Light-Regulation of Tryptophan Synthase by Combining Protein Design and Enzymology
Abstract
:1. Introduction
2. Results and Discussion
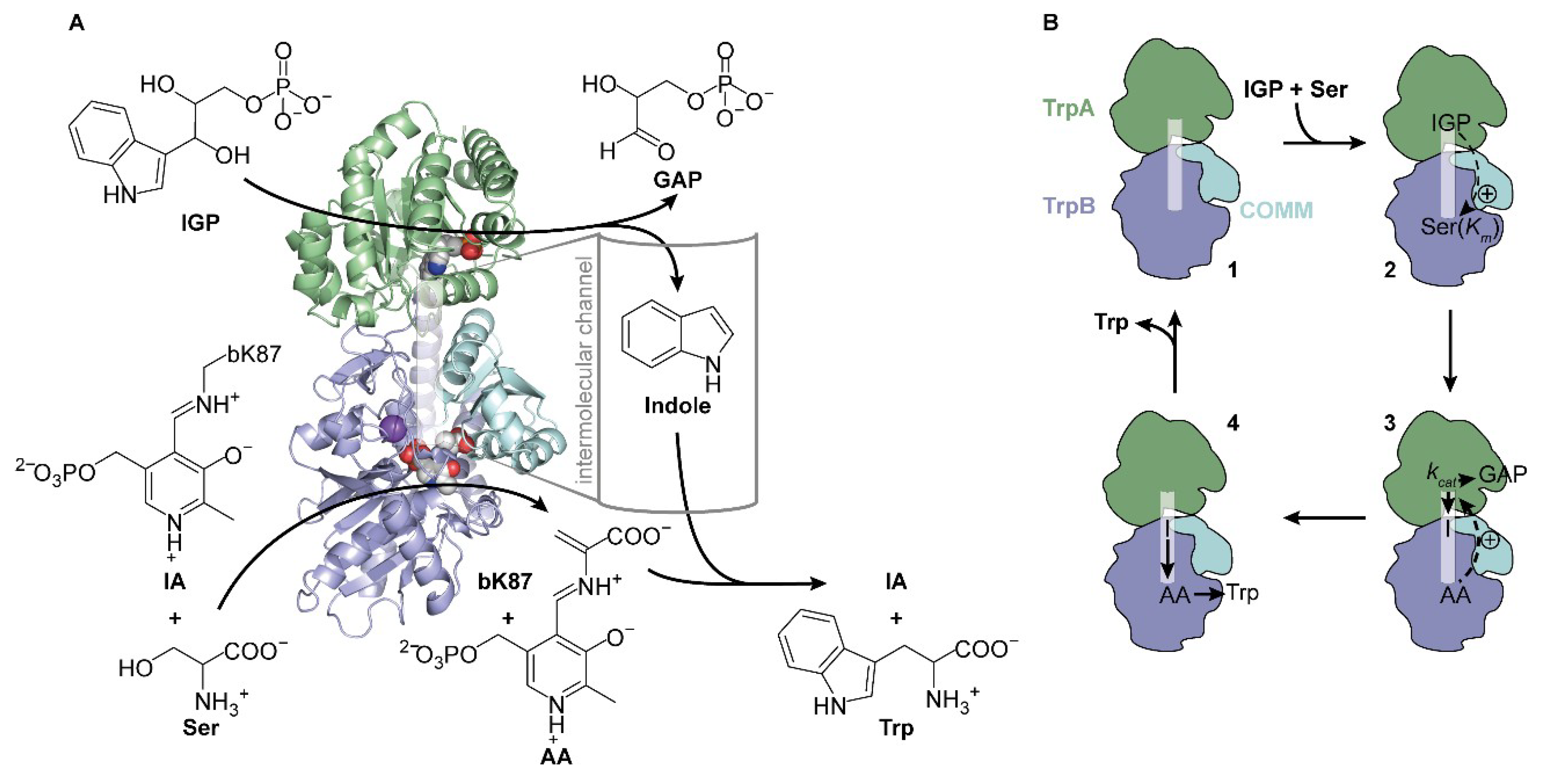
2.1. Kinetic Characterization of TS from Salmonella typhimurium
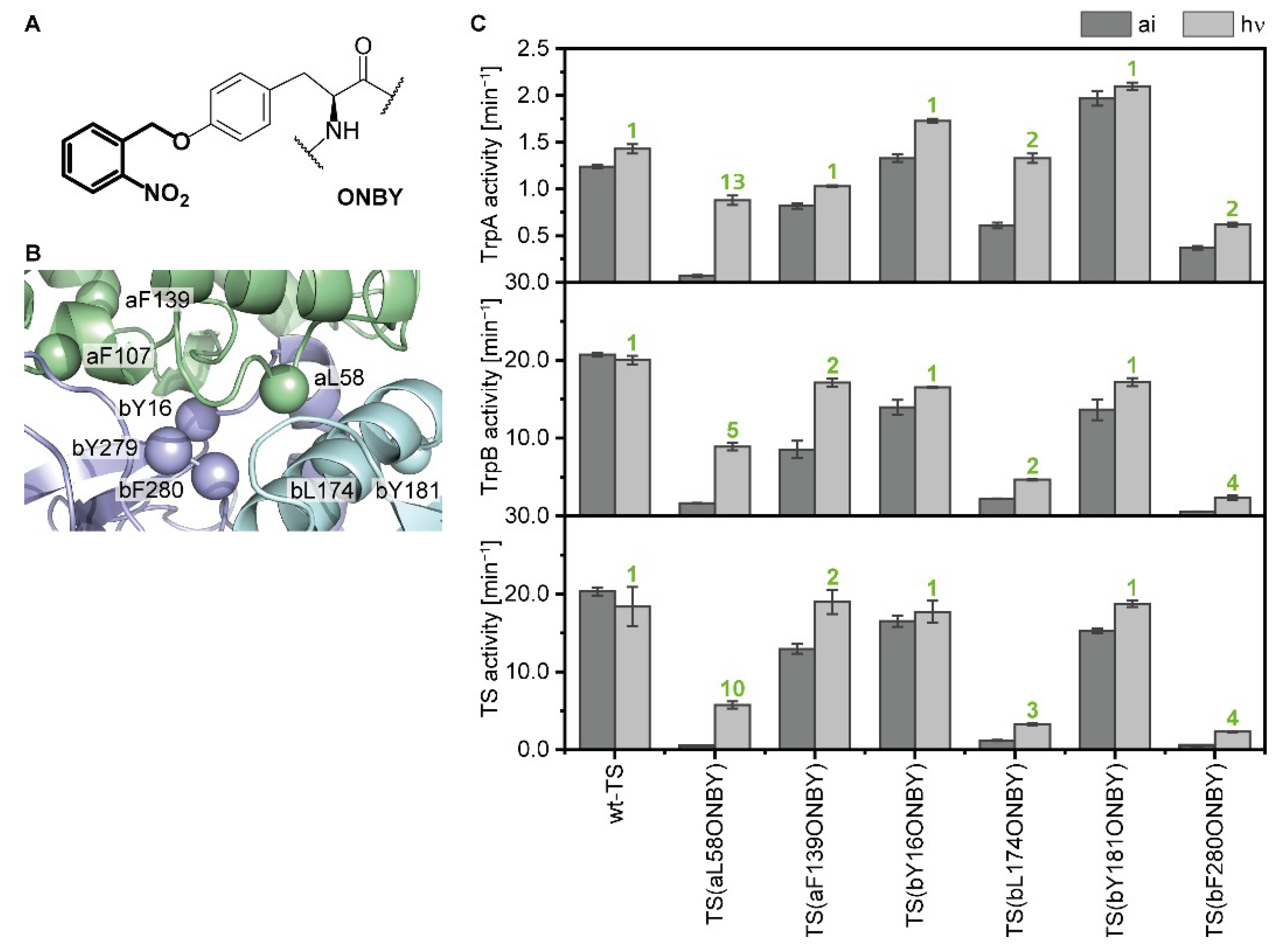
2.2. Identification and Characterization of An Efficient Position for Light-Activation of TS
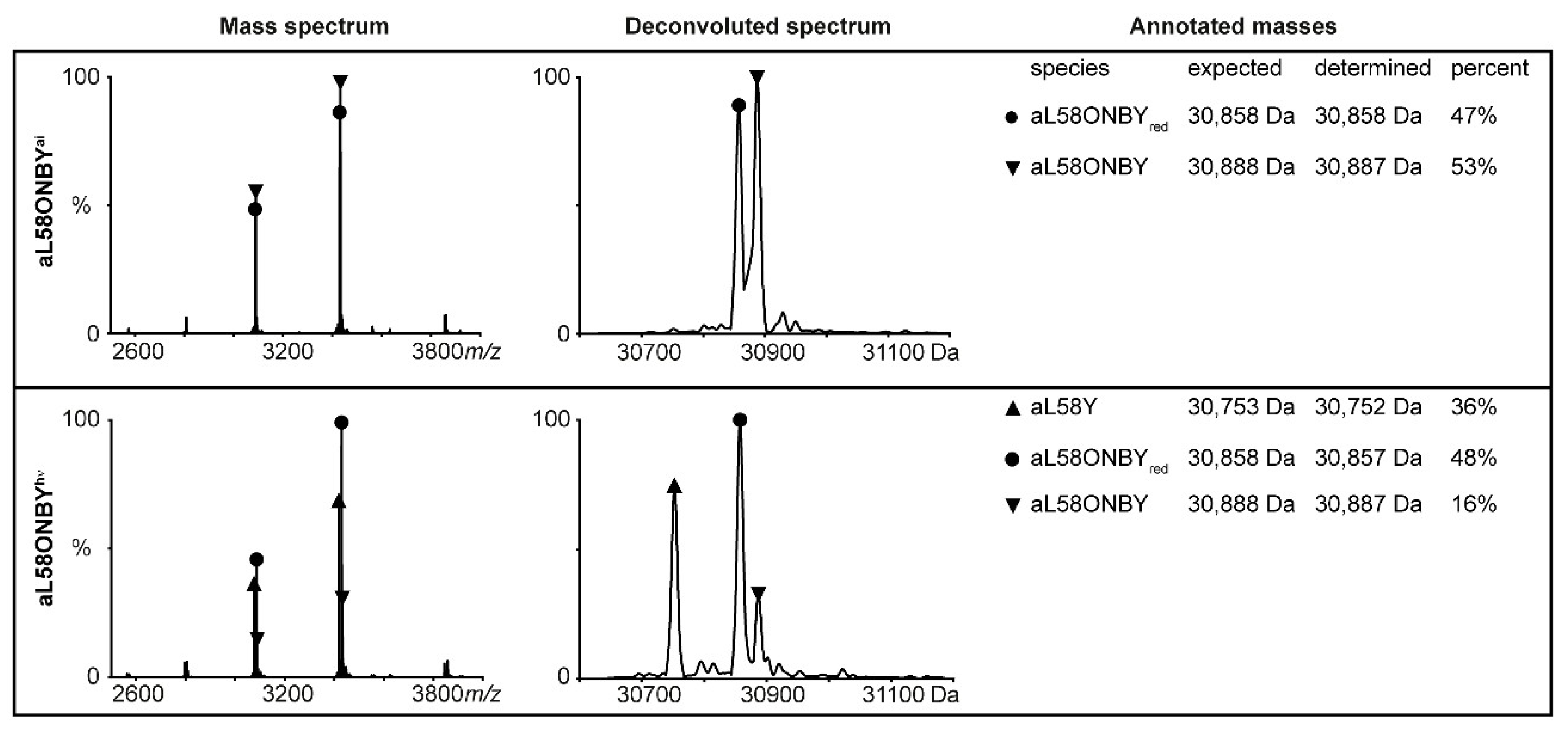
2.3. Inhibition of Catalytic Actvity of TS(aL58ONBY) Can Be Reversed by Light
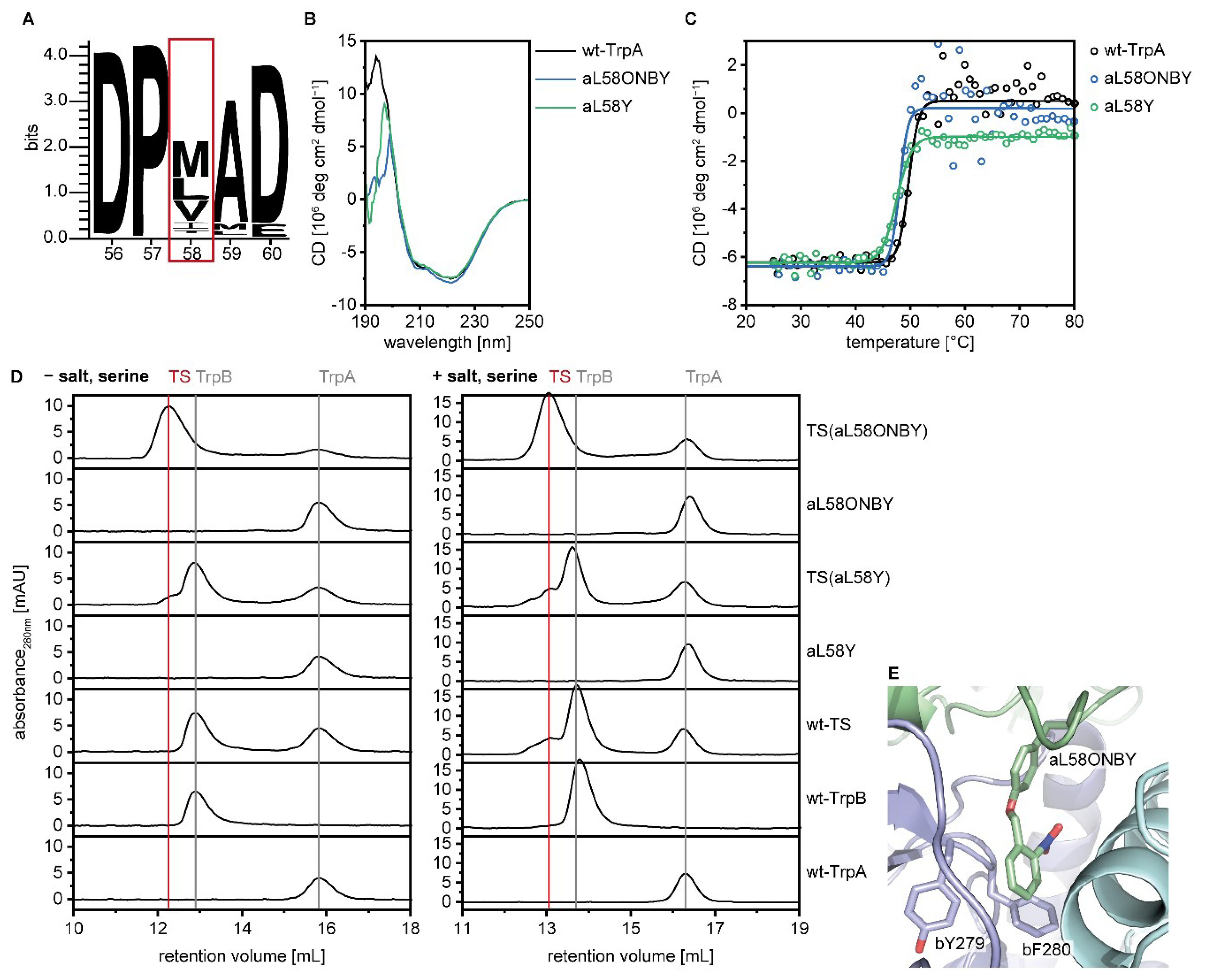
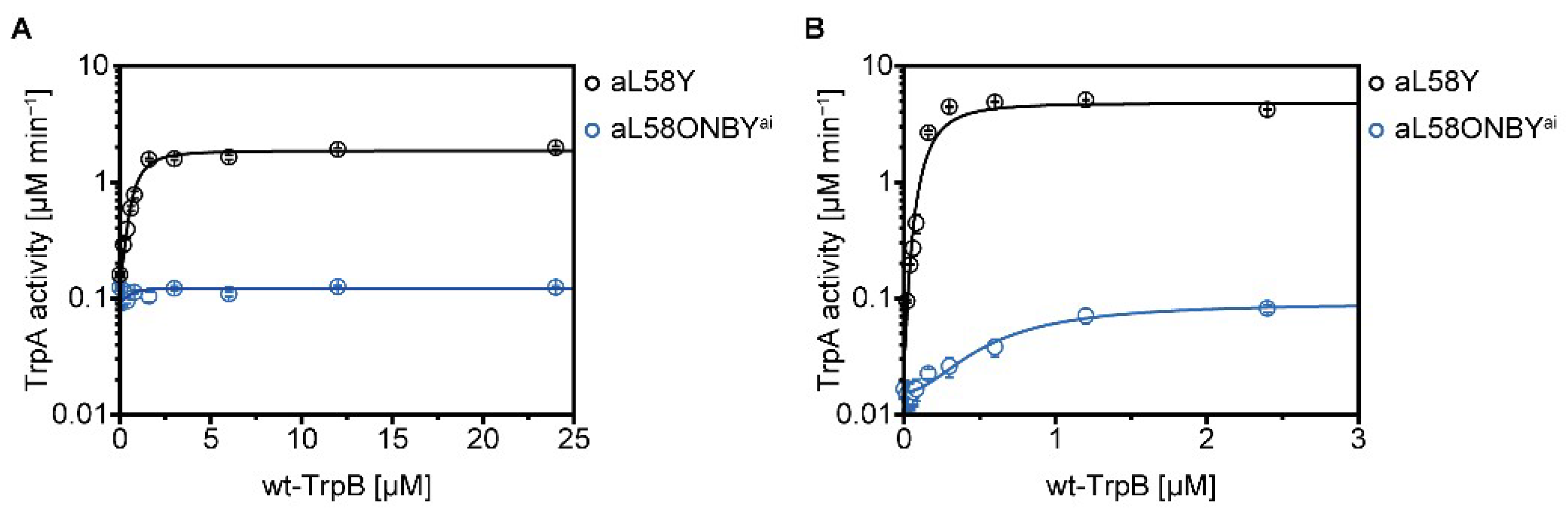
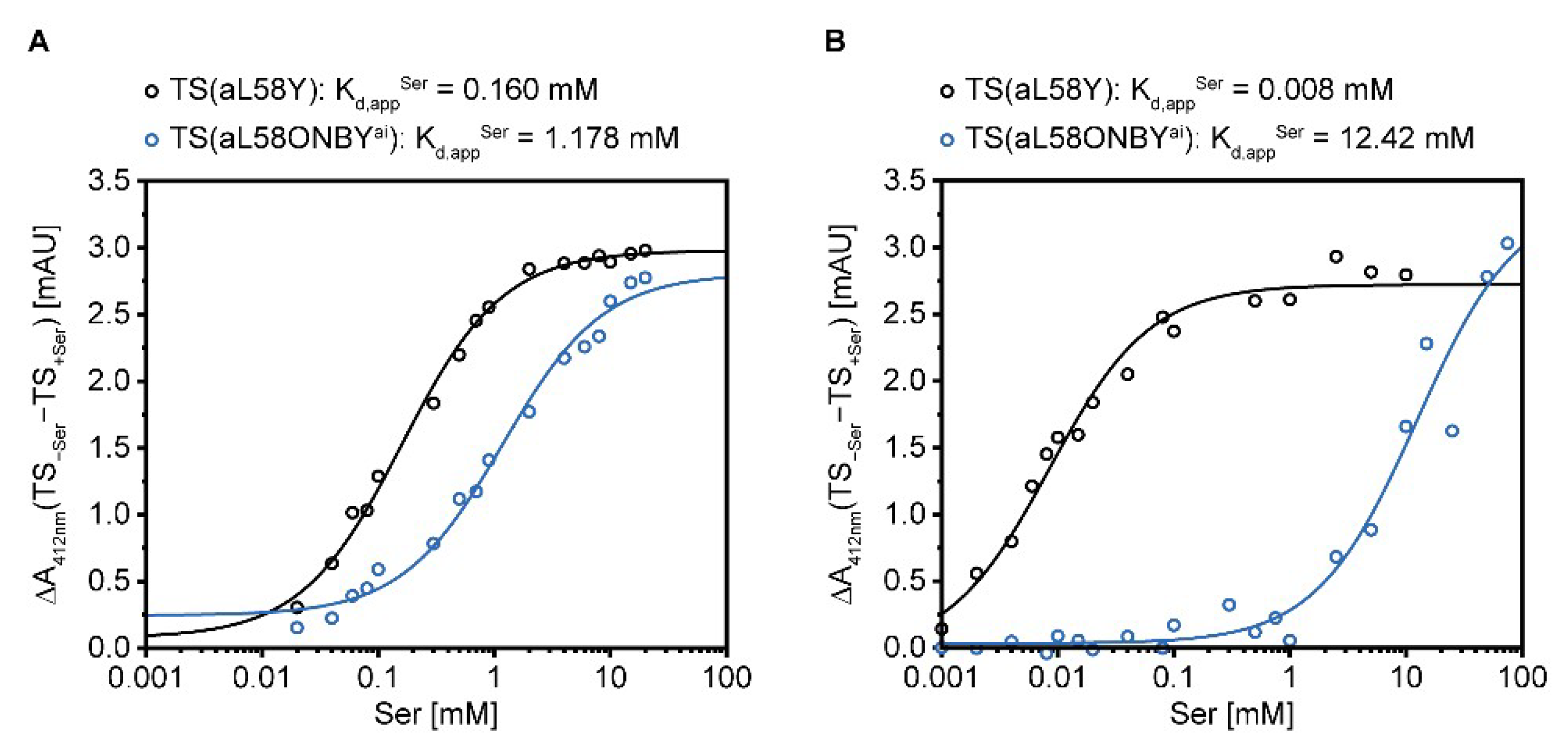
2.4. Allosteric Activation of TrpA and TrpB is Inhibited by aL58ONBY
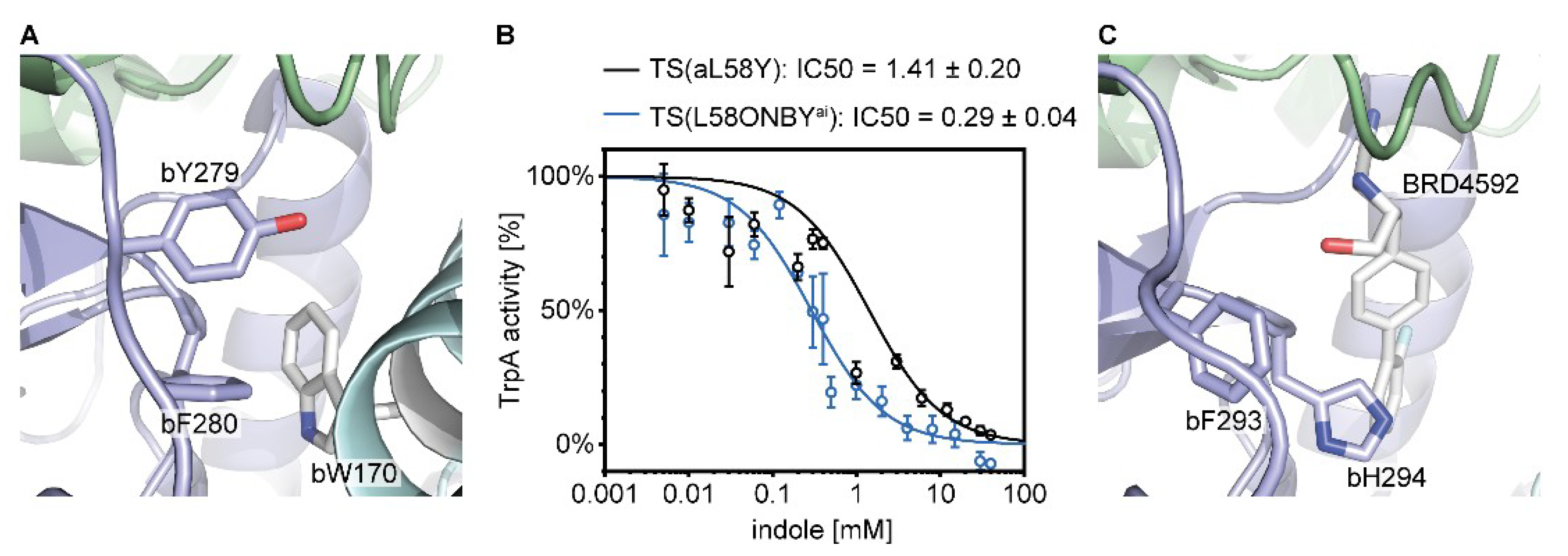
2.5. Optimized Light-Activation of TS by Combination of Kinetic Effects
4. Materials and Methods
4.1. Strains, Enzymes, and Chemicals
4.2. Subcloning of the trpA, ectrpA, ectrpB, and vioA Genes
4.3. Site-Directed Mutagenesis of trpA and trpB
4.4. Expression and Purification of TS
4.5. Expression and Purification of Auxiliary Enzymes
4.6. Activity Measurements
4.7. Tryptic Digest and MS Analysis of Protein Sequences
4.8. MD Simulation
4.9. Generating a Sequence Logo for TrpA Subunits
4.10. Native MS Analysis
4.11. CD Analysis
4.12. Analytical Size-Exclusion Chromatography
4.13. UV/Vis Spectral Analysis of Serine Binding
4.14. Equations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| UAA | Unnatural amino acid |
| aaRS | Aminoacyl-tRNA synthetase |
| ONBY | o-nitrobenzyl-O-tyrosine |
| LAF | Light activation factor |
| TS | Tryptophan synthase from Salmonella typhimurium |
| IGP | Indole glycerol phosphate |
| GAP | Glyceraldehyde-3-phosphate |
| PLP | Pyridoxal phosphate |
| AA | Aminoacrylate |
| IA | Internal aldimine |
| TS | Tryptophan synthase from Saccharomyces cerevisiae |
| ecTS | Tryptophan synthase from Escherichia coli |
| HRP | Horse-radish peroxidase |
| MS | Mass spectrometry |
| MD | Molecular dynamics |
| CD | Circular dichroism |
| ai | “as isolated” |
| hν | Irradiated |
| SEM | Standard error of mean |
| SE | Standard error |
References
- Schmidt-Dannert, C.; Lopez-Gallego, F. A roadmap for biocatalysis–functional and spatial orchestration of enzyme cascades. Microb. Biotechnol. 2016, 9, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T.; Höhne, M. (Eds.) Protein Engineering: Methods and Protocols; Springer: New York, NY, USA, 2018; ISBN 978-1-4939-7366-8. [Google Scholar]
- Arnold, F.H. Directed evolution: Bringing new chemistry to life. Angew. Chem. Int. Ed. 2018, 57, 4143–4148. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xun, G.; Feng, Y. The state-of-the-art strategies of protein engineering for enzyme stabilization. Biotechnol. Adv. 2019, 37, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Woodley, J.M. Accelerating the implementation of biocatalysis in industry. Appl. Microbiol. Biotechnol. 2019, 103, 4733–4739. [Google Scholar] [CrossRef] [PubMed]
- Szymański, W.; Beierle, J.M.; Kistemaker, H.A.V.; Velema, W.A.; Feringa, B.L. Reversible photocontrol of biological systems by the incorporation of molecular photoswitches. Chem. Rev. 2013, 113, 6114–6178. [Google Scholar] [CrossRef] [PubMed]
- Schmermund, L.; Jurkaš, V.; Özgen, F.F.; Barone, G.D.; Büchsenschütz, H.C.; Winkler, C.K.; Schmidt, S.; Kourist, R.; Kroutil, W. Photo-biocatalysis: Biotransformations in the presence of light. ACS Catal. 2019, 9, 4115–4144. [Google Scholar] [CrossRef]
- Losi, A.; Gardner, K.H.; Möglich, A. Blue-light receptors for optogenetics. Chem. Rev. 2018, 118, 10659–10709. [Google Scholar] [CrossRef]
- Rost, B.R.; Schneider-Warme, F.; Schmitz, D.; Hegemann, P. Optogenetic tools for subcellular applications in neuroscience. Neuron 2017, 96, 572–603. [Google Scholar] [CrossRef]
- Hüll, K.; Morstein, J.; Trauner, D. In vivo photopharmacology. Chem. Rev. 2018, 118, 10710–10747. [Google Scholar] [CrossRef]
- Baker, A.S.; Deiters, A. Optical control of protein function through unnatural amino acid mutagenesis and other optogenetic approaches. ACS Chem. Biol. 2014, 9, 1398–1407. [Google Scholar] [CrossRef]
- Bardhan, A.; Deiters, A. Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr. Opin. Struct. Biol. 2019, 57, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [PubMed]
- Deiters, A.; Groff, D.; Ryu, Y.; Xie, J.; Schultz, P.G. A genetically encoded photocaged tyrosine. Angew. Chem. Int. Ed. 2006, 45, 2728–2731. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.; Young, D.D.; Deiters, A. A light-activated DNA polymerase. Angew. Chem. Int. Ed. 2009, 48, 5950–5953. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.; Young, D.D.; Deiters, A. Photocaged T7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells. ChemBioChem 2010, 11, 972–977. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Y.; Liu, Y.; Zheng, S.; Wang, X.; Zhao, J.; Yang, F.; Zhang, G.; Wang, C.; Chen, P.R. Time-resolved protein activation by proximal decaging in living systems. Nature 2019, 569, 509–513. [Google Scholar] [CrossRef]
- Luo, J.; Torres-Kolbus, J.; Liu, J.; Deiters, A. Genetic encoding of photocaged tyrosines with improved light-activation properties for the optical control of protease function. ChemBioChem 2017, 18, 1442–1447. [Google Scholar] [CrossRef]
- Larson, A.S.; Hergenrother, P.J. Light activation of Staphylococcus aureus toxin YoeBSa1 reveals guanosine-specific endoribonuclease activity. Biochemistry 2014, 53, 188–201. [Google Scholar] [CrossRef]
- Zhang, G.; Li, J.; Xie, R.; Fan, X.; Liu, Y.; Zheng, S.; Ge, Y.; Chen, P.R. Bioorthogonal chemical activation of kinases in living systems. ACS Cent. Sci. 2016, 2, 325–331. [Google Scholar] [CrossRef]
- Motlagh, H.N.; Wrabl, J.O.; Li, J.; Hilser, V.J. The ensemble nature of allostery. Nature 2014, 508, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Kastritis, P.L.; Gavin, A.-C. Enzymatic complexes across scales. Essays Biochem. 2018, 62, 501–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makhlynets, O.V.; Raymond, E.A.; Korendovych, I.V. Design of allosterically regulated protein catalysts. Biochemistry 2015, 54, 1444–1456. [Google Scholar] [CrossRef] [PubMed]
- Romney, D.K.; Murciano-Calles, J.; Wehrmüller, J.E.; Arnold, F.H. Unlocking reactivity of TrpB: A general biocatalytic platform for synthesis of tryptophan analogues. J. Am. Chem. Soc. 2017, 139, 10769–10776. [Google Scholar] [CrossRef] [PubMed]
- Miles, E.W. Tryptophan synthase. Structure, function, and protein engineering. Subcell. Biochem. 1995, 24, 207–254. [Google Scholar]
- Raboni, S.; Bettati, S.; Mozzarelli, A. Tryptophan synthase: A mine for enzymologists. Cell. Mol. Life Sci. 2009, 66, 2391–2403. [Google Scholar] [CrossRef]
- Barends, T.R.M.; Dunn, M.F.; Schlichting, I. Tryptophan synthase, an allosteric molecular factory. Curr. Opin. Chem. Biol. 2008, 12, 593–600. [Google Scholar] [CrossRef]
- Weyand, M.; Schlichting, I. Crystal structure of wild-type tryptophan synthase complexed with the natural substrate indole-3-glycerol phosphate. Biochemistry 1999, 38, 16469–16480. [Google Scholar] [CrossRef]
- Hyde, C.C.; Ahmed, S.A.; Padlan, E.A.; Miles, E.W.; Davies, D.R. Three-dimensional structure of the tryptophan synthase α2β2 multienzyme complex from Salmonella typhimurium. J. Biol. Chem. 1988, 263, 17857–17871. [Google Scholar]
- Dunn, M.F. Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Arch. Biochem. Biophys. 2012, 519, 154–166. [Google Scholar] [CrossRef] [Green Version]
- Mozzarelli, A.; Peracchi, A.; Rossi, G.L.; Ahmed, S.A.; Miles, E.W. Microspectrophotometric studies on single crystals of the tryptophan synthase alpha 2 beta 2 complex demonstrate formation of enzyme-substrate intermediates. J. Biol. Chem. 1989, 264, 15774–15780. [Google Scholar]
- Dunn, M.F.; Aguilar, V.; Brzovic, P.; Drewe, W.F.; Houben, K.F.; Leja, C.A.; Roy, M. The tryptophan synthase bienzyme complex transfers indole between the α- and β-sites via a 25–30 Å long tunnel. Biochemistry 1990, 29, 8598–8607. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.S.; Miles, E.W.; Johnson, K.A. Serine modulates substrate channeling in tryptophan synthase. A novel intersubunit triggering mechanism. J. Biol. Chem. 1991, 266, 8020–8033. [Google Scholar] [PubMed]
- Miles, E.W. The tryptophan synthase α2β2 complex: Cleavage of a flexible loop in the α subunit alters allosteric properties. J. Biol. Chem. 1991, 266, 10715–10718. [Google Scholar] [PubMed]
- Peracchi, A.; Mozzarelli, A.; Rossi, G.L. Monovalent cations affect dynamic and functional properties of the tryptophan synthase α2β2 complex. Biochemistry 1995, 34, 9459–9465. [Google Scholar] [CrossRef]
- Woehl, E.; Dunn, M.F. Mechanisms of monovalent cation action in enzyme catalysis: The tryptophan synthase α-, β-, and αβ-reactions. Biochemistry 1999, 38, 7118–7130. [Google Scholar] [CrossRef]
- Miles, E.W. Tryptophan synthase: Structure, function, and subunit interaction. Adv. Enzymol. Relat. Areas Mol. Biol. 1979, 49, 127–186. [Google Scholar]
- Buller, A.R.; Brinkmann-Chen, S.; Romney, D.K.; Herger, M.; Murciano-Calles, J.; Arnold, F.H. Directed evolution of the tryptophan synthase β-subunit for stand-alone function recapitulates allosteric activation. Proc. Natl. Acad. Sci. USA 2015, 112, 14599. [Google Scholar] [CrossRef]
- Buller, A.R.; van Roye, P.; Cahn, J.K.B.; Scheele, R.A.; Herger, M.; Arnold, F.H. Directed evolution mimics allosteric activation by stepwise tuning of the conformational ensemble. J. Am. Chem. Soc. 2018, 140, 7256–7266. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Q.; Ning, X.; Hang, H.; Ma, J.; Yang, X.; Lu, X.; Zhang, J.; Li, Y.; Niu, C.; et al. Synthesis and biological evaluations of a series of thaxtomin analogues. J. Agric. Food Chem. 2015, 63, 3734–3741. [Google Scholar] [CrossRef]
- Zhang, H.; Ning, X.; Hang, H.; Ru, X.; Li, H.; Li, Y.; Wang, L.; Zhang, X.; Yu, S.; Qiao, Y.; et al. Total synthesis of thaxtomin A and its stereoisomers and findings of their biological activities. Org. Lett. 2013, 15, 5670–5673. [Google Scholar] [CrossRef]
- Boville, C.E.; Scheele, R.A.; Koch, P.; Brinkmann-Chen, S.; Buller, A.R.; Arnold, F.H. Engineered biosynthesis of β-alkyl tryptophan analogues. Angew. Chem. Int. Ed. 2018, 57, 14764–14768. [Google Scholar] [CrossRef] [PubMed]
- Francis, D.; Winn, M.; Latham, J.; Greaney, M.F.; Micklefield, J. An engineered tryptophan synthase opens new enzymatic pathways to β-methyltryptophan and derivatives. ChemBioChem 2017, 18, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Dick, M.; Sarai, N.S.; Martynowycz, M.W.; Gonen, T.; Arnold, F.H. Tailoring tryptophan synthase TrpB for selective quaternary carbon bond formation. ChemRxiv 2019. [Google Scholar] [CrossRef]
- Creighton, T.E. A steady-state kinetic investigation of the reaction mechanism of the tryptophan synthetase of Escherichia coli. Eur. J. Biochem. 1970, 13, 1–10. [Google Scholar] [CrossRef]
- Lane, A.N.; Kirschner, K. The catalytic mechanism of tryptophan synthase from Escherichia coli. Eur. J. Biochem. 1983, 129, 571–582. [Google Scholar] [CrossRef]
- Wellington, S.; Nag, P.P.; Michalska, K.; Johnston, S.E.; Jedrzejczak, R.P.; Kaushik, V.K.; Clatworthy, A.E.; Siddiqi, N.; McCarren, P.; Bajrami, B.; et al. A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase. Nat. Chem. Biol. 2017, 13, 943–950. [Google Scholar] [CrossRef]
- Kameya, M.; Onaka, H.; Asano, Y. Selective tryptophan determination using tryptophan oxidases involved in bis-indole antibiotic biosynthesis. Anal. Biochem. 2013, 438, 124–132. [Google Scholar] [CrossRef] [Green Version]
- Lane, A.N.; Kirschner, K. Mechanism of the physiological reaction catalyzed by tryptophan synthase from Escherichia coli. Biochemistry 1991, 30, 479–484. [Google Scholar] [CrossRef]
- Kirschner, K.; Lane, A.N.; Strasser, A.W.M. Reciprocal communication between the lyase and synthase active sites of the tryptophan synthase bienzyme complex. Biochemistry 1991, 30, 472–478. [Google Scholar] [CrossRef]
- McGaughey, G.B.; Gagné, M.; Rappé, A.K. π-Stacking Interactions: Alive and well in proteins. J. Biol. Chem. 1998, 273, 15458–15463. [Google Scholar] [CrossRef]
- Schneider, T.R.; Gerhardt, E.; Lee, M.; Liang, P.-H.; Anderson, K.S.; Schlichting, I. Loop closure and intersubunit communication in tryptophan synthase. Biochemistry 1998, 37, 5394–5406. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.; Parris, K.D.; Ahmed, S.A.; Miles, E.W.; Davies, D.R. Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase α2β2 complex. Biochemistry 1996, 35, 4211–4221. [Google Scholar] [CrossRef] [PubMed]
- Il’ichev, Y.V.; Wirz, J. Rearrangements of 2-nitrobenzyl compounds: 1. Potential energy surface of 2-nitrotoluene and its isomers explored with ab initio and density functional theory methods. J. Phys. Chem. A 2000, 104, 7856–7870. [Google Scholar] [CrossRef]
- Böcker, J.K.; Dörner, W.; Mootz, H.D. Light-control of the ultra-fast Gp41-1 split intein with preserved stability of a genetically encoded photo-caged amino acid in bacterial cells. Chem. Commun. 2019, 55, 1287–1290. [Google Scholar] [CrossRef] [PubMed]
- Ruvinov, S.B.; Yang, X.-J.; Parris, K.D.; Banik, U.; Ahmed, S.A.; Miles, E.W.; Sackett, D.L. Ligand-mediated changes in the tryptophan synthase indole tunnel probed by nile red fluorescence with wild type, mutant, and chemically modified enzymes. J. Biol. Chem. 1995, 270, 6357–6369. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.S.; Kim, A.Y.; Quillen, J.M.; Sayers, E.; Yang, X.-J.; Miles, E.W. Kinetic characterization of channel impaired mutants of tryptophan synthase. J. Biol. Chem. 1995, 270, 29936–29944. [Google Scholar]
- Weyand, M.; Schlichting, I. Structural basis for the impaired channeling and allosteric inter-subunit communication in the βA169L/βC170W mutant of tryptophan synthase. J. Biol. Chem. 2000, 275, 41058–41063. [Google Scholar] [CrossRef]
- Harris, R.M.; Dunn, M.F. Intermediate trapping via a conformational switch in the Na+-activated tryptophan synthase bienzyme complex. Biochemistry 2002, 41, 9982–9990. [Google Scholar] [CrossRef]
- Abrahams, K.A.; Cox, J.A.G.; Fütterer, K.; Rullas, J.; Ortega-Muro, F.; Loman, N.J.; Moynihan, P.J.; Pérez-Herrán, E.; Jiménez, E.; Esquivias, J.; et al. Inhibiting mycobacterial tryptophan synthase by targeting the inter-subunit interface. Sci. Rep. 2017, 7, 9430. [Google Scholar] [CrossRef]
- Kneuttinger, A.C.; Straub, K.; Bittner, P.; Simeth, N.A.; Bruckmann, A.; Busch, F.; Rajendran, C.; Hupfeld, E.; Wysocki, V.H.; Horinek, D.; et al. Light regulation of enzyme allostery through photo-responsive unnatural amino acids. Cell Chem. Biol. 2019. [Google Scholar] [CrossRef]
- Slanina, T.; Shrestha, P.; Palao, E.; Kand, D.; Peterson, J.A.; Dutton, A.S.; Rubinstein, N.; Weinstain, R.; Winter, A.H.; Klán, P. In search of the perfect photocage: Structure–reactivity relationships in meso-methyl BODIPY photoremovable protecting groups. J. Am. Chem. Soc. 2017, 139, 15168–15175. [Google Scholar] [CrossRef]
- Walton, D.P.; Dougherty, D.A. A general strategy for visible-light decaging based on the quinone trimethyl lock. J. Am. Chem. Soc. 2017, 139, 4655–4658. [Google Scholar] [CrossRef] [PubMed]
- Bose, M.; Groff, D.; Xie, J.; Brustad, E.; Schultz, P.G. The incorporation of a photoisomerizable amino acid into proteins in E. coli. J. Am. Chem. Soc. 2006, 128, 388–389. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Samanta, S.; Convertino, M.; Dokholyan, N.V.; Deiters, A. Reversible and tunable photoswitching of protein function through genetic encoding of azobenzene amino acids in mammalian cells. ChemBioChem 2018, 19, 2178–2185. [Google Scholar] [CrossRef] [PubMed]
- John, A.A.; Ramil, C.P.; Tian, Y.; Cheng, G.; Lin, Q. Synthesis and site-specific incorporation of red-shifted azobenzene amino acids into proteins. Org. Lett. 2015, 17, 6258–6261. [Google Scholar] [CrossRef]
- Busch, F.; Rajendran, C.; Mayans, O.; Löffler, P.; Merkl, R.; Sterner, R. TrpB2 enzymes are O-phospho-L-serine dependent tryptophan synthases. Biochemistry 2014, 53, 6078–6083. [Google Scholar] [CrossRef]
- Leopoldseder, S.; Hettwer, S.; Sterner, R. Evolution of multi-enzyme complexes: The case of tryptophan synthase. Biochemistry 2006, 45, 14111–14119. [Google Scholar] [CrossRef]
- Young, T.S.; Ahmad, I.; Yin, J.A.; Schultz, P.G. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010, 395, 361–374. [Google Scholar] [CrossRef]
- Schlee, S.; Dietrich, S.; Kurćon, T.; Delaney, P.; Goodey, N.M.; Sterner, R. Kinetic mechanism of indole-3-glycerol phosphate synthase. Biochemistry 2013, 52, 132–142. [Google Scholar] [CrossRef]
- Rohweder, B.; Semmelmann, F.; Endres, C.; Sterner, R. Standardized cloning vectors for protein production and generation of large gene libraries in Escherichia coli. BioTechniques 2018, 64, 24–26. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Darden, T.; Nabuurs, S.B.; Finkelstein, A.; Vriend, G. Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins 2004, 57, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.; Chang, H.-Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2014, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- VanAernum, Z.L.; Gilbert, J.D.; Belov, M.E.; Makarov, A.A.; Horning, S.R.; Wysocki, V.H. Surface-induced dissociation of noncovalent protein complexes in an extended mass range orbitrap mass spectrometer. Anal. Chem. 2019, 91, 3611–3618. [Google Scholar] [CrossRef] [PubMed]
- VanAernum, Z.; Busch, F.; Jones, B.J.; Jia, M.; Chen, Z.; Boyken, S.E.; Sahasrabuddhe, A.; Baker, D.; Wysocki, V. Rapid online buffer exchange: A method for screening of proteins, protein complexes, and cell lysates by native mass spectrometry. ChemRxiv 2019. [Google Scholar] [CrossRef]
- Marty, M.T.; Baldwin, A.J.; Marklund, E.G.; Hochberg, T.K.A.; Benesch, J.L.P.; Robinson, C.V. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015, 87, 4370–4376. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Copeland, R.A. Kinetics of single-substrate enzyme reactions. In Enzymes, 2nd ed.; Copeland, R.A., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2002; pp. 109–145. ISBN 0-471-22063-9. [Google Scholar]
- Copeland, R.A. Reversible inhibitors. In Enzymes, 2nd ed.; Copeland, R.A., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2002; pp. 266–304. ISBN 0-471-22063-9. [Google Scholar]
- Finkelstein, A.V.; Badretdinov, A.Y.; Gutin, A.M. Why do protein architectures have boltzmann-like statistics? Proteins 1995, 23, 142–150. [Google Scholar] [CrossRef]

| Reaction | Parameter | Dimension | Value TS | Value ecTS |
|---|---|---|---|---|
| TrpA 1 | kcat | [min−1] | 2.4 ± 0.2 | 1.5 ± 0.1 |
| KmIGP | [µM] | 29.4 ± 8.4 | 17.6 ± 5.4 | |
| kcat/KmIGP | [s−1M−1] | 13.6 · 102 | 14.2 · 102 | |
| TrpB 2 | kcat | [min−1] | 24.1 ± 0.6 | 21.8 ± 0.7 |
| Kmindole | [µM] | 51.0 ± 4.5 | 10.6 ± 1.6 | |
| kcat/Kmindole | [s−1M−1] | 7.9 · 103 | 34.3 · 103 | |
| KmSer | [mM] | 0.53 ± 0.05 | 0.52 ± 0.07 | |
| kcat/KmSer | [s−1M−1] | 7.6 · 102 | 7.0 · 102 | |
| TS 3 | kcat | [min−1] | 21.3 ± 0.6 | 28.3 ± 1.2 |
| KmIGP | [µM] | 25.9 ± 2.6 | 24.9 ± 3.7 | |
| kcat/KmIGP | [s−1M−1] | 13.7 · 103 | 18.9 · 103 | |
| KmSer | [mM] | 0.23 ± 0.03 | 0.35 ± 0.05 | |
| kcat/KmSer | [s−1M−1] | 15.4 · 102 | 13.5 · 102 |
| Reaction | Parameter | Dimension | Value TS(aL58ONBYai) | LAF | Value TS(aL58ONBYhν) | Value TS(L58Y) |
|---|---|---|---|---|---|---|
| TrpA | kcat | [min−1] | 0.19 ± 0.02 | 8 → | 1.50 ± 0.07 | 2.30 ± 0.09 |
| KmIGP | [µM] | 549 ± 92 | 3 ← | 194 ± 23 | 234 ± 20 | |
| kcat/KmIGP | [s−1M−1] | 0.06 · 102 | 22 → | 1.29 · 102 | 1.64 · 102 | |
| TrpB | kcat | [min−1] | 0.9 ± 0.0 | 13 → | 12.1 ± 0.4 | 16.3 ± 0.3 |
| Kmindole | [µM] | 3.4 ± 0.4 | 4 → | 13.9 ± 1.5 | 37.1 ± 2.4 | |
| kcat/Kmindole | [s−1M−1] | 4.41 · 103 | 3 → | 14.51 · 103 | 7.32 · 103 | |
| KmSer | [mM] | 3.9 ± 0.3 | 6 ← | 0.7 ± 0.1 | 0.7 ± 0.0 | |
| kcat/KmSer | [s−1M−1] | 0.04 · 102 | 72 → | 2.88 · 102 | 3.88 · 102 | |
| TS | kcat | [min−1] | 0.3 ± 0.0 | 18 → | 5.4 ± 0.2 | 10.9 ± 0.5 |
| KmIGP | [µM] | 103 ± 18 | 2 ← | 58 ± 10 | 29 ± 6 | |
| kcat/KmIGP | [s−1M−1] | 0.05 · 103 | 31 → | 1.55 · 103 | 6.26 · 103 | |
| KmSer | [mM] | 0.6 ± 0.1 | 3 ← | 0.2 ± 0.0 | 0.2 ± 0.0 | |
| kcat/KmSer | [s−1M−1] | 0.08 · 102 | 56 → | 4.50 · 102 | 9.08 · 102 |
| Reaction | Parameter | Dimension | Value TS(aL58ONBY)ai | Value TS(aL58Y) |
|---|---|---|---|---|
| TrpA (−Ser) | V01 | [µM min−1] | 0.09 ± 0.01 | 0.16 ± 0.01 |
| Vmax2 | [µM min−1] | 0.12 ± 0.00 | 1.86 ± 0.06 | |
| Vmax/V03 | 1 | 12 | ||
| TrpA (+Ser) | V0 | [µM min−1] | 0.02 ± 0.00 | 0.03 ± 0.04 |
| Vmax | [µM min−1] | 0.09 ± 0.00 | 4.80 ± 0.33 | |
| Vmax/V0 | 4 | 160 |
| Reaction | Parameter | Dimension | Value TS(aL58ONBYai) | Value TS(aL58ONBY) | Value TS(aL58Y) |
|---|---|---|---|---|---|
| TrpA (−Ser) | V01 | [min−1] | 0.15 | 0.17 | 1.27 |
| Vhν2 | [min−1] | — | 0.73 | 0.84 | |
| LAF | — | 4 | 0.66 | ||
| TrpA (+Ser) | V0 | [min−1] | 0.20 | 0.20 | 17.83 |
| Vhν | [min−1] | — | 8.46 | 10.01 | |
| LAF | — | 42 | 0.56 | ||
| TS | V0 | [min−1] | <0.03 | <0.02 | 4.95 |
| Vhν | [min−1] | — | 2.83 | 3.10 | |
| LAF | — | >141 | 0.63 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kneuttinger, A.C.; Zwisele, S.; Straub, K.; Bruckmann, A.; Busch, F.; Kinateder, T.; Gaim, B.; Wysocki, V.H.; Merkl, R.; Sterner, R. Light-Regulation of Tryptophan Synthase by Combining Protein Design and Enzymology. Int. J. Mol. Sci. 2019, 20, 5106. https://doi.org/10.3390/ijms20205106
Kneuttinger AC, Zwisele S, Straub K, Bruckmann A, Busch F, Kinateder T, Gaim B, Wysocki VH, Merkl R, Sterner R. Light-Regulation of Tryptophan Synthase by Combining Protein Design and Enzymology. International Journal of Molecular Sciences. 2019; 20(20):5106. https://doi.org/10.3390/ijms20205106
Chicago/Turabian StyleKneuttinger, Andrea C., Stefanie Zwisele, Kristina Straub, Astrid Bruckmann, Florian Busch, Thomas Kinateder, Barbara Gaim, Vicki H. Wysocki, Rainer Merkl, and Reinhard Sterner. 2019. "Light-Regulation of Tryptophan Synthase by Combining Protein Design and Enzymology" International Journal of Molecular Sciences 20, no. 20: 5106. https://doi.org/10.3390/ijms20205106
APA StyleKneuttinger, A. C., Zwisele, S., Straub, K., Bruckmann, A., Busch, F., Kinateder, T., Gaim, B., Wysocki, V. H., Merkl, R., & Sterner, R. (2019). Light-Regulation of Tryptophan Synthase by Combining Protein Design and Enzymology. International Journal of Molecular Sciences, 20(20), 5106. https://doi.org/10.3390/ijms20205106





