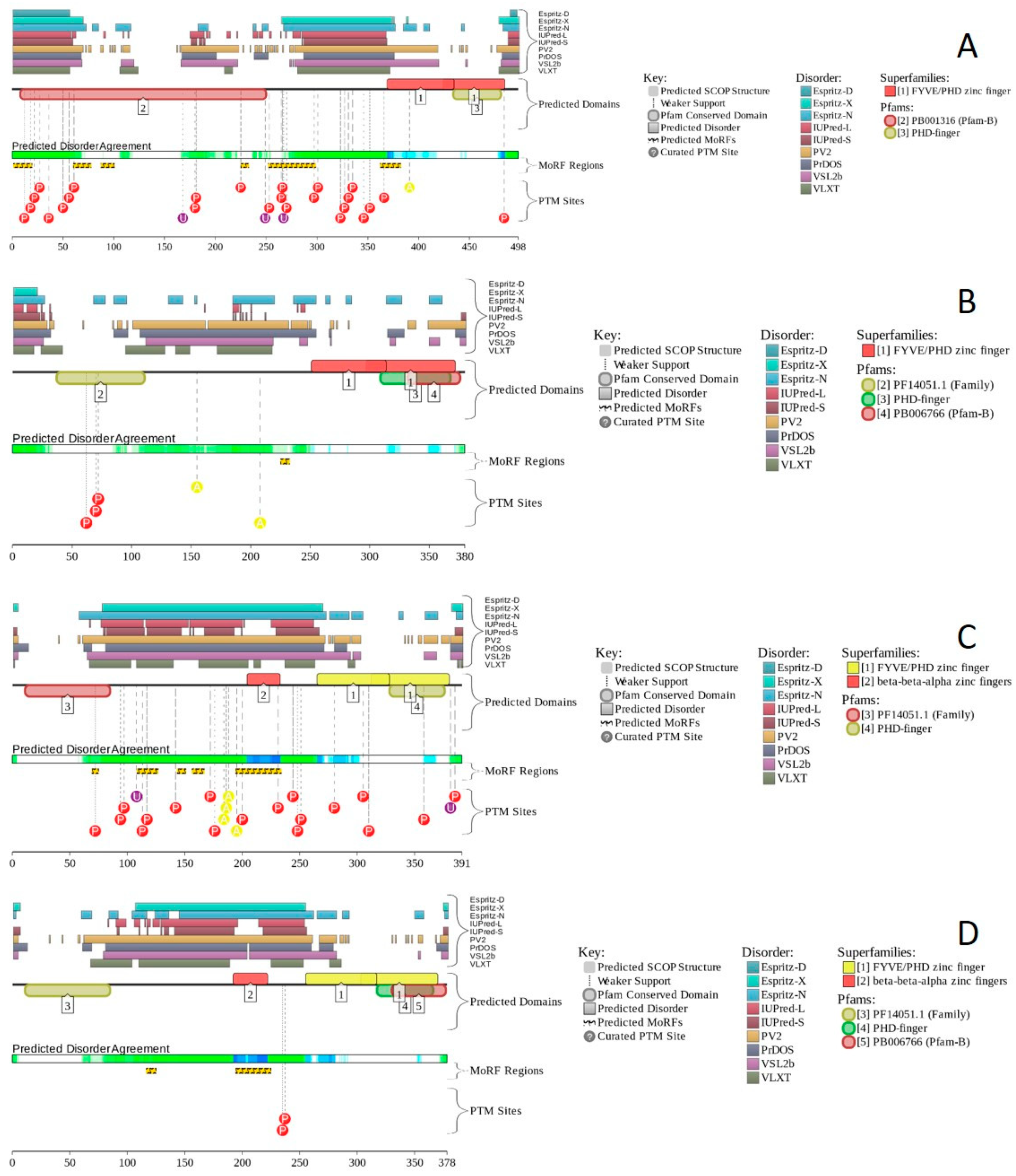
2.17. PHF10/BAF45A (PPID = 53.3%), DPF1/BAF45B (PPID = 35.8%), DPF2/BAF45D (PPID = 59.7%), and DPF3/BAF45C (PPID = 50.5%)
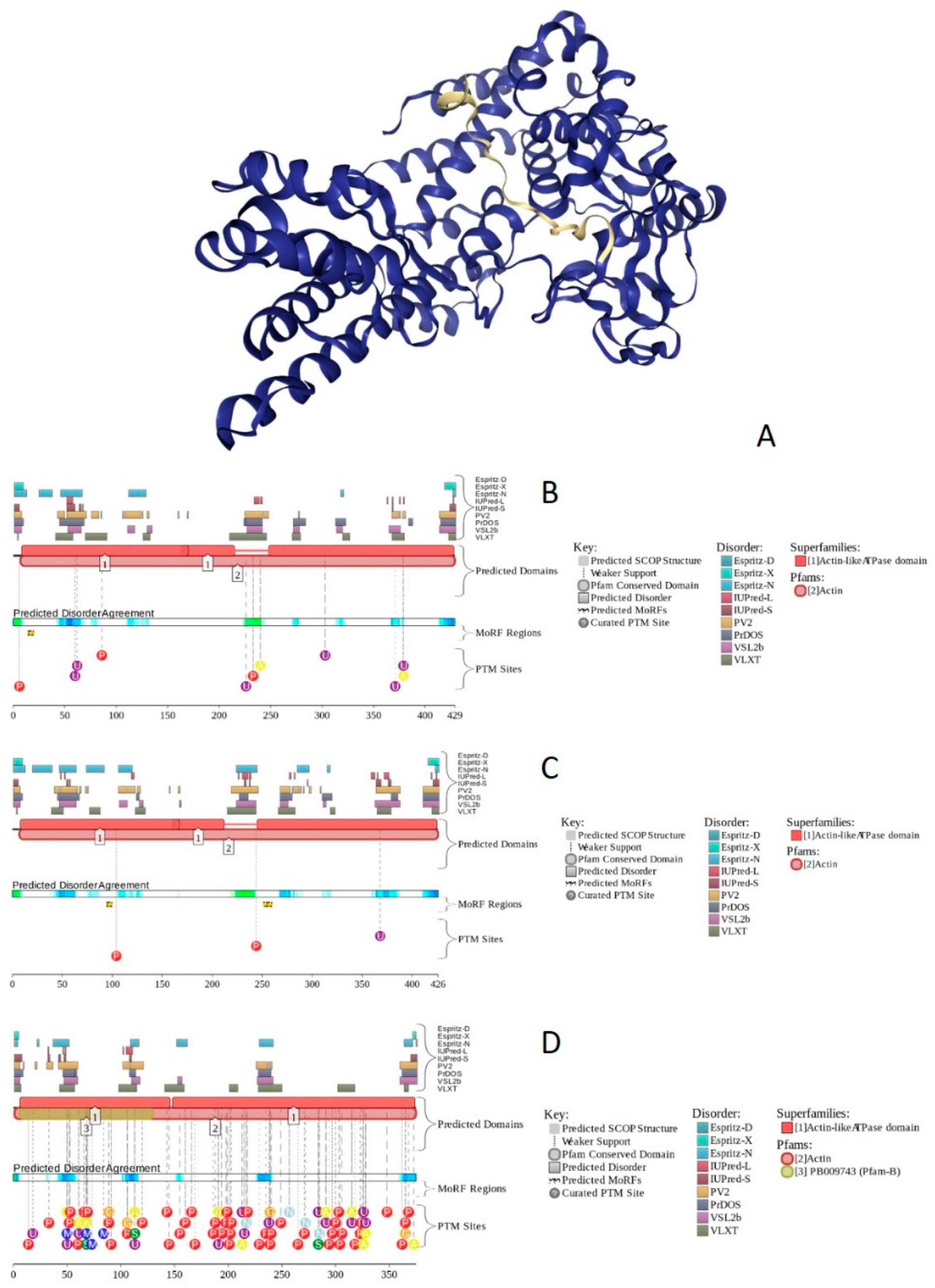
2.18. BAF53A/ACTL6A (PPID = 23.5%), BAF53B/ACTL6B (PPID = 25.4%), and β-Actin/ACTB (PPID = 22.1%)
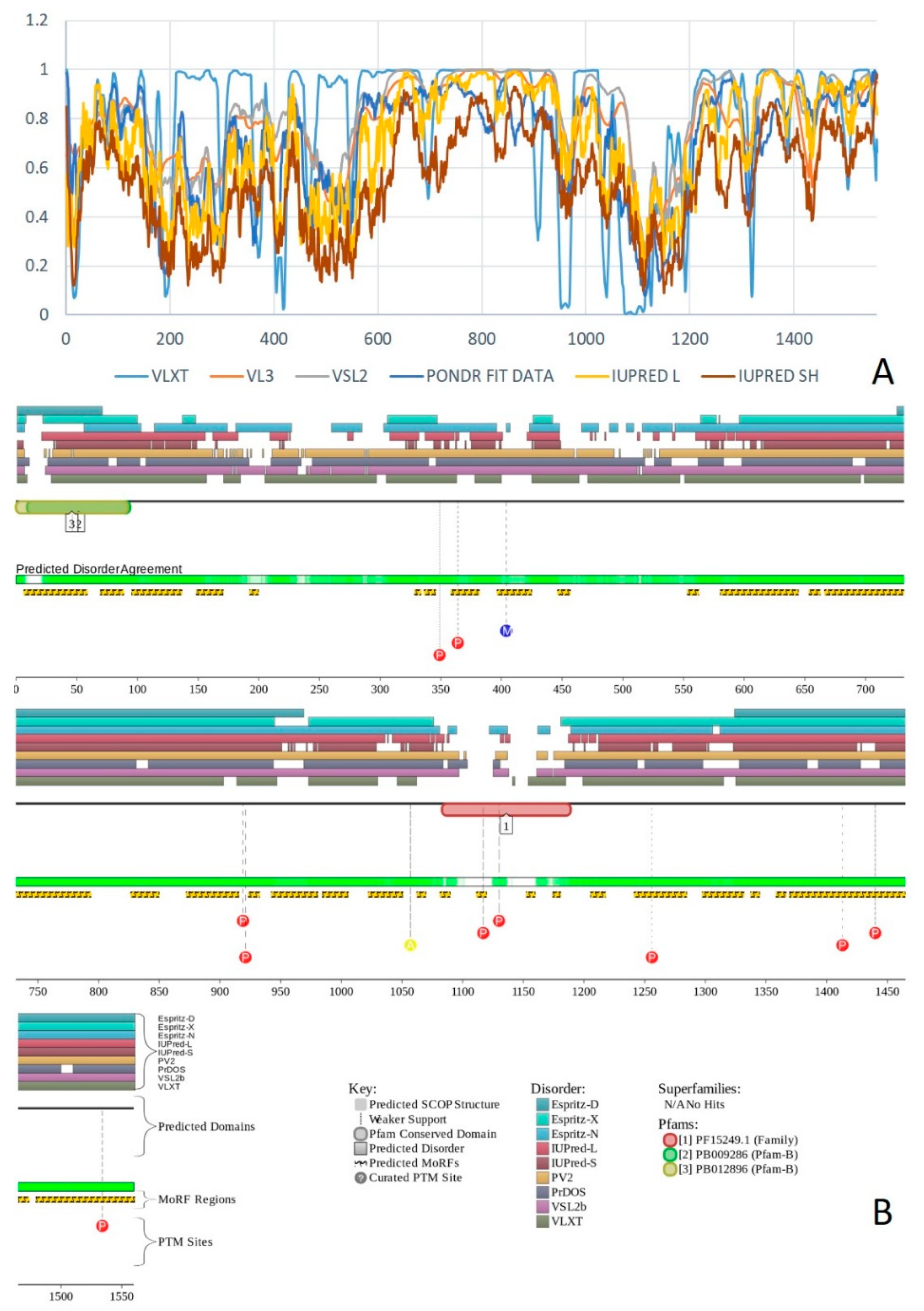
2.19. GLTSCR1/BICRA (PPID = 88.2%) and GLTSCR1L/BICRAL (PPID = 74.8%)
3. Experimental Section
3.1. Datasets
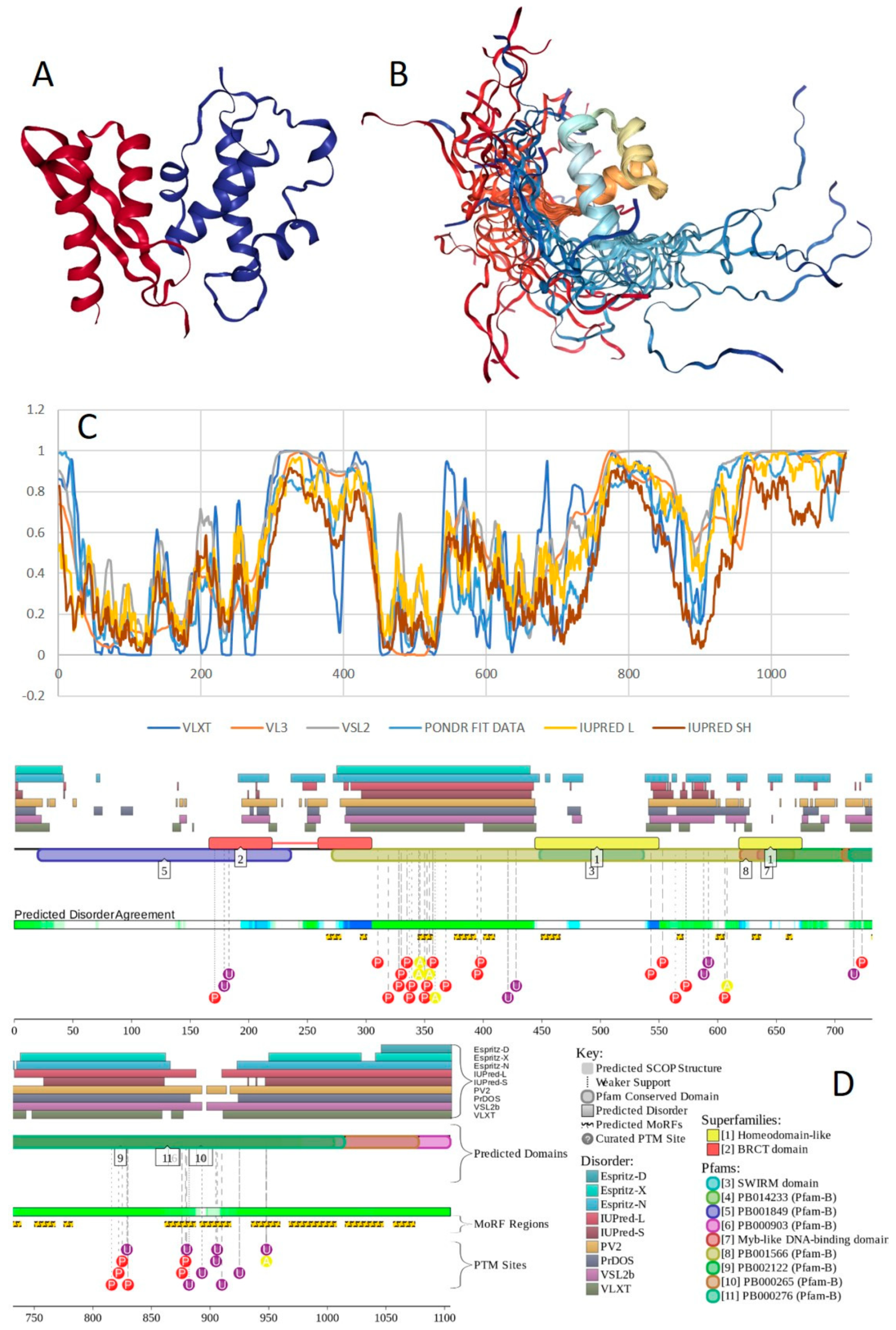
3.2. Computational Characterization of Intrinsic Disorder in the BAF Complex
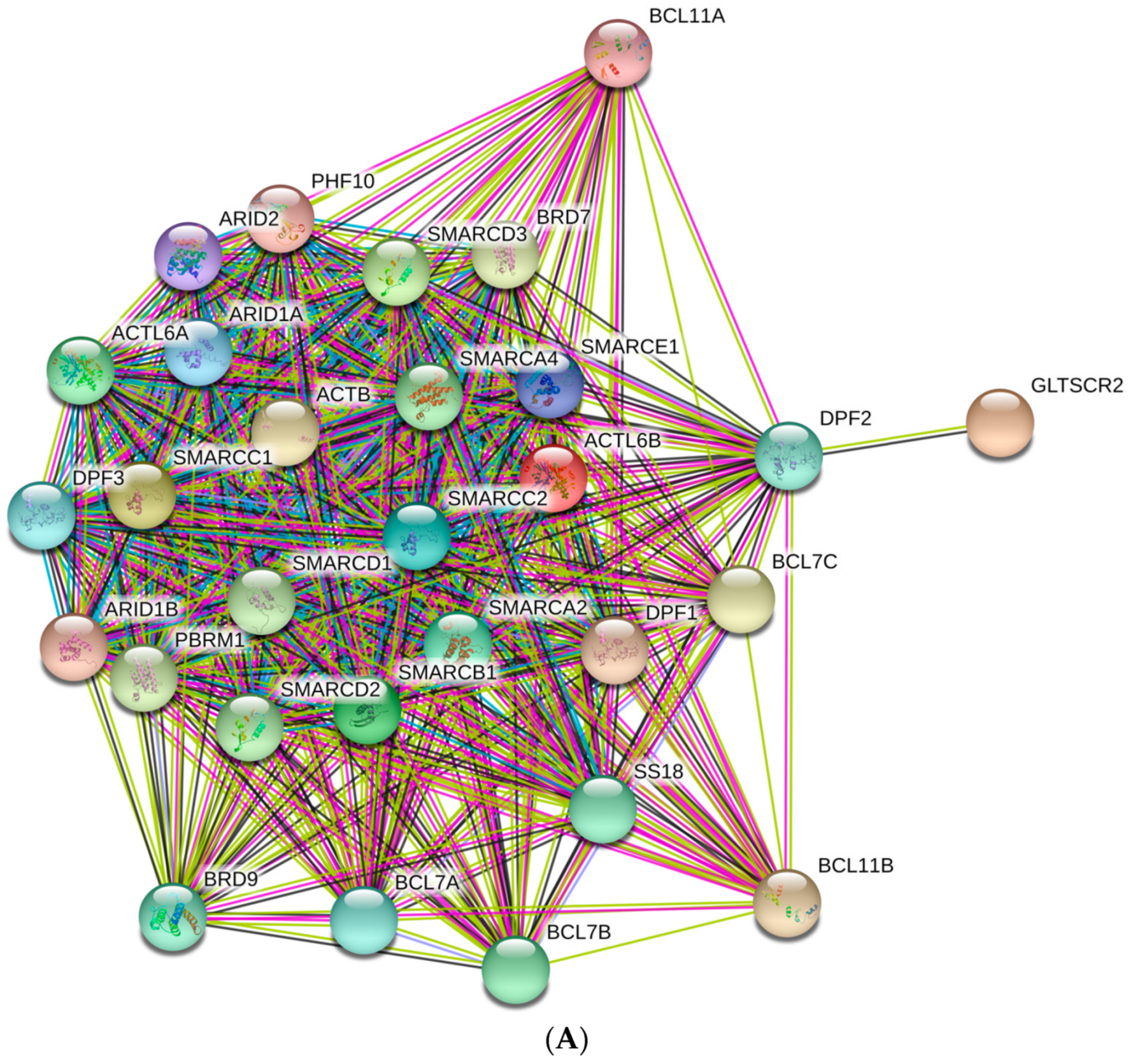
3.3. Computational Evaluation of Interactability of the Human BAF Complex Subunits
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Arnaud, O.; Le Loarer, F.; Tirode, F. BAFfling pathologies: Alterations of BAF complexes in cancer. Cancer Lett. 2018, 419, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Trotter, K.W.; Archer, T.K. Nuclear receptors and chromatin remodeling machinery. Mol. Cell. Endocrinol. 2007, 265–266, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Vignali, M.; Hassan, A.H.; Neely, K.E.; Workman, J.L. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000, 20, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Cui, K.; Murray, D.M.; Ling, C.; Xue, Y.; Gerstein, A.; Parsons, R.; Zhao, K.; Wang, W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005, 19, 1662–1667. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef] [PubMed]
- Lorch, Y.; Maier-Davis, B.; Kornberg, R.D. Mechanism of chromatin remodeling. Proc. Natl. Acad. Sci. USA 2010, 107, 3458–3462. [Google Scholar] [CrossRef]
- Rando, O.J.; Zhao, K.; Janmey, P.; Crabtree, G.R. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA 2002, 99, 2824–2829. [Google Scholar] [CrossRef]
- Panamarova, M.; Cox, A.; Wicher, K.B.; Butler, R.; Bulgakova, N.; Jeon, S.; Rosen, B.; Seong, R.H.; Skarnes, W.; Crabtree, G.; et al. The BAF chromatin remodelling complex is an epigenetic regulator of lineage specification in the early mouse embryo. Development 2016, 143, 1271–1283. [Google Scholar] [CrossRef]
- Cui, K.; Tailor, P.; Liu, H.; Chen, X.; Ozato, K.; Zhao, K. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol. Cell Biol. 2004, 24, 4476–4486. [Google Scholar] [CrossRef]
- Liu, L.; Wan, X.; Zhou, P.; Zhou, X.; Zhang, W.; Hui, X.; Yuan, X.; Ding, X.; Zhu, R.; Meng, G.; et al. The chromatin remodeling subunit Baf200 promotes normal hematopoiesis and inhibits leukemogenesis. J. Hematol. Oncol. 2018, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Nogales, E.; Ciferri, C. Structure and function of SWI/SNF chromatin remodeling complexes and mechanistic implications for transcription. Prog. Biophys. Mol. Biol. 2010, 102, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Singleton, M.R.; Wigley, D.B. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002, 184, 1819–1826. [Google Scholar] [CrossRef] [PubMed]
- Hota, S.K.; Johnson, J.R.; Verschueren, E.; Thomas, R.; Blotnick, A.M.; Zhu, Y.; Sun, X.; Pennacchio, L.A.; Krogan, N.J.; Bruneau, B.G. Dynamic BAF chromatin remodeling complex subunit inclusion promotes temporally distinct gene expression programs in cardiogenesis. Development 2019, 146, dev174086. [Google Scholar] [CrossRef]
- Wang, W.; Xue, Y.; Zhou, S.; Kuo, A.; Cairns, B.R.; Crabtree, G.R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996, 10, 2117–2130. [Google Scholar] [CrossRef]
- Xue, Y.; Canman, J.C.; Lee, C.S.; Nie, Z.; Yang, D.; Moreno, G.T.; Young, M.K.; Salmon, E.D.; Wang, W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13015–13020. [Google Scholar] [CrossRef]
- St Pierre, R.; Kadoch, C. Mammalian SWI/SNF complexes in cancer: Emerging therapeutic opportunities. Curr. Opin. Genet. Dev. 2017, 42, 56–67. [Google Scholar] [CrossRef]
- Alpsoy, A.; Dykhuizen, E.C. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J. Biol. Chem. 2018, 293, 3892–3903. [Google Scholar] [CrossRef]
- Alfert, A.; Moreno, N.; Kerl, K. The BAF complex in development and disease. Epigenetics Chromatin 2019, 12, 19. [Google Scholar] [CrossRef]
- Yan, L.; Xie, S.; Du, Y.; Qian, C. Structural Insights into BAF47 and BAF155 Complex Formation. J. Mol. Biol. 2017, 429, 1650–1660. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, W.; Rando, O.J.; Xue, Y.; Swiderek, K.; Kuo, A.; Crabtree, G.R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 1998, 95, 625–636. [Google Scholar] [CrossRef]
- Lu, C.; Allis, C.D. SWI/SNF complex in cancer. Nat. Genet. 2017, 49, 178–179. [Google Scholar] [CrossRef] [PubMed]
- Heare, T.; Hensley, M.A.; Dell’Orfano, S. Bone tumors: Osteosarcoma and Ewing’s sarcoma. Curr. Opin. Pediatr. 2009, 21, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Burchill, S.A. Ewing’s sarcoma: Diagnostic, prognostic, and therapeutic implications of molecular abnormalities. J. Clin. Pathol. 2003, 56, 96–102. [Google Scholar] [CrossRef]
- Herrero-Martin, D.; Fourtouna, A.; Niedan, S.; Riedmann, L.T.; Schwentner, R.; Aryee, D.N. Factors Affecting EWS-FLI1 Activity in Ewing’s Sarcoma. Sarcoma 2011, 2011, 352580. [Google Scholar] [CrossRef]
- Zilfou, J.T.; Lowe, S.W. Tumor suppressive functions of p53. Cold Spring Harb. Perspect Biol. 2009, 1, a001883. [Google Scholar] [CrossRef]
- Deneen, B.; Denny, C.T. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene 2001, 20, 6731–6741. [Google Scholar] [CrossRef]
- Batra, S.; Reynolds, C.P.; Maurer, B.J. Fenretinide cytotoxicity for Ewing’s sarcoma and primitive neuroectodermal tumor cell lines is decreased by hypoxia and synergistically enhanced by ceramide modulators. Cancer Res. 2004, 64, 5415–5424. [Google Scholar] [CrossRef]
- van der Schaft, D.W.; Hillen, F.; Pauwels, P.; Kirschmann, D.A.; Castermans, K.; Egbrink, M.G.; Tran, M.G.; Sciot, R.; Hauben, E.; Hogendoorn, P.C.; et al. Tumor cell plasticity in Ewing sarcoma, an alternative circulatory system stimulated by hypoxia. Cancer Res. 2005, 65, 11520–11528. [Google Scholar] [CrossRef]
- Shi, Y.; Chang, M.; Wang, F.; Ouyang, X.; Jia, Y.; Du, H. Role and mechanism of hypoxia-inducible factor-1 in cell growth and apoptosis of breast cancer cell line MDA-MB-231. Oncol. Lett. 2010, 1, 657–662. [Google Scholar] [CrossRef]
- Herrero-Martin, D.; Osuna, D.; Ordonez, J.L.; Sevillano, V.; Martins, A.S.; Mackintosh, C.; Campos, M.; Madoz-Gurpide, J.; Otero-Motta, A.P.; Caballero, G.; et al. Stable interference of EWS-FLI1 in an Ewing sarcoma cell line impairs IGF-1/IGF-1R signalling and reveals TOPK as a new target. Br. J. Cancer 2009, 101, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.S.; Mackintosh, C.; Martin, D.H.; Campos, M.; Hernandez, T.; Ordonez, J.L.; de Alava, E. Insulin-like growth factor I receptor pathway inhibition by ADW742, alone or in combination with imatinib, doxorubicin, or vincristine, is a novel therapeutic approach in Ewing tumor. Clin. Cancer Res. 2006, 12, 3532–3540. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.J.; Kadoch, C. Disruption of mammalian SWI/SNF and polycomb complexes in human sarcomas: Mechanisms and therapeutic opportunities. J. Pathol. 2018, 244, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.F.; Shorter, J. RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 2017, 474, 1417–1438. [Google Scholar] [CrossRef] [PubMed]
- Lieutaud, P.; Ferron, F.; Uversky, A.V.; Kurgan, L.; Uversky, V.N.; Longhi, S. How disordered is my protein and what is its disorder for? A guide through the "dark side" of the protein universe. Intrinsically Disord. Proteins 2016, 4, e1259708. [Google Scholar] [CrossRef]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 1231–1264. [Google Scholar] [CrossRef]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef]
- Dunker, A.K.; Obradovic, Z.; Romero, P.; Garner, E.C.; Brown, C.J. Intrinsic protein disorder in complete genomes. Genome Inform. Ser. Workshop Genome Inform. 2000, 11, 161–171. [Google Scholar]
- Peng, Z.; Yan, J.; Fan, X.; Mizianty, M.J.; Xue, B.; Wang, K.; Hu, G.; Uversky, V.N.; Kurgan, L. Exceptionally abundant exceptions: Comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol. Life Sci. 2015, 72, 137–151. [Google Scholar] [CrossRef]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly order in protein intrinsic disorder distribution: Disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012, 30, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Tokuriki, N.; Oldfield, C.J.; Uversky, V.N.; Berezovsky, I.N.; Tawfik, D.S. Do viral proteins possess unique biophysical features? Trends Biochem. Sci. 2009, 34, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Williams, R.W.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Archaic chaos: Intrinsically disordered proteins in Archaea. BMC Syst. Biol. 2010, 4 (Suppl. 1), S1. [Google Scholar] [CrossRef]
- Uversky, V.N. The mysterious unfoldome: Structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010, 2010, 568068. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Dosztanyi, Z.; Simon, I. Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J. Proteome Res. 2006, 5, 1996–2000. [Google Scholar] [CrossRef]
- Krasowski, M.D.; Reschly, E.J.; Ekins, S. Intrinsic disorder in nuclear hormone receptors. J. Proteome Res. 2008, 7, 4359–4372. [Google Scholar] [CrossRef]
- Shimizu, K.; Toh, H. Interaction between intrinsically disordered proteins frequently occurs in a human protein-protein interaction network. J. Mol. Biol. 2009, 392, 1253–1265. [Google Scholar] [CrossRef]
- Pentony, M.M.; Jones, D.T. Modularity of intrinsic disorder in the human proteome. Proteins 2010, 78, 212–221. [Google Scholar] [CrossRef]
- Tompa, P.; Kalmar, L. Power law distribution defines structural disorder as a structural element directly linked with function. J. Mol. Biol. 2010, 403, 346–350. [Google Scholar] [CrossRef]
- Schad, E.; Tompa, P.; Hegyi, H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011, 12, R120. [Google Scholar] [CrossRef]
- Dyson, H.J. Expanding the proteome: Disordered and alternatively folded proteins. Q. Rev. Biophys. 2011, 44, 467–518. [Google Scholar] [CrossRef] [PubMed]
- Pancsa, R.; Tompa, P. Structural disorder in eukaryotes. PLoS ONE 2012, 7, e34687. [Google Scholar] [CrossRef] [PubMed]
- Midic, U.; Obradovic, Z. Intrinsic disorder in putative protein sequences. Proteome Sci. 2012, 10 (Suppl. 1), S19. [Google Scholar] [CrossRef]
- Hegyi, H.; Tompa, P. Increased structural disorder of proteins encoded on human sex chromosomes. Mol. Biosyst. 2012, 8, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Korneta, I.; Bujnicki, J.M. Intrinsic disorder in the human spliceosomal proteome. PLoS Comput. Biol. 2012, 8, e1002641. [Google Scholar] [CrossRef] [PubMed]
- Kahali, B.; Ghosh, T.C. Disorderness in Escherichia coli proteome: Perception of folding fidelity and protein-protein interactions. J. Biomol Struct. Dyn. 2013, 31, 472–476. [Google Scholar] [CrossRef]
- Di Domenico, T.; Walsh, I.; Tosatto, S.C. Analysis and consensus of currently available intrinsic protein disorder annotation sources in the MobiDB database. BMC Bioinform. 2013, 14 (Suppl. 7), S3. [Google Scholar] [CrossRef]
- Darling, A.L.; Uversky, V.N. Intrinsic Disorder in Proteins with Pathogenic Repeat Expansions. Molecules 2017, 22, 2027. [Google Scholar] [CrossRef]
- Jorda, J.; Xue, B.; Uversky, V.N.; Kajava, A.V. Protein tandem repeats - the more perfect, the less structured. FEBS J. 2010, 277, 2673–2682. [Google Scholar] [CrossRef]
- Williams, R.M.; Obradovic, Z.; Mathura, V.; Braun, W.; Garner, E.C.; Young, J.; Takayama, S.; Brown, C.J.; Dunker, A.K. The protein non-folding problem: Amino acid determinants of intrinsic order and disorder. Pac. Symp. Biocomput. 2001, 89–100. [Google Scholar]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic Disorder and Functional Proteomics. Biophys. J. 2007, 92, 1439–1456. [Google Scholar] [CrossRef] [PubMed]
- Vacic, V.; Uversky, V.N.; Dunker, A.K.; Lonardi, S. Composition Profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinform. 2007, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Garner, E.; Guilliot, S.; Romero, P.; Albrecht, K.; Hart, J.; Obradovic, Z.; Kissinger, C.; Villafranca, J.E. Protein disorder and the evolution of molecular recognition: Theory, predictions and observations. Pac. Symp Biocomput. 1998, 3, 473–484. [Google Scholar]
- Garner, E.; Cannon, P.; Romero, P.; Obradovic, Z.; Dunker, A.K. Predicting Disordered Regions from Amino Acid Sequence: Common Themes Despite Differing Structural Characterization. Genome Inform. Ser. Workshop Genome Inform. 1998, 9, 201–213. [Google Scholar]
- Campen, A.; Williams, R.M.; Brown, C.J.; Meng, J.; Uversky, V.N.; Dunker, A.K. TOP-IDP-scale: A new amino acid scale measuring propensity for intrinsic disorder. Protein Pept. Lett. 2008, 15, 956–963. [Google Scholar] [CrossRef]
- Uversky, V.N. Paradoxes and wonders of intrinsic disorder: Complexity of simplicity. Intrinsically Disord. Proteins 2016, 4, e1135015. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Daughdrill, G.W.; Pielak, G.J.; Uversky, V.N.; Cortese, M.S.; Dunker, A.K. Natively disordered proteins. In Handbook of Protein Folding; Buchner, J., Kiefhaber, T., Eds.; Wiley-VCH, Verlag GmbH & Co. KGaA: Weinheim, Germany, 2005; pp. 271–353. [Google Scholar]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: Which way to go? Cell Mol. Life Sci. 2003, 60, 1852–1871. [Google Scholar] [CrossRef]
- Zhang, T.; Faraggi, E.; Li, Z.; Zhou, Y. Intrinsically semi-disordered state and its role in induced folding and protein aggregation. Cell Biochem. Biophys. 2013, 67, 1193–1205. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsic disorder-based protein interactions and their modulators. Curr. Pharm. Des. 2013, 19, 4191–4213. [Google Scholar] [CrossRef]
- Jakob, U.; Kriwacki, R.; Uversky, V.N. Conditionally and transiently disordered proteins: Awakening cryptic disorder to regulate protein function. Chem. Rev. 2014, 114, 6779–6805. [Google Scholar] [CrossRef]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef]
- Dunker, A.K.; Brown, C.J.; Obradovic, Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002, 62, 25–49. [Google Scholar]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. The case for intrinsically disordered proteins playing contributory roles in molecular recognition without a stable 3D structure. F1000 Biol. Rep. 2013, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015, 282, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- DeForte, S.; Uversky, V.N. Order, Disorder, and Everything in Between. Molecules 2016, 21, 1090. [Google Scholar] [CrossRef]
- Peng, Z.; Xue, B.; Kurgan, L.; Uversky, V.N. Resilience of death: Intrinsic disorder in proteins involved in the programmed cell death. Cell Death Differ. 2013, 20, 1257–1267. [Google Scholar] [CrossRef]
- Rautureau, G.J.P.; Day, C.L.; Hinds, M.G. Intrinsically Disordered Proteins in Bcl-2 Regulated Apoptosis. Int. J. Mol. Sci. 2010, 11, 1808–1824. [Google Scholar] [CrossRef]
- Hinds, M.G.; Smits, C.; Fredericks-Short, R.; Risk, J.M.; Bailey, M.; Huang, D.C.S.; Day, C.L. Bim, Bad and Bmf: Intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ. 2007, 14, 128–136. [Google Scholar] [CrossRef]
- Djulbegovic, M.B.; Uversky, V.N. Ferroptosis—An iron- and disorder-dependent programmed cell death. Int. J. Biol. Macromol. 2019, 135, 1052–1069. [Google Scholar] [CrossRef]
- DeForte, S.; Uversky, V.N. Not an exception to the rule: The functional significance of intrinsically disordered protein regions in enzymes. Mol. Biosyst. 2017, 13, 463–469. [Google Scholar] [CrossRef]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [PubMed]
- Fuxreiter, M.; Toth-Petroczy, A.; Kraut, D.A.; Matouschek, A.; Lim, R.Y.; Xue, B.; Kurgan, L.; Uversky, V.N. Disordered proteinaceous machines. Chem. Rev. 2014, 114, 6806–6843. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett. 2015, 589, 2498–2506. [Google Scholar] [CrossRef] [PubMed]
- Coelho Ribeiro Mde, L.; Espinosa, J.; Islam, S.; Martinez, O.; Thanki, J.J.; Mazariegos, S.; Nguyen, T.; Larina, M.; Xue, B.; Uversky, V.N. Malleable ribonucleoprotein machine: Protein intrinsic disorder in the Saccharomyces cerevisiae spliceosome. PeerJ 2013, 1, e2. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Romero, P.R.; Noutsou, M.; Maurice, M.M.; Rudiger, S.G.; William, A.M., Jr.; Mizianty, M.J.; Kurgan, L.; Uversky, V.N.; Dunker, A.K. Stochastic machines as a colocalization mechanism for scaffold protein function. FEBS Lett. 2013, 587, 1587–1591. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef]
- Pejaver, V.; Hsu, W.L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014, 23, 1077–1093. [Google Scholar] [CrossRef]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; Legall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339. [Google Scholar] [CrossRef]
- Li, P.; Banjade, S.; Cheng, H.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Snaidero, N.; Pahler, G.; Frey, S.; Sanchez, P.; Zweckstetter, M.; Janshoff, A.; Schneider, A.; Weil, M.T.; Schaap, I.A.; et al. Myelin membrane assembly is driven by a phase transition of myelin basic proteins into a cohesive protein meshwork. PLoS Biol. 2013, 11, e1001577. [Google Scholar] [CrossRef] [PubMed]
- Feric, M.; Brangwynne, C.P. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 2013, 15, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 2017, 239, 97–114. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: Discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol. Biosyst. 2008, 4, 328–340. [Google Scholar] [CrossRef]
- Xue, B.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. CDF it all: Consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett. 2009, 583, 1469–1474. [Google Scholar] [CrossRef]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pac. Symp. Biocomput. 2012, 128–139. [Google Scholar]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Trotter, K.W.; Archer, T.K. The BRG1 transcriptional coregulator. Nucl. Recept Signal. 2008, 6, e004. [Google Scholar] [CrossRef] [PubMed]
- Phelan, M.L.; Sif, S.; Narlikar, G.J.; Kingston, R.E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 1999, 3, 247–253. [Google Scholar] [CrossRef]
- Belandia, B.; Parker, M.G. Nuclear receptors: A rendezvous for chromatin remodeling factors. Cell 2003, 114, 277–280. [Google Scholar] [CrossRef]
- Xu, W.; Cho, H.; Kadam, S.; Banayo, E.M.; Anderson, S.; Yates, J.R., 3rd; Emerson, B.M.; Evans, R.M. A methylation-mediator complex in hormone signaling. Genes Dev. 2004, 18, 144–156. [Google Scholar] [CrossRef]
- Underhill, C.; Qutob, M.S.; Yee, S.P.; Torchia, J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 2000, 275, 40463–40470. [Google Scholar] [CrossRef]
- Kadam, S.; Emerson, B.M. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 2003, 11, 377–389. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [CrossRef]
- Reisman, D.N.; Sciarrotta, J.; Wang, W.; Funkhouser, W.K.; Weissman, B.E. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: Correlation with poor prognosis. Cancer Res. 2003, 63, 560–566. [Google Scholar]
- Lee, D.; Kim, J.W.; Seo, T.; Hwang, S.G.; Choi, E.J.; Choe, J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 2002, 277, 22330–22337. [Google Scholar] [CrossRef] [PubMed]
- Bochar, D.A.; Wang, L.; Beniya, H.; Kinev, A.; Xue, Y.; Lane, W.S.; Wang, W.; Kashanchi, F.; Shiekhattar, R. BRCA1 is associated with a human SWI/SNF-related complex: Linking chromatin remodeling to breast cancer. Cell 2000, 102, 257–265. [Google Scholar] [CrossRef]
- Reyes, J.C.; Barra, J.; Muchardt, C.; Camus, A.; Babinet, C.; Yaniv, M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 1998, 17, 6979–6991. [Google Scholar] [CrossRef] [PubMed]
- Savas, S.; Skardasi, G. The SWI/SNF complex subunit genes: Their functions, variations, and links to risk and survival outcomes in human cancers. Crit. Rev. Oncol. Hematol. 2018, 123, 114–131. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.I.; Chudnovsky, Y.; Duggan, B.; Zajchowski, D.; Greenbowe, J.; Ross, J.S.; Gay, L.M.; Ali, S.M.; Elvin, J.A. Comprehensive genomic profiling reveals inactivating SMARCA4 mutations and low tumor mutational burden in small cell carcinoma of the ovary, hypercalcemic-type. Gynecol. Oncol. 2017, 147, 626–633. [Google Scholar] [CrossRef]
- Kascak, P.; Zamecnik, M.; Bystricky, B. Small Cell Carcinoma of the Ovary (Hypercalcemic Type): Malignant Rhabdoid Tumor. Case Rep. Oncol. 2016, 9, 305–311. [Google Scholar] [CrossRef]
- Errichiello, E.; Mustafa, N.; Vetro, A.; Notarangelo, L.D.; de Jonge, H.; Rinaldi, B.; Vergani, D.; Giglio, S.R.; Morbini, P.; Zuffardi, O. SMARCA4 inactivating mutations cause concomitant Coffin-Siris syndrome, microphthalmia and small-cell carcinoma of the ovary hypercalcaemic type. J. Pathol. 2017, 243, 9–15. [Google Scholar] [CrossRef]
- Love, C.; Sun, Z.; Jima, D.; Li, G.; Zhang, J.; Miles, R.; Richards, K.L.; Dunphy, C.H.; Choi, W.W.; Srivastava, G.; et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012, 44, 1321–1325. [Google Scholar] [CrossRef]
- Jones, D.T.; Jager, N.; Kool, M.; Zichner, T.; Hutter, B.; Sultan, M.; Cho, Y.J.; Pugh, T.J.; Hovestadt, V.; Stutz, A.M.; et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 2012, 488, 100–105. [Google Scholar] [CrossRef]
- Roy, N.; Malik, S.; Villanueva, K.E.; Urano, A.; Lu, X.; Von Figura, G.; Seeley, E.S.; Dawson, D.W.; Collisson, E.A.; Hebrok, M. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015, 29, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Kashyap, L.; Kakkar, A.; Sarkar, C.; Julka, P.K. Atypical teratoid/rhabdoid tumors: Challenges and search for solutions. Cancer Manag. Res. 2016, 8, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Roberts, C.W. Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer Genet. 2014, 207, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Heck, J.E.; Lombardi, C.A.; Cockburn, M.; Meyers, T.J.; Wilhelm, M.; Ritz, B. Epidemiology of rhabdoid tumors of early childhood. Pediatr Blood Cancer 2013, 60, 77–81. [Google Scholar] [CrossRef]
- Levy, P.; Baraitser, M. Coffin-Siris syndrome. J. Med. Genet. 1991, 28, 338–341. [Google Scholar] [CrossRef]
- Sokpor, G.; Xie, Y.; Rosenbusch, J.; Tuoc, T. Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front. Mol. Neurosci 2017, 10, 243. [Google Scholar] [CrossRef]
- He, S.; Pirity, M.K.; Wang, W.L.; Wolf, L.; Chauhan, B.K.; Cveklova, K.; Tamm, E.R.; Ashery-Padan, R.; Metzger, D.; Nakai, A.; et al. Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenetics Chromatin 2010, 3, 21. [Google Scholar] [CrossRef]
- Van Houdt, J.K.; Nowakowska, B.A.; Sousa, S.B.; van Schaik, B.D.; Seuntjens, E.; Avonce, N.; Sifrim, A.; Abdul-Rahman, O.A.; van den Boogaard, M.J.; Bottani, A.; et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat. Genet. 2012, 44, 445–449. [Google Scholar] [CrossRef]
- Koga, M.; Ishiguro, H.; Yazaki, S.; Horiuchi, Y.; Arai, M.; Niizato, K.; Iritani, S.; Itokawa, M.; Inada, T.; Iwata, N.; et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum. Mol. Genet. 2009, 18, 2483–2494. [Google Scholar] [CrossRef]
- Papillon, J.P.N.; Nakajima, K.; Adair, C.D.; Hempel, J.; Jouk, A.O.; Karki, R.G.; Mathieu, S.; Mobitz, H.; Ntaganda, R.; Smith, T.; et al. Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers. J. Med. Chem. 2018, 61, 10155–10172. [Google Scholar] [CrossRef]
- Keppler, B.R.; Archer, T.K. Ubiquitin-dependent and ubiquitin-independent control of subunit stoichiometry in the SWI/SNF complex. J. Biol. Chem. 2010, 285, 35665–35674. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Archer, T.K. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol. Cell Biol. 2005, 25, 9016–9027. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shan, Z.; Xu, Y.; Yang, D.; Wu, J.; Men, C. BAF155 inhibits proliferation and migration by up-regulation of p16 and inactivation of P13K/AKT and Wnt/B-catenin pathways in PC3 cells. Int. J. Clin. Exp. Pathol 2017, 10, 2644–2651. [Google Scholar]
- DelBove, J.; Rosson, G.; Strobeck, M.; Chen, J.; Archer, T.K.; Wang, W.; Knudsen, E.S.; Weissman, B.E. Identification of a core member of the SWI/SNF complex, BAF155/SMARCC1, as a human tumor suppressor gene. Epigenetics 2011, 6, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.L.; Considine, S.W.; Thomas, A.Z.; Lynch, T.H.; Manecksha, R.P. Digital rectal examination in primary care is important for early detection of prostate cancer: A retrospective cohort analysis study. Br. J. Gen. Pract 2014, 64, e783–e787. [Google Scholar] [CrossRef]
- Kim, S.; Zhang, Z.; Upchurch, S.; Isern, N.; Chen, Y. Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition. J. Biol. Chem. 2004, 279, 16670–16676. [Google Scholar] [CrossRef]
- Kowenz-Leutz, E.; Pless, O.; Dittmar, G.; Knoblich, M.; Leutz, A. Crosstalk between C/EBPbeta phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 2010, 29, 1105–1115. [Google Scholar] [CrossRef]
- Zhai, N.; Zhao, Z.L.; Cheng, M.B.; Di, Y.W.; Yan, H.X.; Cao, C.Y.; Dai, H.; Zhang, Y.; Shen, Y.F. Human PIH1 associates with histone H4 to mediate the glucose-dependent enhancement of pre-rRNA synthesis. J. Mol. Cell Biol. 2012, 4, 231–241. [Google Scholar] [CrossRef][Green Version]
- Adler, H.T.; Chinery, R.; Wu, D.Y.; Kussick, S.J.; Payne, J.M.; Fornace, A.J., Jr.; Tkachuk, D.C. Leukemic HRX fusion proteins inhibit GADD34-induced apoptosis and associate with the GADD34 and hSNF5/INI1 proteins. Mol. Cell Biol. 1999, 19, 7050–7060. [Google Scholar] [CrossRef]
- Wu, D.Y.; Tkachuck, D.C.; Roberson, R.S.; Schubach, W.H. The human SNF5/INI1 protein facilitates the function of the growth arrest and DNA damage-inducible protein (GADD34) and modulates GADD34-bound protein phosphatase-1 activity. J. Biol. Chem. 2002, 277, 27706–27715. [Google Scholar] [CrossRef]
- Cheng, S.W.; Davies, K.P.; Yung, E.; Beltran, R.J.; Yu, J.; Kalpana, G.V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 1999, 22, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Sohn, H.; Kalpana, G.V.; Choe, J. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature 1999, 399, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.Y.; Kalpana, G.V.; Goff, S.P.; Schubach, W.H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 1996, 70, 6020–6028. [Google Scholar] [PubMed]
- Li, X.S.; Trojer, P.; Matsumura, T.; Treisman, J.E.; Tanese, N. Mammalian SWI/SNF--a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol. Cell Biol. 2010, 30, 1673–1688. [Google Scholar] [CrossRef]
- Sandhya, S.; Maulik, A.; Giri, M.; Singh, M. Domain architecture of BAF250a reveals the ARID and ARM-repeat domains with implication in function and assembly of the BAF remodeling complex. PLoS ONE 2018, 13, e0205267. [Google Scholar] [CrossRef]
- Nie, Z.; Xue, Y.; Yang, D.; Zhou, S.; Deroo, B.J.; Archer, T.K.; Wang, W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell Biol. 2000, 20, 8879–8888. [Google Scholar] [CrossRef]
- Lei, I.; Gao, X.; Sham, M.H.; Wang, Z. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J. Biol. Chem. 2012, 287, 24255–24262. [Google Scholar] [CrossRef]
- Shu, C.A.; Zhou, Q.; Jotwani, A.R.; Iasonos, A.; Leitao, M.M., Jr.; Konner, J.A.; Aghajanian, C.A. Ovarian clear cell carcinoma, outcomes by stage: The MSK experience. Gynecol. Oncol. 2015, 139, 236–241. [Google Scholar] [CrossRef]
- Samartzis, E.P.; Noske, A.; Dedes, K.J.; Fink, D.; Imesch, P. ARID1A mutations and PI3K/AKT pathway alterations in endometriosis and endometriosis-associated ovarian carcinomas. Int. J. Mol. Sci. 2013, 14, 18824–18849. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Tse, K.; Zeng, T.; Senz, J.; McConechy, M.K.; Anglesio, M.S.; Kalloger, S.E.; et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010, 363, 1532–1543. [Google Scholar] [CrossRef]
- Otto, J.E.; Kadoch, C. A Two-Faced mSWI/SNF Subunit: Dual Roles for ARID1A in Tumor Suppression and Oncogenicity in the Liver. Cancer Cell 2017, 32, 542–543. [Google Scholar] [CrossRef] [PubMed]
- Mathur, R.; Alver, B.H.; San Roman, A.K.; Wilson, B.G.; Wang, X.; Agoston, A.T.; Park, P.J.; Shivdasani, R.A.; Roberts, C.W. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 2017, 49, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.N.; Roberts, C.W. ARID1A mutations in cancer: Another epigenetic tumor suppressor? Cancer Discov. 2013, 3, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Khursheed, M.; Kolla, J.N.; Kotapalli, V.; Gupta, N.; Gowrishankar, S.; Uppin, S.G.; Sastry, R.A.; Koganti, S.; Sundaram, C.; Pollack, J.R.; et al. ARID1B, a member of the human SWI/SNF chromatin remodeling complex, exhibits tumour-suppressor activities in pancreatic cancer cell lines. Br. J. Cancer 2013, 108, 2056–2062. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, J.; Han, Y.; Huang, Z.; Ying, J.; Bi, X.; Zhao, J.; Fang, Y.; Zhou, H.; Zhou, J.; et al. ARID2: A new tumor suppressor gene in hepatocellular carcinoma. Oncotarget 2011, 2, 886–891. [Google Scholar] [CrossRef]
- Haggar, F.A.; Boushey, R.P. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009, 22, 191–197. [Google Scholar] [CrossRef]
- Fletcher, R.H. The diagnosis of colorectal cancer in patients with symptoms: Finding a needle in a haystack. BMC Med. 2009, 7, 18. [Google Scholar] [CrossRef]
- Maconi, G.; Manes, G.; Porro, G.B. Role of symptoms in diagnosis and outcome of gastric cancer. World J. Gastroenterol. 2008, 14, 1149–1155. [Google Scholar] [CrossRef]
- Weledji, E.P.; Enow Orock, G.; Ngowe, M.N.; Nsagha, D.S. How grim is hepatocellular carcinoma? Ann. Med. Surg. (Lond) 2014, 3, 71–76. [Google Scholar] [CrossRef]
- Lomeli, H.; Castillo-Robles, J. The developmental and pathogenic roles of BAF57, a special subunit of the BAF chromatin-remodeling complex. FEBS Lett. 2016, 590, 1555–1569. [Google Scholar] [CrossRef]
- Hah, N.; Kolkman, A.; Ruhl, D.D.; Pijnappel, W.W.; Heck, A.J.; Timmers, H.T.; Kraus, W.L. A role for BAF57 in cell cycle-dependent transcriptional regulation by the SWI/SNF chromatin remodeling complex. Cancer Res. 2010, 70, 4402–4411. [Google Scholar] [CrossRef] [PubMed]
- Battaglioli, E.; Andres, M.E.; Rose, D.W.; Chenoweth, J.G.; Rosenfeld, M.G.; Anderson, M.E.; Mandel, G. REST repression of neuronal genes requires components of the hSWI.SNF complex. J. Biol. Chem. 2002, 277, 41038–41045. [Google Scholar] [CrossRef] [PubMed]
- Kerr, K.; Qualmann, K.; Esquenazi, Y.; Hagan, J.; Kim, D.H. Familial Syndromes Involving Meningiomas Provide Mechanistic Insight Into Sporadic Disease. Neurosurgery 2018, 83, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Chung, C.K.; Jahng, T.A. Surgical outcome of spinal canal meningiomas. J. Korean Neurosurg Soc. 2007, 42, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, S.; Comstock, C.E.; Ertel, A.; Jeong, K.W.; Stallcup, M.R.; Addya, S.; McCue, P.A.; Ostrander, W.F., Jr.; Augello, M.A.; Knudsen, K.E. Aberrant BAF57 signaling facilitates prometastatic phenotypes. Clin. Cancer Res. 2013, 19, 2657–2667. [Google Scholar] [CrossRef]
- Kagami, S.; Kurita, T.; Kawagoe, T.; Toki, N.; Matsuura, Y.; Hachisuga, T.; Matsuyama, A.; Hashimoto, H.; Izumi, H.; Kohno, K. Prognostic significance of BAF57 expression in patients with endometrial carcinoma. Histol. Histopathol. 2012, 27, 593–599. [Google Scholar] [CrossRef]
- Varela, I.; Tarpey, P.; Raine, K.; Huang, D.; Ong, C.K.; Stephens, P.; Davies, H.; Jones, D.; Lin, M.L.; Teague, J.; et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011, 469, 539–542. [Google Scholar] [CrossRef]
- Xia, W.; Nagase, S.; Montia, A.G.; Kalachikov, S.M.; Keniry, M.; Su, T.; Memeo, L.; Hibshoosh, H.; Parsons, R. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008, 68, 1667–1674. [Google Scholar] [CrossRef]
- Lee, H.; Dai, F.; Zhuang, L.; Xiao, Z.D.; Kim, J.; Zhang, Y.; Ma, L.; You, M.J.; Wang, Z.; Gan, B. BAF180 regulates cellular senescence and hematopoietic stem cell homeostasis through p21. Oncotarget 2016, 7, 19134–19146. [Google Scholar] [CrossRef]
- Ljungberg, B.; Hanbury, D.C.; Kuczyk, M.A.; Merseburger, A.S.; Mulders, P.F.; Patard, J.J.; Sinescu, I.C. European Association of Urology Guideline Group for renal cell, c. Renal cell carcinoma guideline. Eur. Urol. 2007, 51, 1502–1510. [Google Scholar] [CrossRef]
- Sobanko, J.F.; Meijer, L.; Nigra, T.P. Epithelioid sarcoma: A review and update. J. Clin. Aesthet Dermatol. 2009, 2, 49–54. [Google Scholar] [PubMed]
- Li, L.; Fan, X.S.; Xia, Q.Y.; Rao, Q.; Liu, B.; Yu, B.; Shi, Q.L.; Lu, Z.F.; Zhou, X.J. Concurrent loss of INI1, PBRM1, and BRM expression in epithelioid sarcoma: Implications for the cocontributions of multiple SWI/SNF complex members to pathogenesis. Hum. Pathol. 2014, 45, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.M.; Parmar, A.D.; Geller, D.A. Intrahepatic cholangiocarcinoma. Surg. Oncol. Clin. N. Am. 2014, 23, 231–246. [Google Scholar] [CrossRef]
- Misumi, K.; Hayashi, A.; Shibahara, J.; Arita, J.; Sakamoto, Y.; Hasegawa, K.; Kokudo, N.; Fukayama, M. Intrahepatic cholangiocarcinoma frequently shows loss of BAP1 and PBRM1 expression, and demonstrates specific clinicopathological and genetic characteristics with BAP1 loss. Histopathology 2017, 70, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Kage-Nakadai, E.; Yoshina, S.; Imae, R.; Mitani, S. The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway. PLoS Genet. 2015, 11, e1004921. [Google Scholar] [CrossRef]
- Lei, Z.; JunHui, L.; PeiFeng, L. Candidate genes mediated by estrogen-related receptor gamma in pancreatic beta cells. J. Biochem. Mol. Toxicol. 2019, 33, e22390. [Google Scholar] [CrossRef]
- Zani, V.J.; Asou, N.; Jadayel, D.; Heward, J.M.; Shipley, J.; Nacheva, E.; Takasuki, K.; Catovsky, D.; Dyer, M.J. Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood 1996, 87, 3124–3134. [Google Scholar] [CrossRef]
- Jadayel, D.M.; Osborne, L.R.; Coignet, L.J.; Zani, V.J.; Tsui, L.C.; Scherer, S.W.; Dyer, M.J. The BCL7 gene family: Deletion of BCL7B in Williams syndrome. Gene 1998, 224, 35–44. [Google Scholar] [CrossRef]
- Natter, S.; Seiberler, S.; Hufnagl, P.; Binder, B.R.; Hirschl, A.M.; Ring, J.; Abeck, D.; Schmidt, T.; Valent, P.; Valenta, R. Isolation of cDNA clones coding for IgE autoantigens with serum IgE from atopic dermatitis patients. FASEB J. 1998, 12, 1559–1569. [Google Scholar] [CrossRef]
- Prasad, A.; Rabionet, R.; Espinet, B.; Zapata, L.; Puiggros, A.; Melero, C.; Puig, A.; Sarria-Trujillo, Y.; Ossowski, S.; Garcia-Muret, M.P.; et al. Identification of Gene Mutations and Fusion Genes in Patients with Sezary Syndrome. J. Invest. Dermatol. 2016, 136, 1490–1499. [Google Scholar] [CrossRef]
- Mathies, L.D.; Aliev, F.; Investigators, C.; Davies, A.G.; Dick, D.M.; Bettinger, J.C. Variation in SWI/SNF Chromatin Remodeling Complex Proteins is Associated with Alcohol Dependence and Antisocial Behavior in Human Populations. Alcohol. Clin. Exp. Res. 2017, 41, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.; Estruch, S.B.; Graham, S.A.; McRae, J.; Sawiak, S.J.; Hurst, J.A.; Joss, S.K.; Holder, S.E.; Morton, J.E.; Turner, C.; et al. BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription. Am. J. Hum. Genet. 2016, 99, 253–274. [Google Scholar] [CrossRef] [PubMed]
- Bauer, D.E.; Orkin, S.H. Hemoglobin switching’s surprise: The versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr. Opin. Genet. Dev. 2015, 33, 62–70. [Google Scholar] [CrossRef]
- Basak, A.; Sankaran, V.G. Regulation of the fetal hemoglobin silencing factor BCL11A. Ann. N. Y. Acad. Sci. 2016, 1368, 25–30. [Google Scholar] [CrossRef]
- Obata, M.; Kominami, R.; Mishima, Y. BCL11B tumor suppressor inhibits HDM2 expression in a p53-dependent manner. Cell Signal. 2012, 24, 1047–1052. [Google Scholar] [CrossRef]
- Cismasiu, V.B.; Ghanta, S.; Duque, J.; Albu, D.I.; Chen, H.M.; Kasturi, R.; Avram, D. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood 2006, 108, 2695–2702. [Google Scholar] [CrossRef] [PubMed]
- Nishiguchi, Y.; Ohmoto, M.; Koki, J.; Enomoto, T.; Kominami, R.; Matsumoto, I.; Hirota, J. Bcl11b/Ctip2 is required for development of lingual papillae in mice. Dev. Biol. 2016, 416, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Punwani, D.; Zhang, Y.; Yu, J.; Cowan, M.J.; Rana, S.; Kwan, A.; Adhikari, A.N.; Lizama, C.O.; Mendelsohn, B.A.; Fahl, S.P.; et al. Multisystem Anomalies in Severe Combined Immunodeficiency with Mutant BCL11B. N. Engl. J. Med. 2016, 375, 2165–2176. [Google Scholar] [CrossRef] [PubMed]
- Chiaretti, S.; Foa, R. T-cell acute lymphoblastic leukemia. Haematologica 2009, 94, 160–162. [Google Scholar] [CrossRef]
- Paul, S.; Kantarjian, H.; Jabbour, E.J. Adult Acute Lymphoblastic Leukemia. Mayo Clin. Proc. 2016, 91, 1645–1666. [Google Scholar] [CrossRef]
- Terwilliger, T.; Abdul-Hay, M. Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer J. 2017, 7, e577. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, T.; Koibuchi, N.; Chin, W.W. Synovial sarcoma translocation (SYT) encodes a nuclear receptor coactivator. Endocrinology 2005, 146, 3892–3899. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.-J.; Chu, C.-H.; Shieh, S.-J. Groin synovial sarcoma with intraluminal femoral sheath involvement. Formosan J. Surg. 2014, 47, 36–41. [Google Scholar] [CrossRef]
- Banito, A.; Li, X.; Laporte, A.N.; Roe, J.S.; Sanchez-Vega, F.; Huang, C.H.; Dancsok, A.R.; Hatzi, K.; Chen, C.C.; Tschaharganeh, D.F.; et al. The SS18-SSX Oncoprotein Hijacks KDM2B-PRC1.1 to Drive Synovial Sarcoma. Cancer Cell 2018, 34, 346–348. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.J.; Pulice, J.L.; Beird, H.C.; Ingram, D.R.; D’Avino, A.R.; Shern, J.F.; Charville, G.W.; Hornick, J.L.; Nakayama, R.T.; Garcia-Rivera, E.M.; et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell 2018, 33, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Crew, A.J.; Clark, J.; Fisher, C.; Gill, S.; Grimer, R.; Chand, A.; Shipley, J.; Gusterson, B.A.; Cooper, C.S. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 1995, 14, 2333–2340. [Google Scholar] [CrossRef]
- He, L.; Tian, X.; Zhang, H.; Hu, T.; Huang, X.; Zhang, L.; Wang, Z.; Zhou, B. BAF200 is required for heart morphogenesis and coronary artery development. PLoS ONE 2014, 9, e109493. [Google Scholar] [CrossRef]
- Li, M.; Zhao, H.; Zhang, X.; Wood, L.D.; Anders, R.A.; Choti, M.A.; Pawlik, T.M.; Daniel, H.D.; Kannangai, R.; Offerhaus, G.J.; et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 2011, 43, 828–829. [Google Scholar] [CrossRef]
- Garbe, C.; Peris, K.; Hauschild, A.; Saiag, P.; Middleton, M.; Bastholt, L.; Grob, J.J.; Malvehy, J.; Newton-Bishop, J.; Stratigos, A.J.; et al. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline - Update 2016. Eur. J. Cancer 2016, 63, 201–217. [Google Scholar] [CrossRef]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A landscape of driver mutations in melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef]
- Dela Cruz, C.S.; Tanoue, L.T.; Matthay, R.A. Lung cancer: Epidemiology, etiology, and prevention. Clin. Chest Med. 2011, 32, 605–644. [Google Scholar] [CrossRef] [PubMed]
- Bilello, K.S.; Murin, S.; Matthay, R.A. Epidemiology, etiology, and prevention of lung cancer. Clin. Chest Med. 2002, 23, 1–25. [Google Scholar] [CrossRef]
- Walling, A.M.; Weeks, J.C.; Kahn, K.L.; Tisnado, D.; Keating, N.L.; Dy, S.M.; Arora, N.K.; Mack, J.W.; Pantoja, P.M.; Malin, J.L. Symptom prevalence in lung and colorectal cancer patients. J. Pain Symptom Manage. 2015, 49, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Manceau, G.; Letouze, E.; Guichard, C.; Didelot, A.; Cazes, A.; Corte, H.; Fabre, E.; Pallier, K.; Imbeaud, S.; Le Pimpec-Barthes, F.; et al. Recurrent inactivating mutations of ARID2 in non-small cell lung carcinoma. Int. J. Cancer 2013, 132, 2217–2221. [Google Scholar] [CrossRef] [PubMed]
- Flynn, E.M.; Huang, O.W.; Poy, F.; Oppikofer, M.; Bellon, S.F.; Tang, Y.; Cochran, A.G. A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications. Structure 2015, 23, 1801–1814. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Muller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Zhou, J.; Liu, H.Y.; Zhou, M.; Wang, L.L.; Zhang, Q.H.; Yang, Y.X.; Xiong, W.; Shen, S.R.; Li, X.L.; et al. The transcriptional regulation role of BRD7 by binding to acetylated histone through bromodomain. J. Cell Biochem. 2006, 97, 882–892. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Park, J.; Chung, J. BP75, bromodomain-containing M(r) 75,000 protein, binds dishevelled-1 and enhances Wnt signaling by inactivating glycogen synthase kinase-3 beta. Cancer Res. 2003, 63, 4792–4795. [Google Scholar]
- Drost, J.; Mantovani, F.; Tocco, F.; Elkon, R.; Comel, A.; Holstege, H.; Kerkhoven, R.; Jonkers, J.; Voorhoeve, P.M.; Agami, R.; et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat. Cell Biol. 2010, 12, 380–389. [Google Scholar] [CrossRef]
- Burrows, A.E.; Smogorzewska, A.; Elledge, S.J. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc. Natl. Acad. Sci. USA 2010, 107, 14280–14285. [Google Scholar] [CrossRef]
- Koszewski, N.J.; Henry, K.W.; Lubert, E.J.; Gravatte, H.; Noonan, D.J. Use of a modified yeast one-hybrid screen to identify BAF60a interactions with the Vitamin D receptor heterodimer. J. Steroid Biochem. Mol. Biol. 2003, 87, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, P.W.; Fryer, C.J.; Trotter, K.W.; Wang, W.; Archer, T.K. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell Biol. 2003, 23, 6210–6220. [Google Scholar] [CrossRef] [PubMed]
- Witzel, M.; Petersheim, D.; Fan, Y.; Bahrami, E.; Racek, T.; Rohlfs, M.; Puchalka, J.; Mertes, C.; Gagneur, J.; Ziegenhain, C.; et al. Chromatin-remodeling factor SMARCD2 regulates transcriptional networks controlling differentiation of neutrophil granulocytes. Nat. Genet. 2017, 49, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Aasland, R.; Gibson, T.J.; Stewart, A.F. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995, 20, 56–59. [Google Scholar] [CrossRef]
- Huber, F.M.; Greenblatt, S.M.; Davenport, A.M.; Martinez, C.; Xu, Y.; Vu, L.P.; Nimer, S.D.; Hoelz, A. Histone-binding of DPF2 mediates its repressive role in myeloid differentiation. Proc. Natl. Acad. Sci. USA 2017, 114, 6016–6021. [Google Scholar] [CrossRef]
- Xiong, X.; Panchenko, T.; Yang, S.; Zhao, S.; Yan, P.; Zhang, W.; Xie, W.; Li, Y.; Zhao, Y.; Allis, C.D.; et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 2016, 12, 1111–1118. [Google Scholar] [CrossRef]
- Tando, T.; Ishizaka, A.; Watanabe, H.; Ito, T.; Iida, S.; Haraguchi, T.; Mizutani, T.; Izumi, T.; Isobe, T.; Akiyama, T.; et al. Requiem protein links RelB/p52 and the Brm-type SWI/SNF complex in a noncanonical NF-kappaB pathway. J. Biol. Chem. 2010, 285, 21951–21960. [Google Scholar] [CrossRef]
- Vasileiou, G.; Vergarajauregui, S.; Endele, S.; Popp, B.; Buttner, C.; Ekici, A.B.; Gerard, M.; Bramswig, N.C.; Albrecht, B.; Clayton-Smith, J.; et al. Mutations in the BAF-Complex Subunit DPF2 Are Associated with Coffin-Siris Syndrome. Am. J. Hum. Genet. 2018, 102, 468–479. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, C.; Bian, C.; Tempel, W.; Crombet, L.; MacKenzie, F.; Min, J.; Liu, Z.; Qi, C. Crystal structure of the Cys2His2-type zinc finger domain of human DPF2. Biochem. Biophys. Res. Commun. 2011, 413, 58–61. [Google Scholar] [CrossRef]
- Zeng, L.; Zhang, Q.; Li, S.; Plotnikov, A.N.; Walsh, M.J.; Zhou, M.M. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 2010, 466, 258–262. [Google Scholar] [CrossRef]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Cote, J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell Biol. 2004, 24, 1884–1896. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jin, J.; Tomomori-Sato, C.; Sato, S.; Sorokina, I.; Parmely, T.J.; Conaway, R.C.; Conaway, J.W. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 2003, 278, 42733–42736. [Google Scholar] [CrossRef] [PubMed]
- Ikura, T.; Ogryzko, V.V.; Grigoriev, M.; Groisman, R.; Wang, J.; Horikoshi, M.; Scully, R.; Qin, J.; Nakatani, Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 2000, 102, 463–473. [Google Scholar] [CrossRef]
- Doyon, Y.; Cote, J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004, 14, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M.; Gerber, J.; Drapkin, R.; Sif, S.; Ikura, T.; Ogryzko, V.; Lane, W.S.; Nakatani, Y.; Livingston, D.M. The p400 complex is an essential E1A transformation target. Cell 2001, 106, 297–307. [Google Scholar] [CrossRef]
- Kadoch, C.; Crabtree, G.R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv. 2015, 1, e1500447. [Google Scholar] [CrossRef]
- Park, J.; Wood, M.A.; Cole, M.D. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol. Cell Biol. 2002, 22, 1307–1316. [Google Scholar] [CrossRef]
- Jin, J.; Cai, Y.; Yao, T.; Gottschalk, A.J.; Florens, L.; Swanson, S.K.; Gutierrez, J.L.; Coleman, M.K.; Workman, J.L.; Mushegian, A.; et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 2005, 280, 41207–41212. [Google Scholar] [CrossRef]
- Wu, S.; Shi, Y.; Mulligan, P.; Gay, F.; Landry, J.; Liu, H.; Lu, J.; Qi, H.H.; Wang, W.; Nickoloff, J.A.; et al. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat. Struct Mol. Biol. 2007, 14, 1165–1172. [Google Scholar] [CrossRef]
- Yao, T.; Song, L.; Jin, J.; Cai, Y.; Takahashi, H.; Swanson, S.K.; Washburn, M.P.; Florens, L.; Conaway, R.C.; Cohen, R.E.; et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell 2008, 31, 909–917. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Jin, J.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C. Subunit organization of the human INO80 chromatin remodeling complex: An evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 2011, 286, 11283–11289. [Google Scholar] [CrossRef]
- Marom, R.; Jain, M.; Burrage, L.C.; Song, I.W.; Graham, B.H.; Brown, C.W.; Stevens, S.J.C.; Stegmann, A.P.A.; Gunter, A.T.; Kaplan, J.D.; et al. Heterozygous variants in ACTL6A, encoding a component of the BAF complex, are associated with intellectual disability. Hum. Mutat. 2017, 38, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Euskirchen, G.; Auerbach, R.K.; Snyder, M. SWI/SNF chromatin-remodeling factors: Multiscale analyses and diverse functions. J. Biol. Chem. 2012, 287, 30897–30905. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Aksnes, H.; Marie, M.; Boczkowska, M.; Varland, S.; Timmerman, E.; Foyn, H.; Glomnes, N.; Rebowski, G.; Impens, F.; et al. NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc. Natl. Acad. Sci. USA 2018, 115, 4399–4404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Buder, K.; Burkhardt, C.; Schlott, B.; Gorlach, M.; Grosse, F. Nuclear DNA helicase II/RNA helicase A binds to filamentous actin. J. Biol. Chem. 2002, 277, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.V.; Schulke, J.P.; Liebl, C.; Stiess, M.; Avrabos, C.; Bock, J.; Wochnik, G.M.; Davies, H.A.; Zimmermann, N.; Scharf, S.H.; et al. Tumor suppressor down-regulated in renal cell carcinoma 1 (DRR1) is a stress-induced actin bundling factor that modulates synaptic efficacy and cognition. Proc. Natl. Acad. Sci. USA 2011, 108, 17213–17218. [Google Scholar] [CrossRef]
- Mu, P.; Akashi, T.; Lu, F.; Kishida, S.; Kadomatsu, K. A novel nuclear complex of DRR1, F-actin and COMMD1 involved in NF-kappaB degradation and cell growth suppression in neuroblastoma. Oncogene 2017, 36, 5745–5756. [Google Scholar] [CrossRef]
- Procaccio, V.; Salazar, G.; Ono, S.; Styers, M.L.; Gearing, M.; Davila, A.; Jimenez, R.; Juncos, J.; Gutekunst, C.A.; Meroni, G.; et al. A mutation of beta -actin that alters depolymerization dynamics is associated with autosomal dominant developmental malformations, deafness, and dystonia. Am. J. Hum. Genet. 2006, 78, 947–960. [Google Scholar] [CrossRef]
- Riviere, J.B.; van Bon, B.W.; Hoischen, A.; Kholmanskikh, S.S.; O’Roak, B.J.; Gilissen, C.; Gijsen, S.; Sullivan, C.T.; Christian, S.L.; Abdul-Rahman, O.A.; et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 2012, 44, 440–444. [Google Scholar] [CrossRef]
- Marston, S. The Molecular Mechanisms of Mutations in Actin and Myosin that Cause Inherited Myopathy. Int. J. Mol. Sci. 2018, 19, 2020. [Google Scholar] [CrossRef]
- Zech, M.; Jech, R.; Wagner, M.; Mantel, T.; Boesch, S.; Nocker, M.; Jochim, A.; Berutti, R.; Havrankova, P.; Fecikova, A.; et al. Molecular diversity of combined and complex dystonia: Insights from diagnostic exome sequencing. Neurogenetics 2017, 18, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.; Salas, F.; Mokhtari, R.; Dolstra, H.; Pedrosa, E.; Lachman, H.M. Modeling the neuropsychiatric manifestations of Lowe syndrome using induced pluripotent stem cells: Defective F-actin polymerization and WAVE-1 expression in neuronal cells. Mol. Autism 2018, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, I.M.; Povarova, O.I.; Uversky, V.N.; Turoverov, K.K. Native globular actin has a thermodynamically unstable quasi-stationary structure with elements of intrinsic disorder. FEBS J. 2016, 283, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Povarova, O.I.; Uversky, V.N.; Kuznetsova, I.M.; Turoverov, K.K. Actinous enigma or enigmatic actin: Folding, structure, and functions of the most abundant eukaryotic protein. Intrinsically Disord. Proteins 2014, 2, e34500. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Sowa, M.E.; Ottinger, M.; Smith, J.A.; Shi, Y.; Harper, J.W.; Howley, P.M. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell Biol. 2011, 31, 2641–2652. [Google Scholar] [CrossRef]
- Donati, B.; Lorenzini, E.; Ciarrocchi, A. BRD4 and Cancer: Going beyond transcriptional regulation. Mol. Cancer 2018, 17, 164. [Google Scholar] [CrossRef]
- Michel, B.C.; D’Avino, A.R.; Cassel, S.H.; Mashtalir, N.; McKenzie, Z.M.; McBride, M.J.; Valencia, A.M.; Zhou, Q.; Bocker, M.; Soares, L.M.M.; et al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat. Cell Biol. 2018, 20, 1410–1420. [Google Scholar] [CrossRef]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 03, 35–60. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting intrinsic disorder in proteins: An overview. Cell Res. 2009, 19, 929–949. [Google Scholar] [CrossRef]
- Huang, F.; Oldfield, C.J.; Xue, B.; Hsu, W.L.; Meng, J.; Liu, X.; Shen, L.; Romero, P.; Uversky, V.N.; Dunker, A. Improving protein order-disorder classification using charge-hydropathy plots. BMC Bioinform. 2014, 15 (Suppl. 17), S4. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins Struct. Funct. Bioinform. 2005, 61, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Uversky, V.N. p53 Proteoforms and Intrinsic Disorder: An Illustration of the Protein Structure-Function Continuum Concept. Int. J. Mol. Sci. 2016, 17, 1874. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L.; Consortium for Top Down, P. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019, 166, 1–17. [Google Scholar] [CrossRef]
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Multi-functionality of proteins involved in GPCR and G protein signaling: Making sense of structure-function continuum with intrinsic disorder-based proteoforms. Cell Mol. Life Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dave, V.; Iakoucheva, L.M.; Malaney, P.; Metallo, S.J.; Pathak, R.R.; Joerger, A.C. Pathological unfoldomics of uncontrolled chaos: Intrinsically disordered proteins and human diseases. Chem. Rev. 2014, 114, 6844–6879. [Google Scholar] [CrossRef] [PubMed]
- Midic, U.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Unfoldomics of human genetic diseases: Illustrative examples of ordered and intrinsically disordered members of the human diseasome. Protein Pept. Lett. 2009, 16, 1533–1547. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front. Biosci. 2009, 14, 5188–5238. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Midic, U.; Xie, H.; Xue, B.; Vucetic, S.; Iakoucheva, L.M.; Obradovic, Z.; Dunker, A.K. Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genomics 2009, 10 (Suppl. 1), S7. [Google Scholar] [CrossRef]
- Uversky, V.N. Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: Another illustration of the D(2) concept. Expert Rev. Proteomics 2010, 7, 543–564. [Google Scholar] [CrossRef]
- Uversky, V.N. Wrecked regulation of intrinsically disordered proteins in diseases: Pathogenicity of deregulated regulators. Front. Mol. Biosci. 2014, 1, 6. [Google Scholar] [CrossRef]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Hadidy, N.; Uversky, V.N. Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development. Int. J. Mol. Sci. 2019, 20, 5260. https://doi.org/10.3390/ijms20215260
El Hadidy N, Uversky VN. Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development. International Journal of Molecular Sciences. 2019; 20(21):5260. https://doi.org/10.3390/ijms20215260
Chicago/Turabian StyleEl Hadidy, Nashwa, and Vladimir N. Uversky. 2019. "Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development" International Journal of Molecular Sciences 20, no. 21: 5260. https://doi.org/10.3390/ijms20215260
APA StyleEl Hadidy, N., & Uversky, V. N. (2019). Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development. International Journal of Molecular Sciences, 20(21), 5260. https://doi.org/10.3390/ijms20215260





