Using Transcriptome Analysis to Screen for Key Genes and Pathways Related to Cytoplasmic Male Sterility in Cotton (Gossypium hirsutum L.)
Abstract
:1. Introduction
2. Results
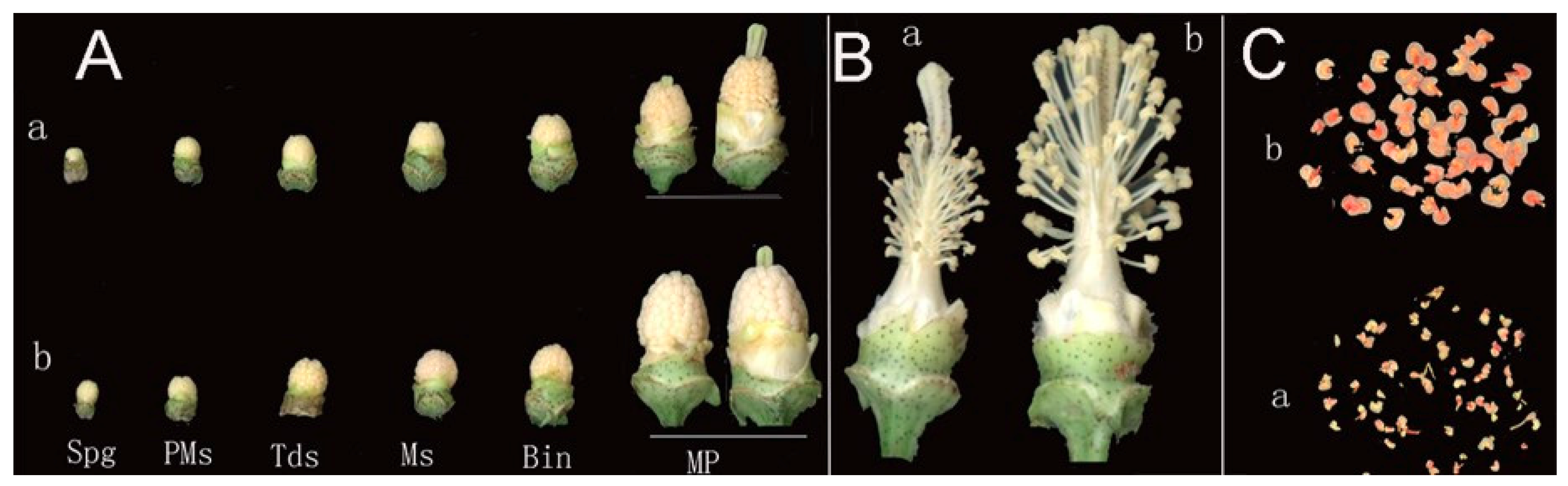
2.1. Morphological Characteristics of the CMS Line C2P5A and the Maintainer Line C2P5B
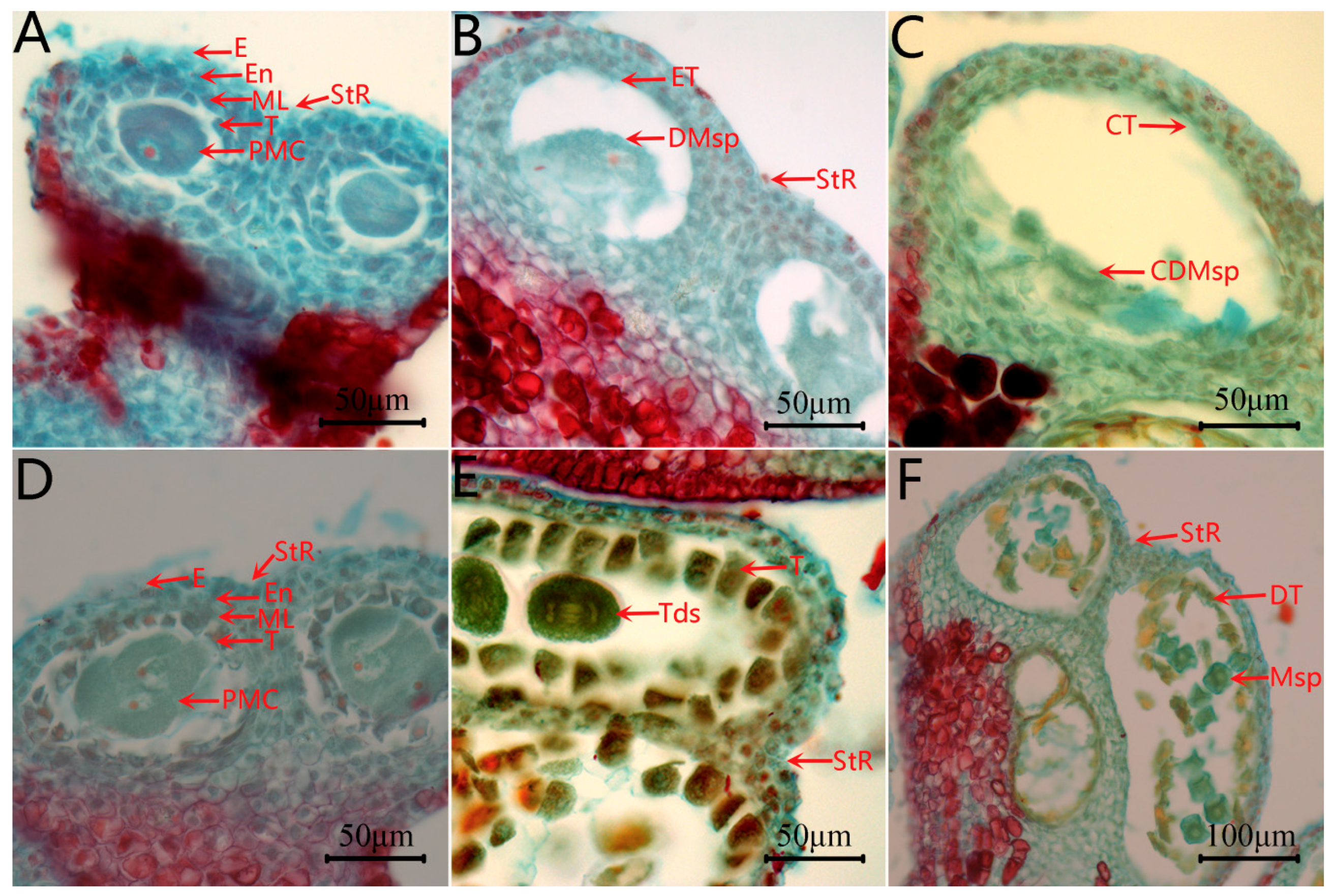
2.2. Microstructure of Anther at Different Developmental Stages of the CMS Line C2P5A
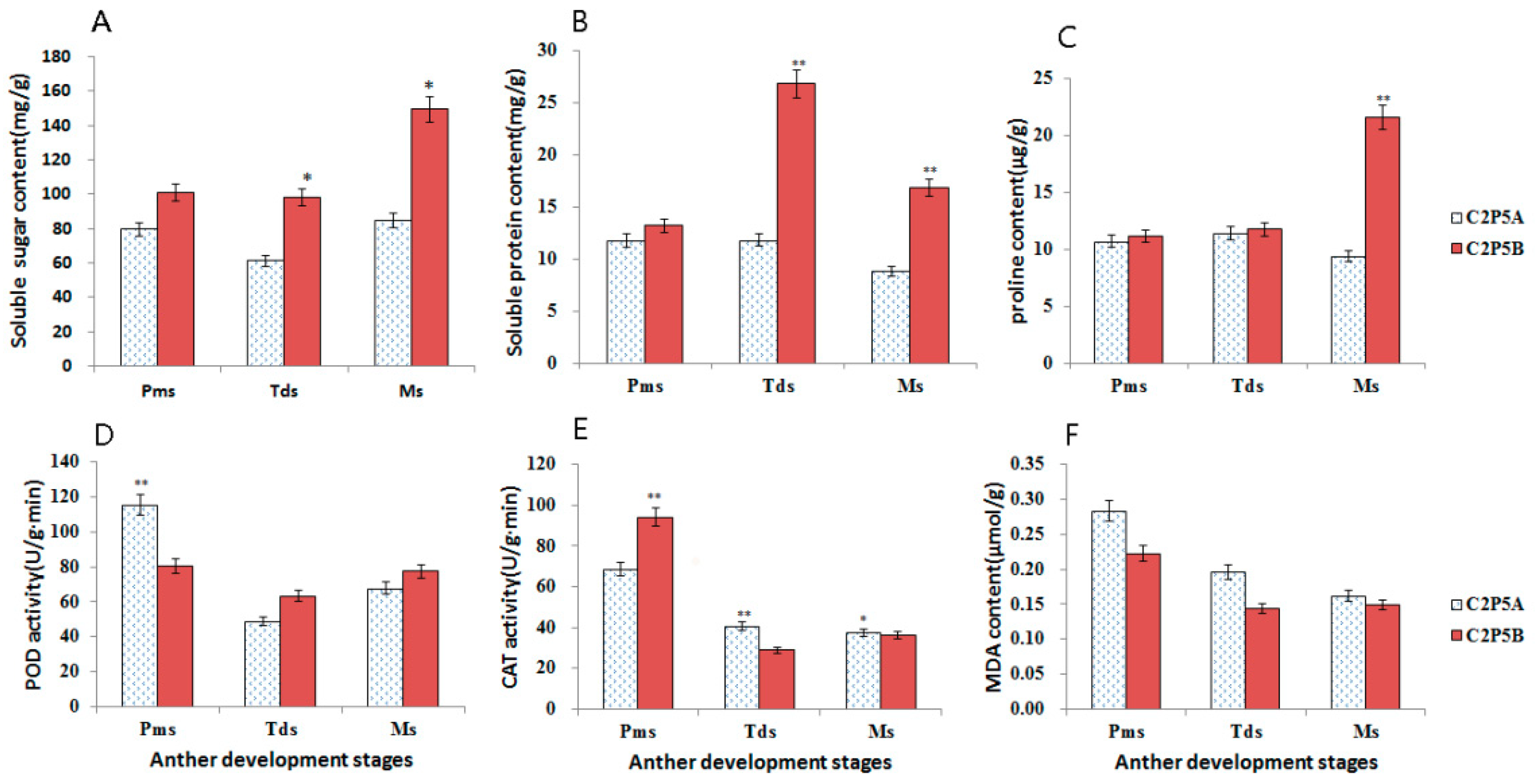
2.3. Measurements of Physiological Indices in the CMS Line C2P5A
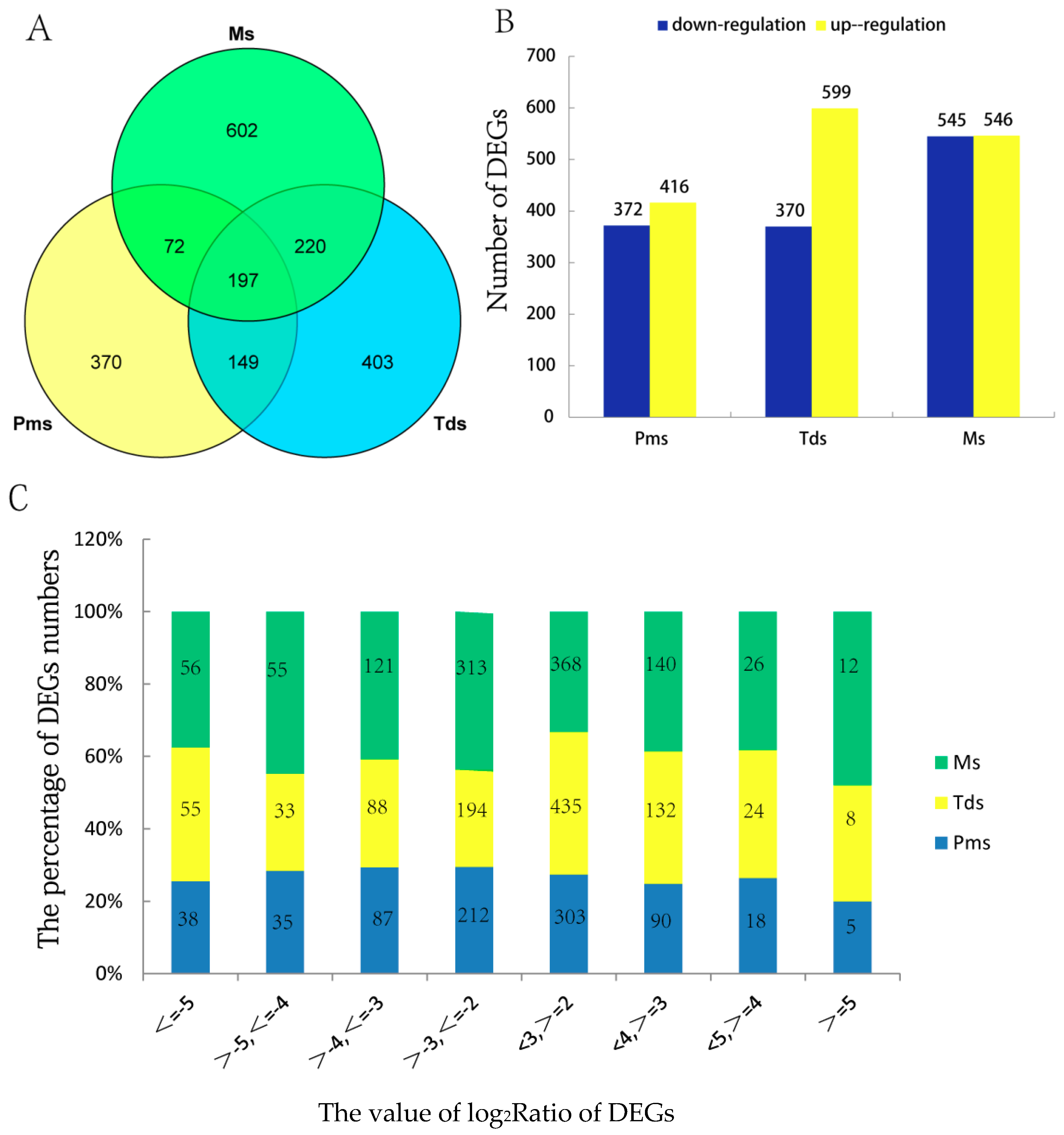
2.4. Identification of Differentially Expressed Genes (DEGs)
2.5. Gene Ontology Annotation and Pathway Enrichment Analysis of DEGs
2.6. Correlation Network Analysis with WGCNA
2.7. Major Pathways and Genes Associated with CMS
2.8. Validation of DEGs by Quantitative Reverse-Transcription PCR (qRT-PCR)
3. Discussion
3.1. The Crucial Period of Abortion in the CMS Line C2P5A
3.2. Major Genes Regulate Anther Development in Cotton
3.3. Major Metabolic Pathways Regulating Anther Development in Cotton
4. Materials and Methods
4.1. Plant Materials
4.2. Histological Analysis
4.3. Measurement of Physiological Indices
4.4. Screening of DEGs and Co-expression Network Analysis of Unigenes
4.5. RT-PCR Analysis for Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CMS | Cytoplasmic male sterility |
| GMS | Genic male sterility |
| MS | Male sterility |
| DEG | Differentially expressed genes |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| RSEM | RNA-Seq by Expectation Maximization |
| RNA-seq | RNA-sequencing |
| qRT-PCR | Quantitative Reverse-Transcription PCR |
| DGE | Digital gene expression |
| WGCNA | Weighted correlation network |
| TTC | Triphenyltetrazolium chloride |
| SEM | Scanning electron microscopy |
| PCD | Programmed cell death |
References
- Zhang, Z.; Li, J.; Muhammad, J.; Cai, J.; Jia, F.; Shi, Y.; Gong, J.; Shang, H.; Liu, A.; Chen, T.; et al. High Resolution Consensus Mapping of Quantitative Trait Loci for Fiber Strength, Length and Micronaire on Chromosome 25 of the Upland Cotton (Gossypium hirsutum L.). PLoS ONE 2015, 10, e0135430. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Scheffler, B.E.; Dennis, E.; Triplett, B.A.; Zhang, T.; Guo, W.; Chen, X.; Stelly, D.M.; Rabinowicz, P.D.; Town, C.D.; et al. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007, 145, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raska, D.A.; Stelly, D.M. Breeding and Genetics Upland Cotton (Gossypium hirsutum L.) x Hawaiian Cotton (G. tomentosum Nutt. ex Seem.) F1 Hybrid Hypoaneuploid Chromosome Substitution Series. J. Cotton Sci. 2006, 10, 263–272. [Google Scholar]
- Samuel Yang, S.; Cheung, F.; Lee, J.J.; Ha, M.; Wei, N.E.; Sze, S.H.; Stelly, D.M.; Thaxton, P.; Triplett, B.; Town, C.D.; et al. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 2006, 47, 761–775. [Google Scholar] [CrossRef] [Green Version]
- Saxena, K.B.; Kumar, R.V.; Tikle, A.N.; Saxena, M.K.; Gautam, V.S.; Rao, S.K.; Khare, D.K.; Chauhan, Y.S.; Saxena, R.K.; Reddy, B.V.S. ICPH 2671—The world’s first commercial food legume hybrid. Plant Breed. 2013, 132, 479–485. [Google Scholar] [CrossRef]
- Bvs, R.; Ramesh, S.; Reddy, P.S. Sorghum breeding research at ICRISAT--goals, strategies, methods and accomplishments. Int. Sorghum Millets Newsl. 2004, 2000, 131–140. [Google Scholar]
- Longin, C.F.H.; Mühleisen, J.; Maurer, H.P.; Zhang, H.; Gowda, M.; Reif, J.C. Hybrid breeding in autogamous cereals. Appl. Genet. 2012, 125, 1087–1096. [Google Scholar] [CrossRef]
- Whitford, R.; Fleury, D.; Reif, J.C.; Garcia, M.; Okada, T.; Korzun, V.; Langridge, P. Hybrid breeding in wheat: Technologies to improve hybrid wheat seed production. J. Exp. Bot. 2013, 64, 5411–5428. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Bohra, A.; Jha, U.C.; Adhimoolam, P.; Bisht, D.; Singh, N.P. Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell. Rep. 2016, 35, 967–993. [Google Scholar] [CrossRef] [PubMed]
- Mühleisen, J.; Maurer, H.P.; Stiewe, G.; Bury, P.; Reif, J.C. Hybrid Breeding in Barley. Crop. Sci. 2013, 53, 819–824. [Google Scholar] [CrossRef]
- Yu, S.; Fan, S.; Wang, H.; Wei, H.; Pang, C. Progresses in Research on Cotton High Yield Breeding in China. Chin. Agric. Sci. 2016, 49, 3465–3476. [Google Scholar]
- Yamagishi, H.; Bhat, S.R. Cytoplasmic male sterility in Brassicaceae crops. Breed. Sci. 2014, 64, 38–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Bao, S.; Zhou, X.; Liu, J.; Zhuang, Y. The key genes and pathways related to male sterility of eggplant revealed by comparative transcriptome analysis. BMC Plant Biol. 2018, 18, 209. [Google Scholar] [CrossRef]
- Kaul, M.L.H. Male Sterility in Higher Plants; Springer: Berlin/Heidelberg, Germany, 1988. [Google Scholar]
- Carlsson, J.; Leino, M.; Sohlberg, J.; Sundström, J.F.; Glimelius, K. Mitochondrial regulation of flower development. Mitochondrion 2008, 8, 74–86. [Google Scholar] [CrossRef]
- McCauley, D.E.; Bailey, M.F. Recent advances in the study of gynodioecy: The interface of theory and empiricism. Ann. Bot. 2009, 104, 611–620. [Google Scholar] [CrossRef]
- Ma, J.; Wei, H.; Song, M.; Pang, C.; Liu, J.; Wang, L.; Zhang, J.; Fan, S.; Yu, S. Transcriptome profiling analysis reveals that flavonoid and ascorbate-glutathione cycle are important during anther development in Upland cotton. PLoS ONE 2012, 7, e49244. [Google Scholar] [CrossRef]
- Wei, M.; Song, M.; Fan, S.; Yu, S. Transcriptomic analysis of differentially expressed genes during anther development in genetic male sterile and wild type cotton by digital gene-expression profiling. BMC Genom. 2013, 14, 97. [Google Scholar] [CrossRef]
- Engelke, T.; Hirsche, J.; Roitsch, T. Anther-specific carbohydrate supply and restoration of metabolically engineered male sterility. J. Exp. Bot. 2010, 61, 2693–2706. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, J.; Liu, J.; Xia, M.; Wang, W.; Shen, F. Transcriptome Analysis of Early Anther Development of Cotton Revealed Male Sterility Genes for Major Metabolic Pathways. J. Plant Growth Regul. 2015, 34, 223–232. [Google Scholar] [CrossRef]
- Edstam, M.M.; Edqvist, J. Involvement of GPI-anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana. Physiol. Plant 2014, 152, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Min, L.; Wu, Z.; Yang, L.; Zhu, L.; Yang, X.; Yuan, D.; Guo, X.; Zhang, X. Defective pollen wall contributes to male sterility in the male sterile line 1355A of cotton. Sci. Rep. 2015, 5, 9608–9615. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Foyer, C.H. Ascorbate and Glutathione: Keeping Active Oxygen Under Control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, Y.; Zhang, M.; Zhang, J.; Stewart, J.M.; Xing, C.; Wu, J.; Jin, S. Transcriptome, cytological and biochemical analysis of cytoplasmic male sterility and maintainer line in CMS-D8 cotton. Plant Mol. Biol. 2018, 97, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Wheeler, G.L. Ascorbic acid in plants: Biosynthesis and function. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 291–314. [Google Scholar] [CrossRef]
- Filkowski, J.; Kovalchuk, O.; Kovalchuk, I. Genome stability of vtc1, tt4, and tt5 Arabidopsis thaliana mutants impaired in protection against oxidative stress. Plant J. 2004, 38, 60–69. [Google Scholar] [CrossRef]
- Hsieh, K.; Huang, A.H. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell. 2007, 19, 582–596. [Google Scholar] [CrossRef]
- Toshiro, I.; Frank, W.; Hao, Y.; Pradeep, D.; Natsuko, I.; Márcio, A.F.; José Luis, R.; Meyerowitz, E.M. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 2004, 430, 356–360. [Google Scholar]
- Shu-Lan, Y.; Li-Fen, X.; Hui-Zhu, M.; Ching San, P.; Wei-Cai, Y.; Lixi, J.; Venkatesan, S.; De, Y. Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell. 2003, 15, 2792–2804. [Google Scholar]
- Shu-Lan, Y.; Lixi, J.; Ching San, P.; Li-Fen, X.; Xue-Qin, Z.; Li-Qun, C.; Wei-Cai, Y.; De, Y. Overexpression of TAPETUM DETERMINANT1 alters the cell fates in the Arabidopsis carpel and tapetum via genetic interaction with excess microsporocytes1/extra sporogenous cells. Plant Physiol. 2005, 139, 186–191. [Google Scholar]
- Da-Zhong, Z.; Guan-Fang, W.; Brooke, S.; Hong, M. The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 2002, 16, 2021–2031. [Google Scholar]
- Catherine, A.; Eugenia, R.; Valerie, H.; Erik, B.; Sacco, D.V. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell. 2005, 17, 3337–3349. [Google Scholar]
- Hord, C.; Chen, C.; DeYoung, B.; Clark, S.; Ma, H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell. 2006, 18, 1667–1680. [Google Scholar] [CrossRef]
- Liu, X.; Huang, J.; Parameswaran, S.; Ito, T.; Seubert, B.; Auer, M.; Rymaszewski, A.; Jia, G.; Owen, H.A.; Zhao, D. The SPOROCYTELESS/NOZZLE Gene Is Involved in Controlling Stamen Identity in Arabidopsis. Plant Physiol. 2009, 151, 1401–1411. [Google Scholar] [CrossRef]
- Hord, C.L.; Sun, Y.J.; Pillitteri, L.J.; Torii, K.U.; Wang, H.; Zhang, S.; Ma, H. Regulation of Arabidopsis Early Anther Development by the Mitogen-Activated Protein Kinases, MPK3 and MPK6, and the ERECTA and Related Receptor-Like Kinases. Mol. Plant 2008, 1, 645–658. [Google Scholar] [CrossRef]
- Wei, Z.; Yujin, S.; Ljudmilla, T.; Changbin, C.; Ueli, G.; Hong, M. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 2006, 133, 3085–3095. [Google Scholar]
- Baomin, F.; Dihong, L.; Xuan, M.; Yiben, P.; Yujin, S.; Gang, N.; Hong, M. Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. Cell. Mol. Biol. 2012, 72, 612–624. [Google Scholar]
- Mizuno, S.; Osakabe, Y.; Maruyama, K.; Ito, T.; Osakabe, K.; Sato, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Receptor-like protein kinase 2 (RPK 2) is a novel factor controlling anther development in Arabidopsis thaliana. Plant J. 2010, 50, 751–766. [Google Scholar] [CrossRef]
- Na, L.; Da-Sheng, Z.; Hai-Sheng, L.; Chang-Song, Y.; Xiao-Xing, L.; Wan-Qi, L.; Zheng, Y.; Ben, X.; Huang-Wei, C.; Jia, W. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006, 18, 2999–3014. [Google Scholar]
- Da-Sheng, Z.; Wan-Qi, L.; Zheng, Y.; Na, L.; Jing, S.; Jue, W.; Yu-Min, L.; Wen-Juan, Y.; Da-Bing, Z. Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 2008, 1, 599–610. [Google Scholar]
- Fu, Z.; Yu, J.; Cheng, X.; Zong, X.; Xu, J.; Chen, M.; Li, Z.; Zhang, D.; Liang, W. The Rice Basic Helix-Loop-Helix Transcription Factor TDR INTERACTING PROTEIN2 Is a Central Switch in Early Anther Development. Plant Cell. 2014, 26, 1512–1524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lilan, H.; Ding, T.; Yi, S.; Qing, H.; Kejian, W.; Ming, L.; Tiegang, L.; Zhukuan, C. MIL2 (MICROSPORELESS2) regulates early cell differentiation in the rice anther. New Phytol. 2012, 196, 402–413. [Google Scholar]
- Lilan, H.; Ding, T.; Keming, Z.; Kejian, W.; Ming, L.; Zhukuan, C. Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell. 2012, 24, 577–588. [Google Scholar]
- Zhu, E.; You, C.; Wang, S.; Cui, J.; Niu, B.; Wang, Y.; Qi, J.; Ma, H.; Chang, F. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. Cell. Mol. Biol. 2015, 83, 976–990. [Google Scholar] [CrossRef]
- Lifang, H.; Wanqi, L.; Changsong, Y.; Xiao, C.; Jie, Z.; Xing, W.; Jianping, H.; Dabing, Z. Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell. 2011, 23, 515–533. [Google Scholar]
- Li, H.; Yuan, Z.; Vizcay-Barrena, G.; Yang, C.; Liang, W.; Zong, J.; Wilson, Z.A.; Zhang, D. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 2011, 156, 615–630. [Google Scholar] [CrossRef]
- Niu, N.; Liang, W.; Yang, X.; Jin, W.; Wilson, Z.A.; Hu, J.; Zhang, D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef]
- Li, X.; Gao, X.; Wei, Y.; Deng, L.; Ouyang, Y.; Chen, G.; Li, X.; Zhang, Q.; Wu, C. Rice APOPTOSIS INHIBITOR5 coupled with two DEAD-box adenosine 5′-triphosphate-dependent RNA helicases regulates tapetum degeneration. Plant Cell. 2011, 23, 1416–1434. [Google Scholar] [CrossRef]
- Luo, D.; Xu, H.; Liu, Z.; Guo, J.; Li, H.; Chen, L.; Fang, C.; Zhang, Q.; Bai, M.; Yao, N.; et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 2013, 45, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Li, X.B. The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. Plant J. 2009, 57, 473–486. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, J.; Du, M.L.; Li, L.; Wang, X.L.; Li, X.B. A cotton gene encoding MYB-like transcription factor is specifically expressed in pollen and is involved in regulation of late anther/pollen development. Plant Cell. Physiol. 2013, 54, 893–906. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Semwal, V.K.; Singh, B.; Khanna-Chopra, R. Delayed expression of SAGs correlates with longevity in CMS wheat plants compared to its fertile plants. Physiol. Mol. Biol. Plants 2014, 20, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Liang, C.Y.; Zhang, M.Y. The relationship between membrane lipid peroxidation and cytoplasmic male sterility in maize. Plant Physiol. Commun. 1996, 32, 331–334. [Google Scholar]
- Jiang, P.; Zhang, X.; Zhu, Y.; Zhu, W.; Xie, H.; Wang, X. Metabolism of reactive oxygen species in cotton cytoplasmic male sterility and its restoration. Plant Cell. Rep. 2007, 26, 1627–1634. [Google Scholar] [CrossRef]
- Yi, J.; Moon, S.; Lee, Y.S.; Zhu, L.; Liang, W.; Zhang, D.; Jung, K.H.; An, G. Defective Tapetum Cell Death 1 (DTC1) Regulates ROS Levels by Binding to Metallothionein during Tapetum Degeneration. Plant Physiol. 2016, 170, 1611–1623. [Google Scholar] [CrossRef]
- Chase, C.D. Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007, 23, 81–90. [Google Scholar] [CrossRef]
- Suzuki, H.; Rodriguez-Uribe, L.; Xu, J.; Zhang, J. Transcriptome analysis of cytoplasmic male sterility and restoration in CMS-D8 cotton. Plant Cell. Rep. 2013, 32, 1531–1542. [Google Scholar] [CrossRef]
- Chen, Z.; Nan, Z.; Li, S.; Grover, C.E.; Nie, H.; Wendel, J.F.; Hua, J. Plant Mitochondrial Genome Evolution and Cytoplasmic Male Sterility. Crit. Rev. Plant Sci. 2017, 36, 1–15. [Google Scholar] [CrossRef]
- He, S.; Lyznik, A.; Mackenzie, S. Pollen fertility restoration by nuclear gene Fr in CMS bean: Nuclear-directed alteration of a mitochondrial population. Genetics 1995, 139, 955–962. [Google Scholar] [PubMed]
- Yi, P.; Wang, L.; Sun, Q.; Zhu, Y. Discovery of mitochondrial chimeric-gene associated with cytoplasmic male sterility of HL-rice. Chin. Sci. Bull. 2002, 47, 744–747. [Google Scholar] [CrossRef]
- Dewey, R.E.; Timothy, D.H.; Iii, C.S.L. Chimeric mitochondrial genes expressed in the C male-sterile cytoplasm of maize. Curr. Genet. 1991, 20, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, Z.; Zhao, N.; Wang, Y.; Nie, H.; Hua, J. The comparison of four mitochondrial genomes reveals cytoplasmic male sterility candidate genes in cotton. BMC Genom. 2018, 19, 775. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar]
- Hollender, C.A.; Kang, C.; Darwish, O.; Geretz, A.; Matthews, B.F.; Slovin, J.; Alkharouf, N.; Liu, Z. Floral Transcriptomes in Woodland Strawberry Uncover Developing Receptacle and Anther Gene Networks. Plant Physiol. 2014, 165, 1062–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunbo, W.; Zhiwen, W.; Fuguang, L.; Wuwei, Y.; Junyi, W.; Guoli, S.; Zhen, Y.; Lin, C.; Haihong, S.; Shilin, Z. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar]
- Fuguang, L.; Guangyi, F.; Kunbo, W.; Fengming, S.; Youlu, Y.; Guoli, S.; Qin, L.; Zhiying, M.; Cairui, L.; Changsong, Z. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 3, 567–572. [Google Scholar]
- Kong, X.; Liu, D.; Liao, X.; Zheng, J.; Diao, Y.; Liu, Y.; Zhou, R. Comparative Analysis of the Cytology and Transcriptomes of the Cytoplasmic Male Sterility Line H276A and Its Maintainer Line H276B of Cotton (Gossypium barbadense L.). Int. J. Mol. Sci. 2017, 18, 2240. [Google Scholar] [CrossRef]
- Liu, J.; Pang, C.; Wei, H.; Song, M.; Meng, Y.; Ma, J.; Fan, S.; Yu, S. iTRAQ-facilitated proteomic profiling of anthers from a photosensitive male sterile mutant and wild-type cotton (Gossypium hirsutum L.). J. Proteom. 2015, 126, 68–81. [Google Scholar] [CrossRef]
- Rao, Y. Cytohistology of cytoplasmic male sterile lines in hybrid rice. Hybrid rice Proceedings of an international symposium, Changsha, China, October 1986; pp. 115–128. [Google Scholar]
- Wang, X.; Pan, J. Genetic basis of fertility restoration to cytoplasmic male sterile lines available in China in upland cotton II. Interactive effects between restorer genes and fertility enhancer gene. Acta. Genet. Sin. 1997, 24, 276–277. [Google Scholar]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; Mcintire, K.N.; Hsu, Y.C.; Pei, Y.L.; Mai, T.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Budar, F.; Pelletier, G. Male sterility in plants: Occurrence, determinism, significance and use. C R Acad. Sci. Iii. 2001, 324, 543–550. [Google Scholar] [CrossRef]
- Linke, B.; Borner, T. Mitochondrial effects on flower and pollen development. Mitochondrion 2005, 5, 389–402. [Google Scholar] [CrossRef]
- Touzet, P.; Budar, F. Unveiling the molecular arms race between two conflicting genomes in cytoplasmic male sterility? Trends Plant Sci. 2004, 9, 568–570. [Google Scholar] [CrossRef]
- Green, D.R.; Reed, J.C. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.; Panda, P.; Sahoo, L.; Panda, S.K. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 2013, 8, e23681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Dai, X.; Li, L.; Jiao, Z.; Huang, Q. Metabolism of reactive oxygen species in cytoplasmic male sterility of rice by marking upmost pulvinus interval. Appl. Biochem. Biotechnol. 2015, 175, 1263–1269. [Google Scholar] [CrossRef]
- Ming-Hua, D.; Jin-Fen, W.; Jin-Long, H.; Hai-Shan, Z.; Xiong-Ze, D.; Zhu-Qing, Z.; Xue-Xiao, Z. Relationship of metabolism of reactive oxygen species with cytoplasmic male sterility in pepper (Capsicum annuum L.). Sci. Hortic. 2012, 134, 232–236. [Google Scholar]
- Liu, Z.; Shi, X.; Li, S.; Hu, G.; Zhang, L.; Song, X. Tapetal-Delayed Programmed Cell Death (PCD) and Oxidative Stress-Induced Male Sterility of Aegilops uniaristata Cytoplasm in Wheat. Int. J. Mol. Sci. 2018, 19, 1708. [Google Scholar] [CrossRef]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate Peroxidase and Catalase Activities and Their Genetic Regulation in Plants Subjected to Drought and Salinity Stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [Green Version]
- Wan, C.; Li, S.; Wen, L.; Kong, J.; Wang, K.; Zhu, Y. Damage of oxidative stress on mitochondria during microspores development in Honglian CMS line of rice. Plant. Cell. Rep. 2007, 26, 373–382. [Google Scholar] [CrossRef]
- Guo, J.; Wang, P.; Cheng, Q.; Sun, L.; Wang, H.; Wang, Y.; Kao, L.; Li, Y.; Qiu, T.; Yang, W.; et al. Proteomic analysis reveals strong mitochondrial involvement in cytoplasmic male sterility of pepper (Capsicum annuum L.). J. Proteom. 2017, 168, 15–27. [Google Scholar] [CrossRef]
- op den Camp, R.G.; Kuhlemeier, C. Aldehyde dehydrogenase in tobacco pollen. Plant. Mol. Biol. 1997, 35, 355–365. [Google Scholar] [CrossRef]
- Ducos, E.; Touzet, P.; Boutry, M. The male sterile G cytoplasm of wild beet displays modified mitochondrial respiratory complexes. Plant J. 2001, 26, 171–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.H.; Kang, J.G.; Kim, B.D. Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.). Plant Mol. Biol. 2007, 63, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.T.; Yao, N. The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 2004, 6, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Banting, G.S.; Glerum, D.M. Mutational analysis of the Saccharomyces cerevisiae cytochrome c oxidase assembly protein Cox11p. Eukaryot. Cell. 2006, 5, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Veniamin, S.; Sawatzky, L.G.; Banting, G.S.; Glerum, D.M. Characterization of the peroxide sensitivity of COX-deficient yeast strains reveals unexpected relationships between COX assembly proteins. Free Radic. Biol. Med. 2011, 51, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y.; et al. Cytochrome P450 family member CYP704B2 catalyzes the -hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell. 2010, 22, 173–190. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.N.; Zhu, J.; Yu, Y.; Teng, X.D.; Lou, Y.; Xu, X.F.; Liu, J.L.; Yang, Z.N. DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J. 2014, 80, 1005–1013. [Google Scholar] [CrossRef]
- Andriankaja, M.E.; Danisman, S.; Mignolet-Spruyt, L.F.; Claeys, H.; Kochanke, I.; Vermeersch, M.; De Milde, L.; De Bodt, S.; Storme, V.; Skirycz, A.; et al. Transcriptional coordination between leaf cell differentiation and chloroplast development established by TCP20 and the subgroup Ib bHLH transcription factors. Plant Mol. Biol. 2014, 85, 233–245. [Google Scholar] [CrossRef]
- Sorensen, A.M.; Krober, S.; Unte, U.S.; Huijser, P.; Dekker, K.; Saedler, H. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003, 33, 413–423. [Google Scholar] [CrossRef]
- Nakata, M.; Ohme-Takagi, M. Two bHLH-type transcription factors, JA-ASSOCIATED MYC2-LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signal. Behav. 2013, 8, e26473. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.H.; Han, M.J.; Lee, Y.S.; Kim, Y.W.; Hwang, I.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; An, G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 2005, 17, 2705–2722. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.S.; Chan, M.T. The bHLH142 Transcription Factor Coordinates with TDR1 to Modulate the Expression of EAT1 and Regulate Pollen Development in Rice. Plant Cell. 2014, 26, 2486–2504. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Wang, L.; Wang, J.; Zeng, Y.; Xu, Y.; Li, S. The transcription factor OsbHLH138 regulates thermosensitive genic male sterility in rice via activation of TMS5. Appl. Genet. 2019, 132, 1721–1723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.B.; Zhu, J.; Gao, J.F.; Wang, C.; Li, H.; Li, H.; Zhang, H.Q.; Zhang, S.; Wang, D.M.; Wang, Q.X.; et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 2007, 52, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005, 17, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, H.; Li, H.; Gao, J.F.; Jiang, H.; Wang, C.; Guan, Y.F.; Yang, Z.N. Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008, 55, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Mandaokar, A.; Thines, B.; Shin, B.; Lange, B.M.; Choi, G.; Koo, Y.J.; Yoo, Y.J.; Choi, Y.D.; Choi, G.; Browse, J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006, 46, 984–1008. [Google Scholar] [CrossRef]
- Andrews, J.; Keegstra, K. Acyl-CoA Synthetase Is Located in the Outer Membrane and Acyl-CoA Thioesterase in the Inner Membrane of Pea Chloroplast Envelopes. Plant Physiol. 1983, 72, 735–740. [Google Scholar] [CrossRef] [Green Version]
- Ichihara, K.I.; Nakagawa, M.; Tanaka, K. Acyl-CoA Synthetase in Maturing Safflower Seeds. Plant Cell. Physiol. 1993, 34, 557–566. [Google Scholar]
- Judy, S.; Jay, S.; John, B. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004, 16, 629–642. [Google Scholar]
- Fulda, M.; Shockey, J.; Werber, M.; Wolter, F.P.; Heinz, E. Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid beta-oxidation. Plant J. 2002, 32, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Katja, S.; Lucie, K.; Elmon, S.; Thomas, C.; Michael, B.; Otto, M.; Claus, W.; Erich, K.; Hans-Peter, S. A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J. Biol. Chem. 2005, 280, 13962–13972. [Google Scholar]
- Wu, Z.; Cheng, J.; Qin, C.; Hu, Z.; Yin, C.; Hu, K. Differential proteomic analysis of anthers between cytoplasmic male sterile and maintainer lines in Capsicum annuum L. Int. J. Mol. Sci. 2013, 14, 22982–22996. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, M.; Croll, D.; Kronstad, J.W. Maize susceptibility to Ustilago maydis is influenced by genetic and chemical perturbation of carbohydrate allocation. Mol. Plant Pathol. 2017, 18, 1222–1237. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Song, Y.; Guo, J.; Wang, J.; Zhang, L.; Niu, N.; Ma, S.; Zhang, G.; Zhao, H. Physiological and metabolome changes during anther development in wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2018, 132, 18–32. [Google Scholar] [CrossRef]
- Datta, R.; Chamusco, K.C.; Chourey, P.S. Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol. 2002, 130, 1645–1656. [Google Scholar] [CrossRef]
- Mamun, E.A.; Alfred, S.; Cantrill, L.C.; Overall, R.L.; Sutton, B.G. Effects of chilling on male gametophyte development in rice. Cell. Biol. Int. 2006, 30, 583–591. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Y.; Li, Y.; Ma, Y.; Zhao, Y.; Wang, C.; Chi, H.; Chen, M.; Ding, Y.; Guo, X.; et al. Proteomic analysis reveals that sugar and fatty acid metabolisms play a central role in sterility of the male-sterile line 1355A of cotton. J. Biol. Chem. 2019, 294, 7057–7067. [Google Scholar] [CrossRef]
- Goss, J.A. Development, physiology, and biochemistry of corn and wheat pollen. Bot. Rev. 1968, 34, 333–358. [Google Scholar] [CrossRef]
- Mascarenhas, J.P. The Biochemistry of Angiosperm Pollen Development. Bot. Rev. 1975, 41, 259–314. [Google Scholar] [CrossRef]
- Yemm, E.W.; Willis, A.J. The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 1954, 57, 508–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Landi, M. Commentary to: “Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds” by Hodges et al., Planta. (1999) 207:604–611. Planta 2017, 245, 1067. [Google Scholar] [CrossRef] [PubMed]
- Bibi, N.; Yuan, S.; Zhu, Y.; Wang, X. Improvements of Fertility Restoration in Cytoplasmic Male Sterile Cotton by Enhanced Expression of Glutathione S-Transferase (GST) Gene. J. Plant Growth Regul. 2014, 33, 420–429. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
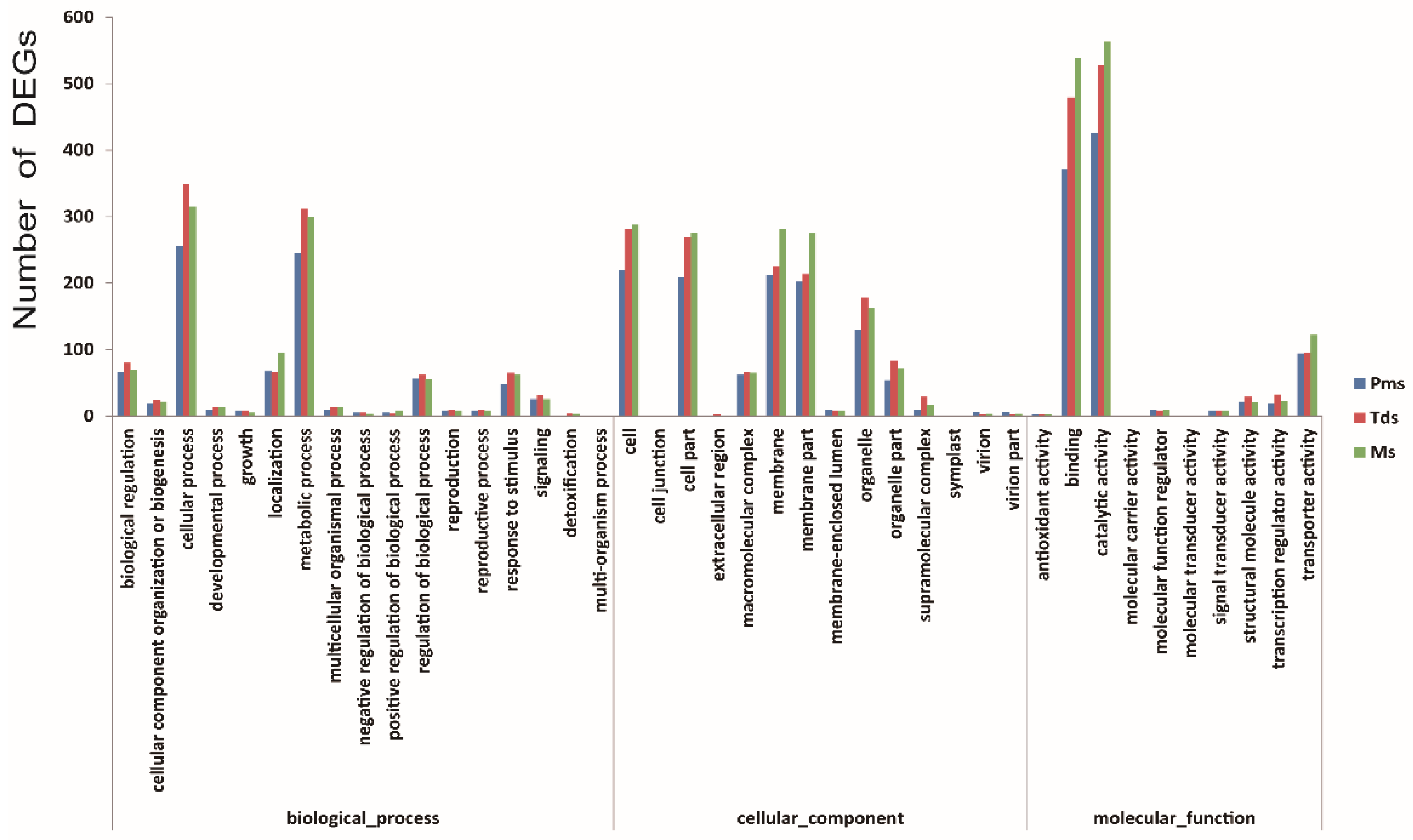
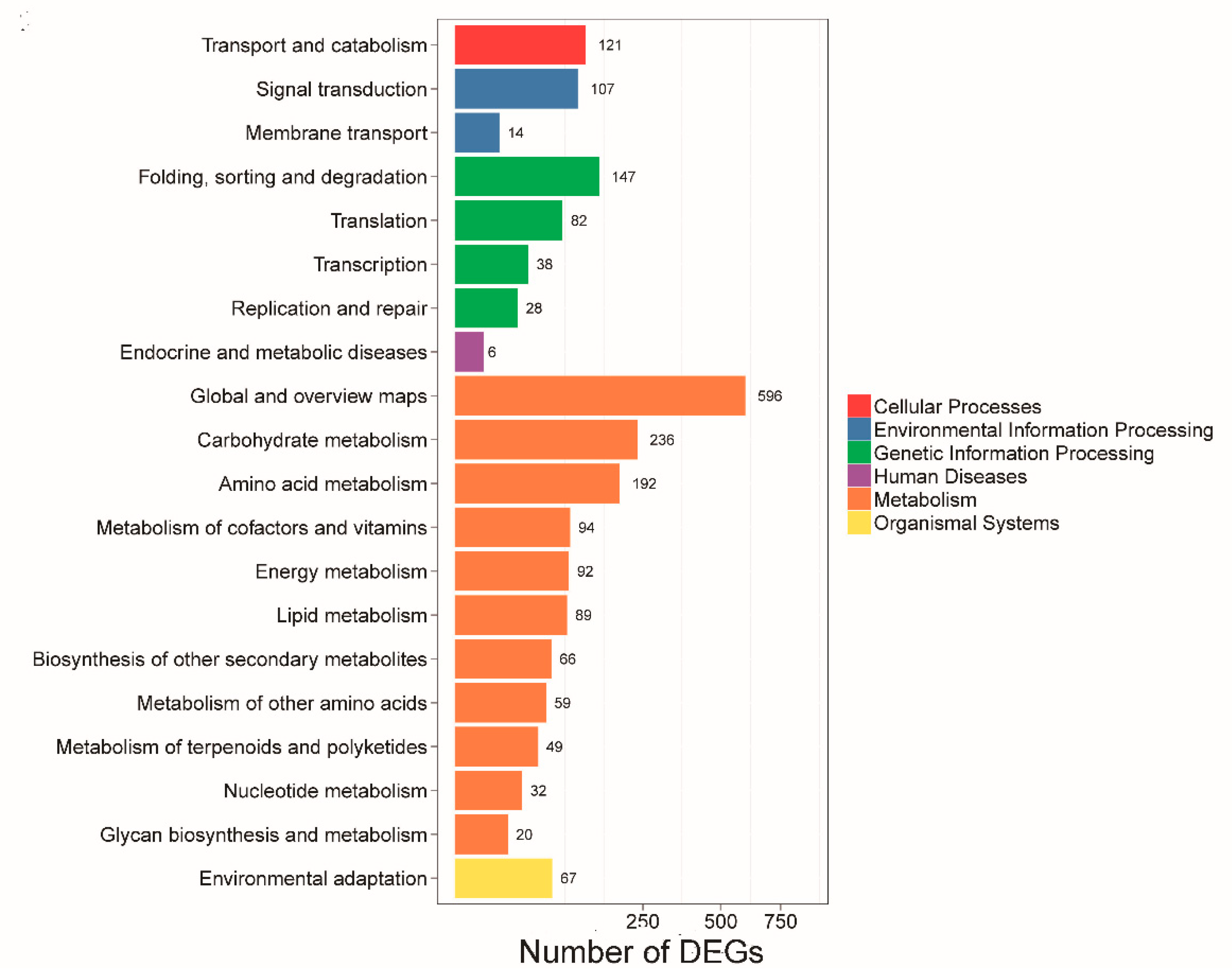
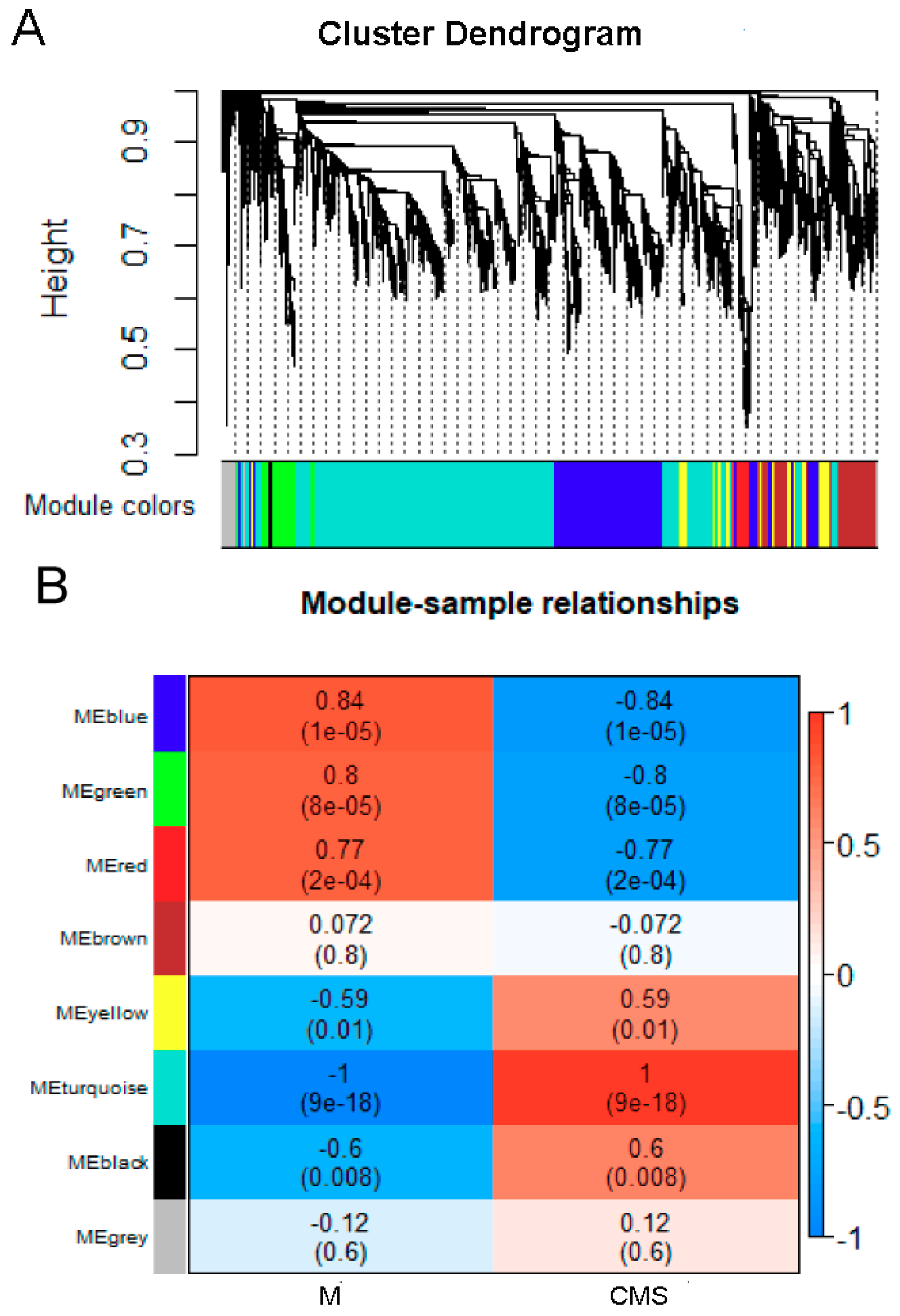
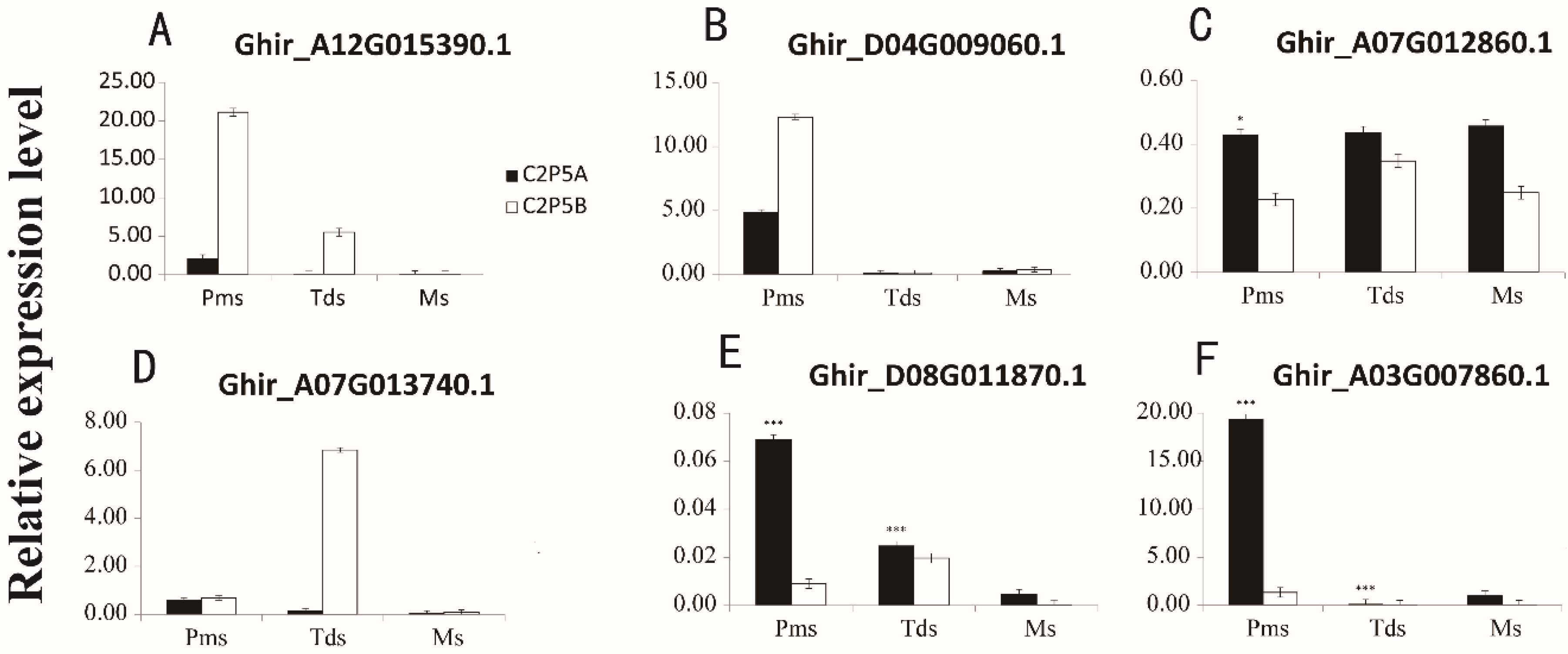
| Sample | Raw Data | Clean Data | GC (%) | Q30(%) | (%)of Total Mapped | Total Gene Number | Total Isoforms | Novel Isoforms |
|---|---|---|---|---|---|---|---|---|
| MsPms1 | 96339898 | 84112026 | 44.33 | 97.65 | 77.42 | 85012 | 126455 | 49311 |
| MsPms2 | 96339060 | 84728466 | 44.43 | 97.84 | 75.51 | 83758 | 122693 | 48159 |
| MsPms3 | 96340692 | 84822750 | 44.52 | 97.81 | 75.44 | 85170 | 125779 | 49293 |
| MsTds1 | 66418638 | 60016542 | 44.22 | 98.33 | 76.38 | 83808 | 122636 | 47699 |
| MsTds2 | 96341884 | 84562392 | 43.57 | 97.68 | 77.36 | 86647 | 129005 | 50208 |
| MsTds3 | 96340878 | 84830410 | 44 | 97.77 | 74.54 | 86626 | 126787 | 50096 |
| MsMs1 | 70226770 | 62112146 | 45.21 | 98.35 | 78.11 | 82246 | 117948 | 45803 |
| MsMs2 | 96340984 | 84269862 | 44.5 | 97.81 | 77.01 | 85808 | 124373 | 49192 |
| MsMs3 | 75363530 | 68729552 | 44.43 | 98.45 | 79.47 | 85289 | 123996 | 48513 |
| MPms1 | 78357630 | 71140510 | 43.98 | 98.42 | 77.1 | 84009 | 123651 | 49187 |
| MPms2 | 93592588 | 83300884 | 43.81 | 97.97 | 76.95 | 84628 | 124284 | 49724 |
| MPms3 | 94122576 | 81326894 | 44.24 | 97.83 | 73.56 | 84969 | 124580 | 49711 |
| MTds1 | 93071774 | 84922682 | 43.79 | 98.38 | 78.87 | 84527 | 124186 | 49781 |
| MTds2 | 79502370 | 72818484 | 43.85 | 98.23 | 80.11 | 83170 | 121809 | 48480 |
| MTds3 | 71210292 | 64700084 | 43.83 | 98.33 | 79.25 | 82748 | 120584 | 48031 |
| MMs1 | 96340778 | 84992800 | 44.36 | 98.03 | 75.37 | 84192 | 124176 | 48798 |
| MMs2 | 96340324 | 84734142 | 43.65 | 97.83 | 77.69 | 84433 | 124979 | 49064 |
| MMs3 | 74569750 | 67998900 | 43.6 | 98.23 | 75.57 | 82250 | 120841 | 47466 |
| Gene ID | Protein Identity | Pms(Fold Change) | Tds(Fold Change) | Ms(Fold Change) | |||
|---|---|---|---|---|---|---|---|
| qRT-PCR | RNA-seq | qRT-PCR | RNA-seq | qRT-PCR | RNA-seq | ||
| Ghir_A10G019450.1 | Cell wall/membrane/envelope biogenesis | −5.41 | −2.68 | 4.51 | 3.15 | 3.47 | 2.37 |
| Ghir_A07G013740.1 | Long chain acyl-CoA synthetase | −1.59 | −2.56 | −3.52 | −2.64 | −0.16 | −2.88 |
| Ghir_A12G015390.1 | Aldehyde dehydrogenase family | −3.88 | −6.73 | −6.10 | −9.37 | −1.05 | −6.86 |
| Ghir_D08G011870.1 | bHLH-MYC transcription factors | 2.87 | 2.13 | 2.00 | 2.16 | 4.67 | 2.52 |
| Ghir_A07G012860.1 | Cytochrome P450 | 0.98 | 2.12 | 0.23 | 2.50 | 0.98 | 2.44 |
| Ghir_D04G009060.1 | Cytochrome oxidase c subunit VIb | −4.53 | −3.49 | −0.29 | −2.37 | 1.87 | 3.11 |
| Ghir_A03G007860.1 | Myb-like DNA-binding domain | 1.61 | 2.56 | 3.70 | 2.23 | 5.73 | 2.19 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Qin, T.; Wei, C.; Sun, J.; Dong, T.; Zhou, R.; Chen, Q.; Wang, Q. Using Transcriptome Analysis to Screen for Key Genes and Pathways Related to Cytoplasmic Male Sterility in Cotton (Gossypium hirsutum L.). Int. J. Mol. Sci. 2019, 20, 5120. https://doi.org/10.3390/ijms20205120
Li Y, Qin T, Wei C, Sun J, Dong T, Zhou R, Chen Q, Wang Q. Using Transcriptome Analysis to Screen for Key Genes and Pathways Related to Cytoplasmic Male Sterility in Cotton (Gossypium hirsutum L.). International Journal of Molecular Sciences. 2019; 20(20):5120. https://doi.org/10.3390/ijms20205120
Chicago/Turabian StyleLi, Yuqing, Tengfei Qin, Chunyan Wei, Jialiang Sun, Tao Dong, Ruiyang Zhou, Quanjia Chen, and Qinglian Wang. 2019. "Using Transcriptome Analysis to Screen for Key Genes and Pathways Related to Cytoplasmic Male Sterility in Cotton (Gossypium hirsutum L.)" International Journal of Molecular Sciences 20, no. 20: 5120. https://doi.org/10.3390/ijms20205120
APA StyleLi, Y., Qin, T., Wei, C., Sun, J., Dong, T., Zhou, R., Chen, Q., & Wang, Q. (2019). Using Transcriptome Analysis to Screen for Key Genes and Pathways Related to Cytoplasmic Male Sterility in Cotton (Gossypium hirsutum L.). International Journal of Molecular Sciences, 20(20), 5120. https://doi.org/10.3390/ijms20205120




