Genome-Wide Identification, Gene Structure and Expression Analysis of the MADS-Box Gene Family Indicate Their Function in the Development of Tobacco (Nicotiana tabacum L.)
Abstract
1. Introduction
2. Results
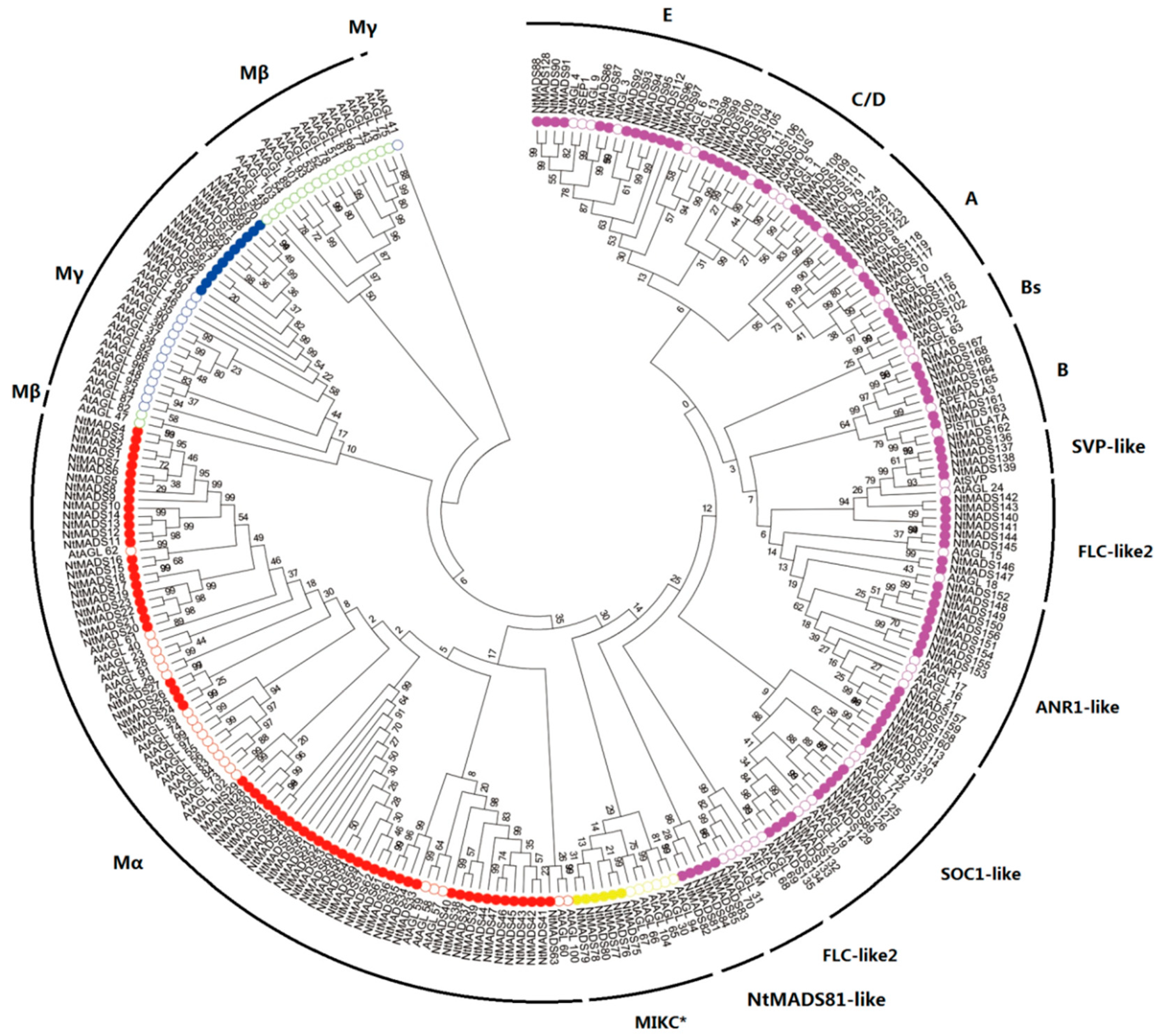
2.1. Identification and Classification of MADS-Box Genes in Tobacco
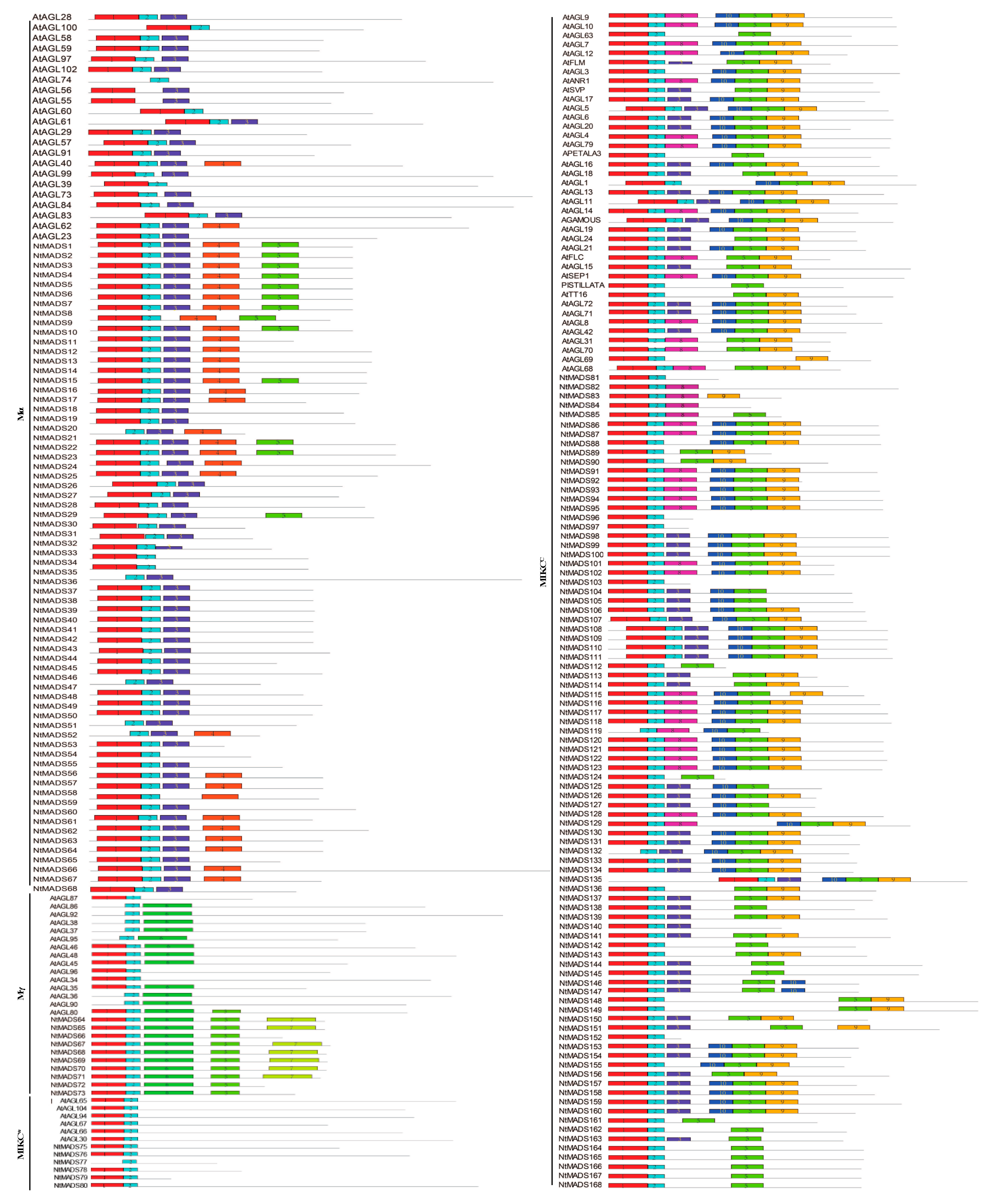
2.2. Structure of MADS-Box Genes in Tobacco
2.3. Motif Analysis of MADS-Box Genes in Tobacco
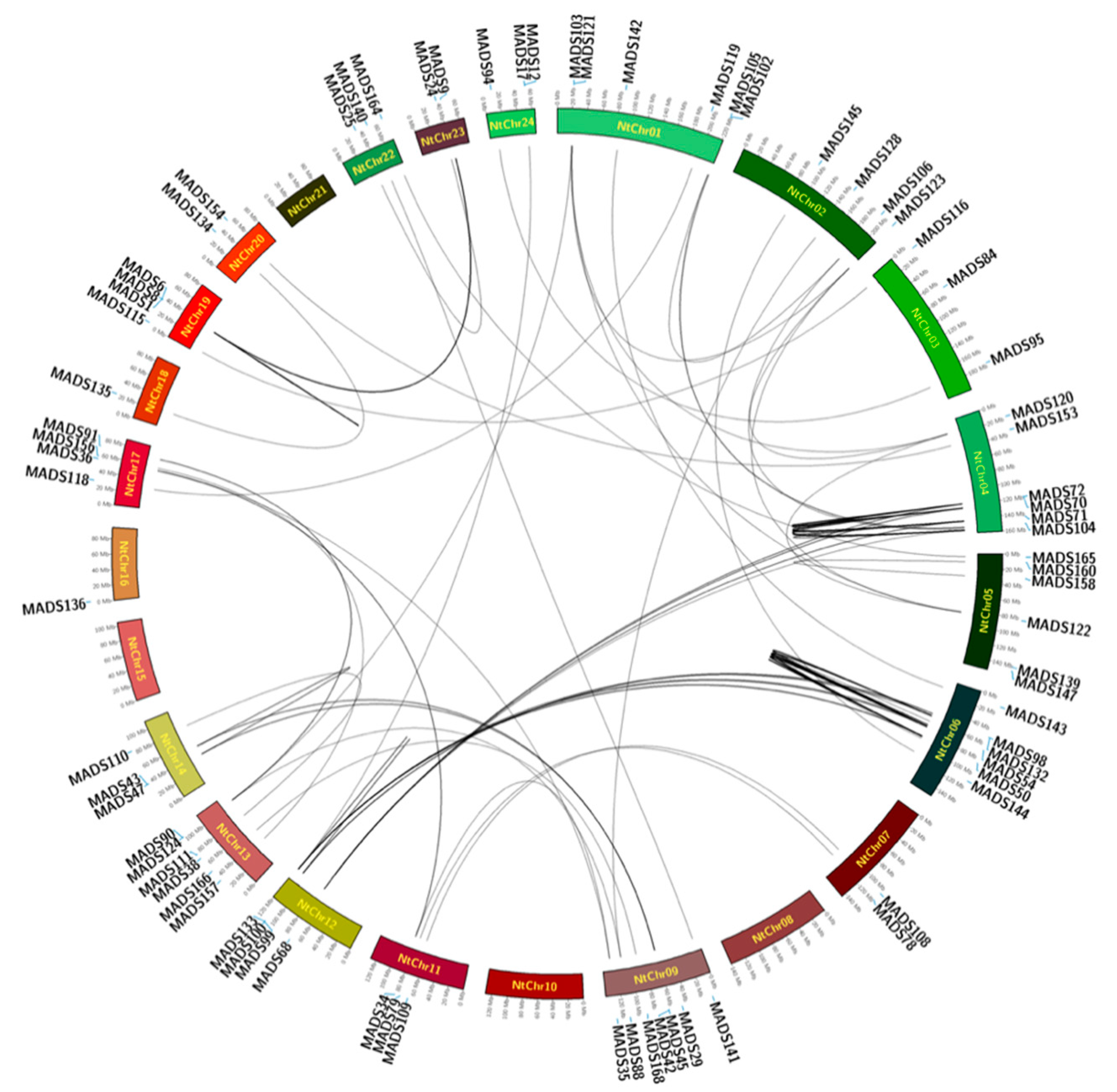
2.4. Location of MADS-Box Genes in Tobacco
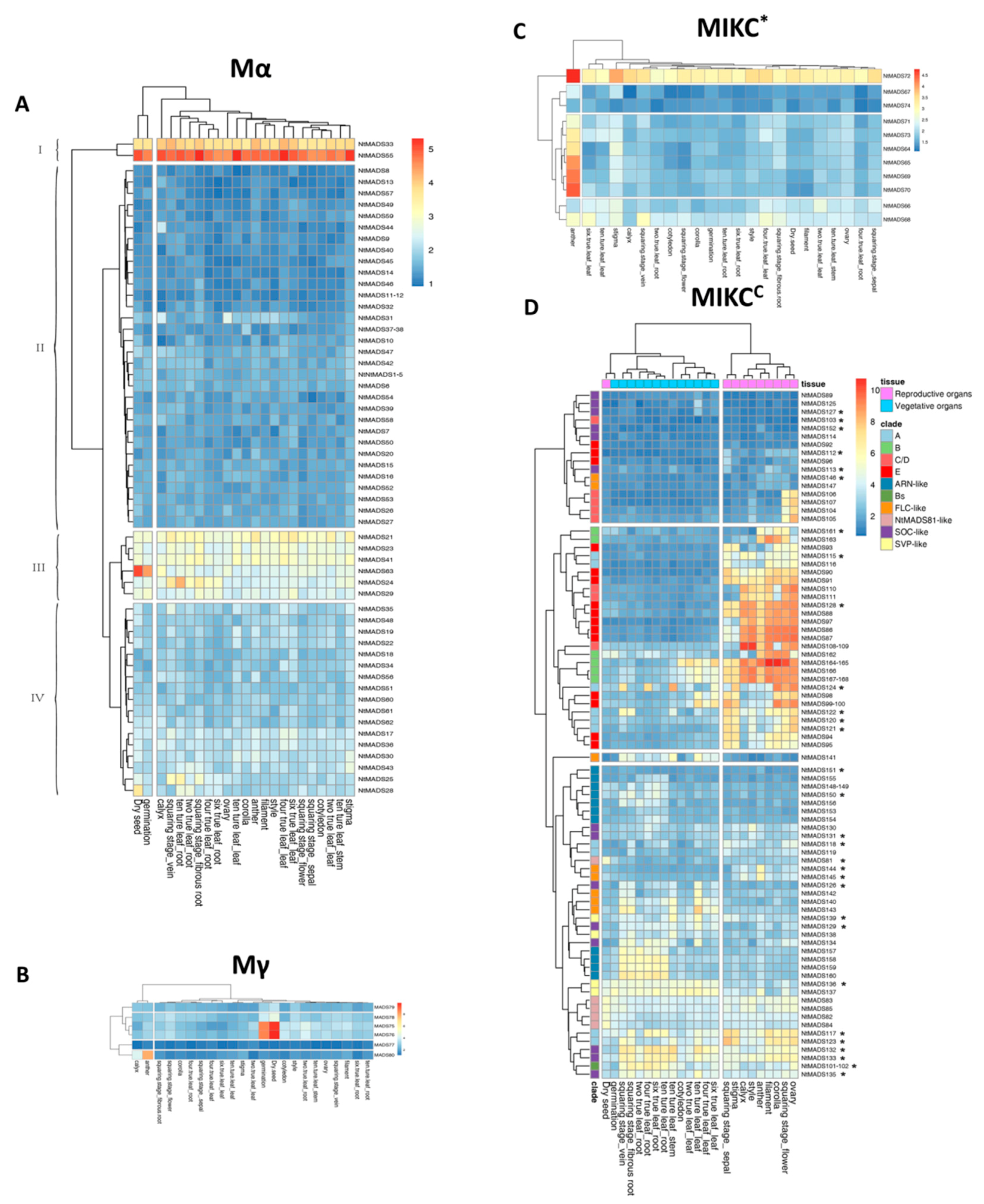
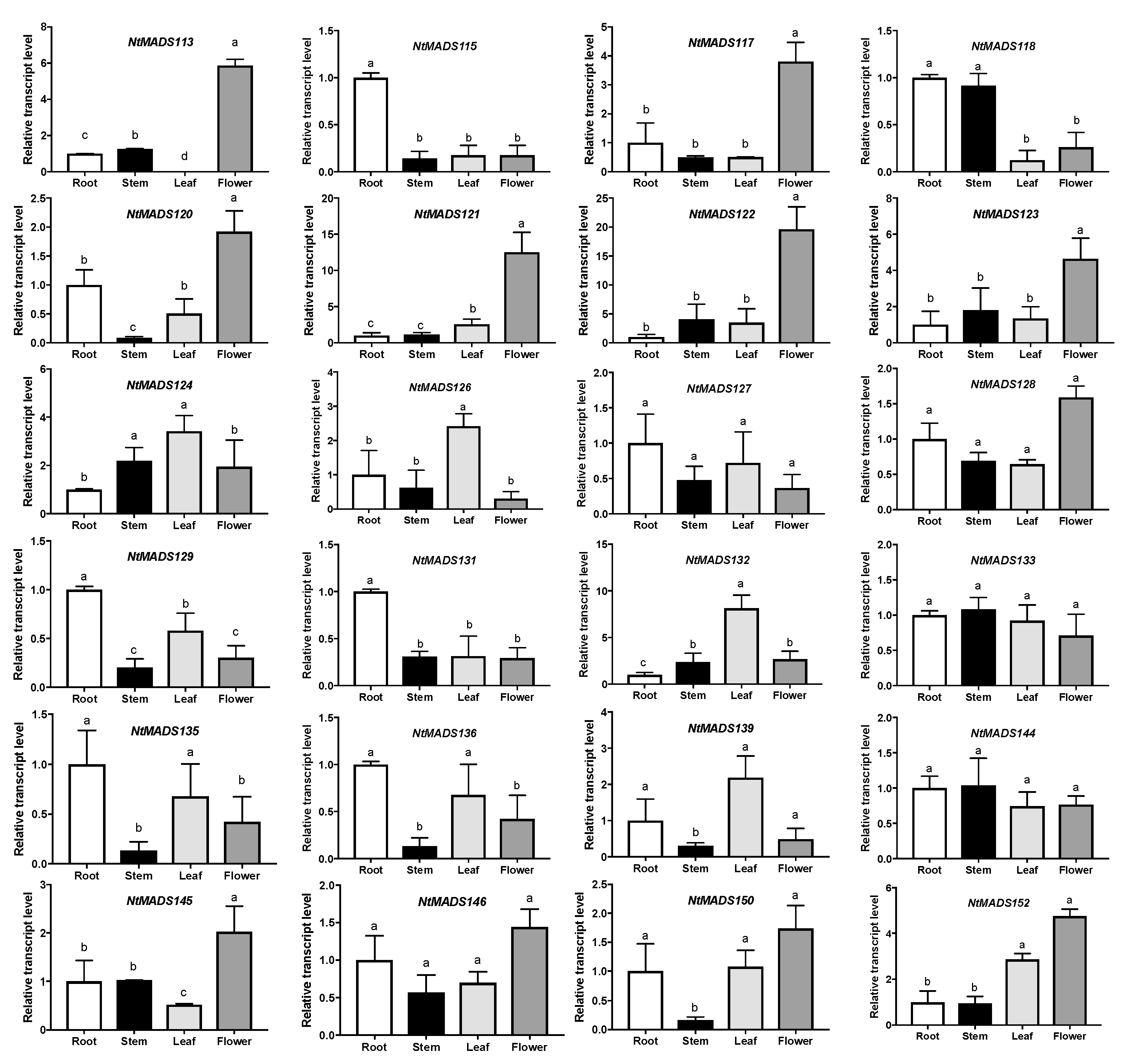
2.5. Expression Patterns of MADS-Box Genes in Tobacco
2.6. Identification of NtSOC1 in Regulating the Flower Time and Development in Tobacco
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phylogenetic and Gene Structure Analyses
4.3. RNA Extraction, cDNA Preparation and Gene Chip
4.4. Chromosomal Location and Gene Duplication
4.5. Quantitative Real-Time PCR of Selected NtMADS-Box Genes
4.6. Plasmid Construction and Tobacco Transgenic Plant
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Availability of Data and Materials
References
- Theissen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.-U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef] [PubMed]
- Passmore, S.; Elble, R.; Tye, B.K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 1989, 3, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Sommer, Z.; Sommer, H. Genetic control of flower development by homeotic genes in antirrhinum majus. Science 1990, 250, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Sommer, H.; Beltran, J.P.; Huijser, P.; Pape, H.; Lonnig, W.E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. Embo J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef]
- Becker, A.; Winter, K.U.; Meyer, B.; Saedler, H.; Theißen, G. MADS-Box gene diversity in seed plants 300 Million years ago. Mol. Biol. Evol. 2000, 17, 1425–1434. [Google Scholar] [CrossRef]
- Shore, P.; Sharrocks, A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995, 229, 87–99. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Ribas de, P.L.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA. 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- De Bodt, S.; Raes, J.; Florquin, K.; Rombauts, S.; Rouze, P.; Theissen, G.; van de Peer, Y. Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J. Mol. Evol. 2003, 56, 573–586. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Parenicova, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Masiero, S.; Colombo, L.; Grini, P.E.; Schnittger, A.; Kater, M.M. The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 2011, 23, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Yang, Z.; Li, X.; Chen, W.; Peng, X.; Cheng, X.; Zhu, S.; Cheng, B. Whole-genome survey and characterization of MADS-box gene family in maize and sorghum. Plant Cell Tissue Organ Cult. 2011, 105, 159–173. [Google Scholar]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Feng, C.; Liu, M.; Wang, J.; Hu, Y. Genome-wide identification, characterization of the MADS-box gene family in Chinese jujube and their involvement in flower development. Sci. Rep. 2017, 7, 1025. [Google Scholar] [CrossRef]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0181443. [Google Scholar] [CrossRef]
- Liu, M.; Fu, Q.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide investigation of the MADS gene family and dehulling genes in tartary buckwheat (Fagopyrum tataricum). Planta 2019, 249, 1301–1318. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Q.; Yang, S.; Lin, S.; Bao, M.; Bendahmane, M.; Wu, Q.; Wang, C.; Fu, Z. Identification and Characterization of the MADS-Box Genes and Their Contribution to Flower Organ in Carnation (Dianthus caryophyllus L.). Genes 2018, 9, 193. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Z.; Li, S.; Hou, M.; Zhou, Y.; Zhao, Y.; Li, G.; Zhao, H.; Ma, H. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A. BMC Genom. 2018, 19, 726. [Google Scholar] [CrossRef] [PubMed]
- Nardeli, S.M.; Artico, S.; Aoyagi, G.M.; Moura, S.M.D.; Silva, T.D.F.; Grossi-De-Sa, M.F.; Romanel, E.; Alves-Ferreira, M. Genome-wide analysis of the MADS-box gene family in polyploid cotton (Gossypium hirsutum) and in its diploid parental species (Gossypium arboreum and Gossypium raimondii). Plant Physiol. Biochem. 2018, 127, 169–184. [Google Scholar] [CrossRef] [PubMed]
- José, D.R.; Diego, L.; Martínez-Zapater, J.M.; María José, C. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol. 2009, 149, 354–369. [Google Scholar]
- Grimplet, J.; Martínez-Zapater, J.M.; Carmona, M.J. Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genom. 2016, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Song, X.; Liu, T.; Huang, Z.; Ren, J.; Hou, X.; Li, Y. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef]
- Wei, X.; Wang, L.; Yu, J.; Zhang, Y.; Li, D.; Zhang, X. Genome-wide identification and analysis of the MADS-box gene family in sesame. Gene 2015, 569, 66–76. [Google Scholar] [CrossRef]
- Wei, B.; Zhang, R.Z.; Guo, J.J.; Liu, D.M.; Li, A.L.; Fan, R.C.; Mao, L.; Zhang, X.Q. Genome-wide analysis of the MADS-box gene family in Brachypodium distachyon. PLoS ONE 2014, 9, e84781. [Google Scholar] [CrossRef]
- Shu, Y.; Yu, D.; Wang, D.; Guo, D.; Guo, C. Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol. Biol. Rep. 2013, 40, 3901–3911. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide analysis of the MADS-Box transcription factor family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Leseberg, C.; Li, A.; Kang, H.; Duvall, M.; Mao, L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 2006, 378, 84–94. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Meyerowitz, E.M. MADS domain proteins in plant development. Biol. Chem. 1997, 378, 1079–1101. [Google Scholar] [PubMed]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Zahn, L.M.; Feng, B.; Ma, H. Beyond the ABC-model: Regulation of floral homeotic genes. Adv. Bot. Res. 2006, 44, 163–207. [Google Scholar]
- Gregis, V.; Sessa, A.; Dorca-Fornell, C.; Kater, M.M. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009, 60, 626–637. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar]
- Colombo, L.; Franken, J.; Koetje, E.; van Went, J.; Dons, H.J.; Angenent, G.C.; van Tunen, A.J. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 1995, 7, 1859–1868. [Google Scholar]
- Ehlers, K.; Bhide, A.S.; Tekleyohans, D.G.; Wittkop, B.; Snowdon, R.J.; Becker, A. The MADS Box Genes ABS, SHP1, and SHP2 Are Essential for the Coordination of Cell Divisions in Ovule and Seed Coat Development and for Endosperm Formation in Arabidopsis thaliana. PLoS ONE 2016, 11, e0165075. [Google Scholar] [CrossRef]
- Theissen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Sung, S.; Amasino, R.M. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 411–416. [Google Scholar] [CrossRef]
- Choi, K.; Kim, J.; Hwang, H.J.; Kim, S.; Park, C.; Kim, S.Y.; Lee, I. The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 2011, 23, 289–303. [Google Scholar] [CrossRef]
- Yuan, W.; Luo, X.; Li, Z.; Yang, W.; Wang, Y.; Liu, R.; Du, J.; He, Y. A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 2016, 48, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Wood, C.C.; Robertson, M.; James Peacock, W.; Dennis, E.S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006, 46, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, O.J.; Kumimoto, R.W.; Wong, B.J.; Riechmann, J.L. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 2003, 15, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Khong, G.N.; Pati, P.K.; Richaud, F.; Parizot, B.; Bidzinski, P.; Mai, C.D.; Bes, M.; Bourrie, I.; Meynard, D.; Beeckman, T.; et al. OsMADS26 Negatively Regulates Resistance to Pathogens and Drought Tolerance in Rice. Plant Physiol. 2015, 169, 2935–2949. [Google Scholar] [CrossRef] [PubMed]
- Puig, J.; Meynard, D.; Khong, G.N.; Pauluzzi, G.; Guiderdoni, E.; Gantet, P. Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr. Patterns 2013, 13, 160–170. [Google Scholar] [CrossRef]
- Yu, C.; Liu, Y.; Zhang, A.; Su, S.; Yan, A.; Huang, L.; Ali, I.; Liu, Y.; Forde, B.G.; Gan, Y. MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS ONE 2015, 10, e0135196. [Google Scholar] [CrossRef]
- Ning, K.; Han, Y.; Chen, Z.; Luo, C.; Wang, S.; Zhang, W.; Li, L.; Zhang, X.; Fan, S.; Wang, Q. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef]
- Saha, G.; Park, J.I.; Jung, H.J.; Ahmed, N.U.; Kayum, M.A.; Chung, M.Y.; Hur, Y.; Cho, Y.G.; Watanabe, M.; Nou, I.S. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genom. 2015, 16, 178. [Google Scholar] [CrossRef]
- Guo, J.; Shi, X.-X.; Zhang, J.-S.; Duan, Y.-H.; Bai, P.-F.; Guan, X.-N.; Kang, Z.-S. A type I MADS-box gene is differentially expressed in wheat in response to infection by the stripe rust fungus. Biol. Plant. 2013, 57, 540–546. [Google Scholar] [CrossRef]
- Jeffreys, A.J.; Harris, S. Processes of gene duplication. Nature 1982, 296, 9–10. [Google Scholar] [CrossRef]
- Liu, C.; Chen, H.; Er, H.L.; Soo, H.M.; Kumar, P.P.; Han, J.H.; Liou, Y.C.; Yu, H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Tadege, M.; Sheldon, C.C.; Helliwell, C.A.; Upadhyaya, N.M.; Dennis, E.S.; Peacock, W.J. Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 2003, 1, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Shitsukawa, N.; Ikari, C.; Mitsuya, T.; Sakiyama, T.; Ishikawa, A.; Takumi, S.; Murai, K. Wheat SOC1 functions independently of WAP1/VRN1, an integrator of vernalization and photoperiod flowering promotion pathways. Physiol. Plant. 2007, 130, 627–636. [Google Scholar] [CrossRef]
- Zhong, X.; Dai, X.; Xv, J.; Wu, H.; Liu, B.; Li, H. Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean. Mol. Biol. Rep. 2012, 39, 6967–6974. [Google Scholar] [CrossRef] [PubMed]
- Hepworth, S.R.; Valverde, F.; Ravenscroft, D.; Mouradov, A.; Coupland, G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002, 21, 4327–4337. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.Y.; Matyasek, R.; Kovarik, A.; Leitch, A.R. Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 2004, 82, 599–606. [Google Scholar] [CrossRef]
- Yuan, Y.-X.; Wu, J.; Sun, R.-F.; Zhang, X.-W.; Xu, D.-H.; Bonnema, G.; Wang, X.-W. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 2009, 60, 1299–1308. [Google Scholar] [CrossRef]
- Wang, D.; Chen, X.; Zhang, Z.; Liu, D.; Song, G.; Kong, X.; Geng, S.; Yang, J.; Wang, B.; Wu, L. A MADS-box gene NtSVP regulates pedicel elongation by directly suppressing a KNAT1-like KNOX gene NtBPL in tobacco (Nicotiana tabacum L.). J. Exp. Bot. 2015, 66, 6233–6244. [Google Scholar] [CrossRef]
- Fedorov, A.; Merican, A.F.; Gilbert, W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc. Natl. Acad. Sci. USA 2002, 99, 128–133. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Wolf, Y.I.; Sorokin, A.V.; Mirkin, B.G.; Koonin, E.V. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003, 13, 1512–1517. [Google Scholar] [CrossRef]
- Gorlova, O.; Fedorov, A.; Logothetis, C.; Amos, C.; Gorlov, I. Genes with a large intronic burden show greater evolutionary conservation on the protein level. BMC Evol. Biol. 2014, 14, 50. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, E.N.; Petrov, D.A.; Hartl, D.L. Genome size and intron size in Drosophila. Mol. Biol. Evol 1998, 15, 770–773. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, M.; Long, M. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999, 27, 3219–3228. [Google Scholar] [PubMed]
- Marais, G.; Nouvellet, P.; Keightley, P.D.; Charlesworth, B. Intron size and exon evolution in Drosophila. Genetics 2005, 170, 481–485. [Google Scholar] [CrossRef][Green Version]
- Dewey, C.N.; Rogozin, I.B.; Koonin, E.V. Compensatory relationship between splice sites and exonic splicing signals depending on the length of vertebrate introns. BMC Genom. 2006, 7, 311. [Google Scholar] [CrossRef]
- Chao, L.; Yan, W.; Liang, X.; Nie, S.; Chen, Y.; Liang, D.; Sun, X.; Karanja, B.K.; Luo, X.; Liu, L. Genome-Wide Characterization of the MADS-Box Gene Family in Radish (Raphanus sativusL.) and Assessment of Its Roles in Flowering and Floral Organogenesis. Front. Plant Sci. 2016, 7, 1390. [Google Scholar]
- Lin, C.S.; Hsu, C.T.; Liao, D.C.; Chang, W.J.; Chou, M.L.; Huang, Y.T.; Chen, J.J.; Ko, S.S.; Chan, M.T.; Shih, M.C. Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol. J. 2016, 14, 284–298. [Google Scholar] [CrossRef]
- Harding, E.W.; Tang, W.; Nichols, K.W.; Fernandez, D.E.; Perry, S.E. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol. 2003, 133, 653–663. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Krober, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef]
- Jang, S.; Torti, S.; Coupland, G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 2009, 60, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Filleur, S.; Rahman, A.; Gotensparre, S.; Forde, B.G. Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 2005, 222, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Tapia-Lopez, R.; Garcia-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Perez-Ruiz, R.V.; Kim, S.H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Nawy, T.; Lee, J.Y.; Colinas, J.; Wang, J.Y.; Thongrod, S.C.; Malamy, J.E.; Birnbaum, K.; Benfey, P.N. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 2005, 17, 1908–1925. [Google Scholar] [CrossRef]
- Uimari, A.; Kotilainen, M.; Elomaa, P.; Yu, D.; Albert, V.A.; Teeri, T.H. Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc. Natl. Acad. Sci. USA 2004, 101, 15817–15822. [Google Scholar] [CrossRef]
- Teeri, T.H.; Uimari, A.; Kotilainen, M.; Laitinen, R.; Help, H.; Elomaa, P.; Albert, V.A. Reproductive meristem fates in Gerbera. J. Exp. Bot. 2006, 57, 3445–3455. [Google Scholar] [CrossRef]
- Berbel, A.; Ferrandiz, C.; Hecht, V.; Dalmais, M.; Lund, O.S.; Sussmilch, F.C.; Taylor, S.A.; Bendahmane, A.; Ellis, T.H.; Beltran, J.P.; et al. VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun. 2012, 3, 797. [Google Scholar] [CrossRef]
- Lee, H.; Suh, S.S.; Park, E.; Cho, E.; Ahn, J.H.; Kim, S.G.; Lee, J.S.; Kwon, Y.M.; Lee, I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000, 14, 2366–2376. [Google Scholar] [CrossRef]
- Ryu, C.H.; Lee, S.; Cho, L.H.; Kim, S.L.; Lee, Y.S.; Choi, S.C.; Jeong, H.J.; Yi, J.; Park, S.J.; Han, C.D.; et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009, 32, 1412–1427. [Google Scholar] [CrossRef]
- Bohlenius, H.; Huang, T.; Charbonnel-Campaa, L.; Brunner, A.M.; Jansson, S.; Strauss, S.H.; Nilsson, O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 2006, 312, 1040–1043. [Google Scholar] [CrossRef] [PubMed]
- Dinkins, R.; Pflipsen, C.; Thompson, A.; Collins, G.B. Ectopic expression of an Arabidopsis single zinc finger gene in tobacco results in dwarf plants. Plant Cell Physiol. 2002, 43, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Ning, G.; Zhang, W.; Jing, Z.; Lv, H.; Bao, M. Overexpression of Petunia SOC1 -like Gene FBP21 in Tobacco Promotes Flowering Without Decreasing Flower or Fruit Quantity. Plant Mol. Biol. Report. 2011, 29, 573–581. [Google Scholar] [CrossRef]
- Liu, S.; Qi, T.T.; Ma, J.J.; Ma, T.; Ma, L.; Lin, X. Ectopic expression of a SOC1 homolog from Phyllostachys violascens alters flowering time and identity of floral organs in Arabidopsis thaliana. Trees 2016, 30, 2203–2215. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Birney, E.; Biswas, M.; Bucher, P.; Cerutti, L.; Corpet, F.; Croning, M.D.; et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001, 29, 37–40. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, G.; Yang, D.-H.; Cao, P.; Yao, H.; Zhang, Y.; Chen, X.; Xiao, B.; Li, F.; Wang, Z.-Y.; Yang, J.; et al. Genome-Wide Identification, Gene Structure and Expression Analysis of the MADS-Box Gene Family Indicate Their Function in the Development of Tobacco (Nicotiana tabacum L.). Int. J. Mol. Sci. 2019, 20, 5043. https://doi.org/10.3390/ijms20205043
Bai G, Yang D-H, Cao P, Yao H, Zhang Y, Chen X, Xiao B, Li F, Wang Z-Y, Yang J, et al. Genome-Wide Identification, Gene Structure and Expression Analysis of the MADS-Box Gene Family Indicate Their Function in the Development of Tobacco (Nicotiana tabacum L.). International Journal of Molecular Sciences. 2019; 20(20):5043. https://doi.org/10.3390/ijms20205043
Chicago/Turabian StyleBai, Ge, Da-Hai Yang, Peijian Cao, Heng Yao, Yihan Zhang, Xuejun Chen, Bingguang Xiao, Feng Li, Zhen-Yu Wang, Jun Yang, and et al. 2019. "Genome-Wide Identification, Gene Structure and Expression Analysis of the MADS-Box Gene Family Indicate Their Function in the Development of Tobacco (Nicotiana tabacum L.)" International Journal of Molecular Sciences 20, no. 20: 5043. https://doi.org/10.3390/ijms20205043
APA StyleBai, G., Yang, D.-H., Cao, P., Yao, H., Zhang, Y., Chen, X., Xiao, B., Li, F., Wang, Z.-Y., Yang, J., & Xie, H. (2019). Genome-Wide Identification, Gene Structure and Expression Analysis of the MADS-Box Gene Family Indicate Their Function in the Development of Tobacco (Nicotiana tabacum L.). International Journal of Molecular Sciences, 20(20), 5043. https://doi.org/10.3390/ijms20205043




