Genome-Wide Identification of Direct Targets of the TTG1–bHLH–MYB Complex in Regulating Trichome Formation and Flavonoid Accumulation in Arabidopsis Thaliana
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Analysis of Downstream Targets Involved in Trichome Formation and Flavonoid Accumulation in ttg1-13 Young Shoots and Expanding True Leaves
2.2. Validation of Downstream Targets Related to Trichome Formation and Flavonoid Accumulation in ttg1-13 Young Shoots and Expanding True Leaves
2.3. Identification of Direct Downstream Targets Contributing to Trichome Formation and Flavonoid Accumulation Regulated by the TTG1-Dependent MBW Complex in Young Shoots and Expanding True Leaves
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Condition
4.2. Generation of Transgenic Plants
4.3. RNA Sequencing (RNA-seq) and Data Analyses
4.4. Gene Expression Analysis
4.5. GR Induction
4.6. ChIP Assay
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Koornneef, M. The complex syndrome of ttg mutants. Arabidopsis Inf. Serv. 1981, 18, 45–51. [Google Scholar]
- Shirley, B.W.; Kubasek, W.L.; Storz, G.; Bruggemann, E.; Koornneef, M.; Ausubel, F.M.; Goodman, H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995, 8, 659–671. [Google Scholar] [CrossRef]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 1999, 11, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Debeaujon, I.; Leon-Kloosterziel, K.M.; Koornneef, M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000, 122, 403–413. [Google Scholar] [CrossRef]
- Western, T.L.; Burn, J.; Tan, W.L.; Skinner, D.J.; Martin-McCaffrey, L.; Moffatt, B.A.; Haughn, G.W. Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 2001, 127, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, Y.; Nambara, E.; Naito, S.; McCourt, P. The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 2004, 37, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, B.; Li, C.; Kulaveerasingam, H.; Chew, F.T.; Yu, H. TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol. 2015, 169, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B.; Chen, B.; Ji, L.; Hao, Y. Site-specific phosphorylation of TRANSPARENT TESTA GLABRA1 mediates carbon partitioning in Arabidopsis seeds. Nat. Commun. 2018, 9, 571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Ann. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Windsor, J.B.; Symonds, V.V.; Mendenhall, J.; Lloyd, A.M. Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J. 2000, 22, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Meissner, R.C.; Shoue, D.A.; Carpita, N.C.; Bevan, M.W. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 2001, 13, 2777–2791. [Google Scholar] [CrossRef] [PubMed]
- Tominaga-Wada, R.; Ishida, T.; Wada, T. New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Bio. 2011, 286, 67–106. [Google Scholar]
- Hoai Nguyen, N.; Jun Hyeok, K.; Woo Young, H.; Ngoc Trinh, N.; Suk-Whan, H.; Hojoung, L. TTG1-mediated flavonols biosynthesis alleviates root growth inhibition in response to ABA. Plant Cell Rep. 2013, 32, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Qi, S.; Li, D.; Jin, C.; Gao, C.; Duan, S.; Feng, B.; Chen, M. TRANSPARENT TESTA GLABRA 1 ubiquitously regulates plant growth and development from Arabidopsis to foxtail millet (Setaria italica). Plant Sci. 2016, 254, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Serna, L.; Martin, C. Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 2006, 11, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Broun, P. Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 2005, 8, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Traw, M.B.; Bergelson, J. Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 2003, 133, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Mauricio, R. Ontogenetics of QTL: The genetic architecture of trichome density over time in Arabidopsis thaliana. Genetica 2005, 123, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jing, S.X.; Luo, S.H.; Li, S.H. Non-volatile natural products in plant glandular trichomes: Chemistry, biological activities and biosynthesis. Nat. prod. Rep. 2019, 17, 626–665. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Schellmann, S.; Hulskamp, M.; Uhrig, J. Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem. Soc. T. 2007, 35, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kurata, T.; Okada, K.; Wada, T. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 2008, 59, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Pesch, M.; Hülskamp, M. One, two, three models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 2009, 12, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Chen, J.G. Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis. Front Plant Sci. 2014, 5, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Morohashi, K.; Hatlestad, G.; Grotewold, E.; Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 2008, 135, 1991–1999. [Google Scholar] [CrossRef] [Green Version]
- Pesch, M.; Schultheiss, I.; Klopffleisch, K.; Uhrig, J.F.; Koegl, M.; Clemen, C.S.; Simon, R.; Weidtkamp-Peters, S.; Hulskamp, M. TRANSPARENT TESTA GLABRA1 and GLABRA1 Compete for Binding to GLABRA3 in Arabidopsis. Plant Physiol. 2015, 168, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Nesi, N.; Jond, C.; Debeaujon, I.; Caboche, M.; Lepiniec, L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 2001, 13, 2099–2114. [Google Scholar] [CrossRef]
- Zhang, B.P.; Schrader, A. TRANSPARENT TESTA GLABRA 1-dependent regulation of flavonoid biosynthesis. Plants 2017, 6, E65. [Google Scholar]
- Mol, J.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar] [CrossRef]
- Feild, T.S.; Lee, D.W.; Holbrook, N.M. Why leaves turn red in autumn. The role of anthocyanins in senescing leaves of red-osier dogwood. Plant Physiol. 2001, 127, 566–574. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Bradshaw, H.D.; Schemske, D.W. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 2003, 426, 176–178. [Google Scholar] [CrossRef] [PubMed]
- Iwashina, T. The flavonoids occurring in plants, and their functions and activities to other organisms. Plant Cell Physiol. 2003, 44, S6. [Google Scholar]
- Hassan, S.; Mathesius, U. The role of flavonoids in root–rhizosphere signalling: Opportunities and challenges for improving plant–microbe interactions. J. Exp. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef] [PubMed]
- Carletti, G.; Nervo, G.; Cattivelli, L. Flavonoids and Melanins: A Common Strategy across Two Kingdoms. Int. J. Biol. Sci. 2014, 10, 1159–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaziano, M.; Riceevans, C. Epidemiology of flavonoids and coronary heart disease. Int. Congress Symp. Ser. R. Soc. Med. 2000, 17, 53–61. [Google Scholar]
- Lee, E.R.; Kang, G.H.; Cho, S.G. Effect of flavonoids on human health: Old subjects but new challenges. Recent Pat. Biotechnol. 2007, 1, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Jadeja, R.N.; Devkar, R.V. Chapter 47–Polyphenols and Flavonoids in Controlling Non-Alcoholic Steatohepatitis. Polyphenols Hum. Health Dis. 2014, 1, 615–623. [Google Scholar]
- Owaga, E.E.; Elbakkoush, A.; Sakhile, K.S.M.; Nyang’Inja, R.A. Nutritional management of mental disorders: Potential role of dietary flavonoids and vitamin E. Food Public Health 2014, 4, 104–109. [Google Scholar]
- Prieto-Domínguez, N.; García-Mediavilla, M.V.; Campos, S.S.; Mauriz, J.L.; González-Gallego, J. Autophagy as a Molecular Target of Flavonoids Underlying their Protective Effects in Human Disease. Curr. Med. Chem. 2017, 25, 814–838. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.; Zhang, F. Am, GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar] [PubMed]
- Zhang, F.; Gonzalez, A.M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2010, 40, 22–34. [Google Scholar] [CrossRef]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Lee, D.W. Anthocyanins in leaves: Distribution, phylogeny and development. Adv. Bot. Res. 2002, 37, 37–53. [Google Scholar]
- Unte, U.S.; Sorensen, A.M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-Box gene that affects pollen sac development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef]
- Kirik, V.; Simon, M.; Huelskamp, M.; Schiefelbein, J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 2004, 268, 506–513. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Manners, J.M. MYC2: The master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef] [PubMed]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Marks, M.D.; Wenger, J.P.; Gilding, E.; Jilk, R.; Dixon, R.A. Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol. Plant 2009, 2, 803–822. [Google Scholar] [CrossRef] [PubMed]
- Kasili, R.; Huang, C.C.; Walker, J.D.; Simmons, L.A.; Zhou, J.; Faulk, C.; Hulskamp, M.; Larkin, J.C. BRANCHLESS TRICHOMES links cell shape and cell cycle control in Arabidopsis trichomes. Development 2011, 138, 2379–2388. [Google Scholar] [CrossRef] [PubMed]
- Khosla, A.; Paper, J.M.; Boehler, A.P.; Bradley, A.M.; Neumann, T.R.; Schrick, K. HD-Zip Proteins GL2 and HDG11 Have Redundant Functions in Arabidopsis Trichomes, and GL2 Activates a Positive Feedback Loop via MYB23. Plant Cell 2014, 26, 2184–2200. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Hattori, S.; Sano, R.; Inoue, K.; Shirano, Y.; Hayashi, H.; Shibata, D.; Sato, S.; Kato, T.; Tabata, S.; et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 2007, 19, 2531–2543. [Google Scholar] [CrossRef] [PubMed]
- Glover, B.J.; Perez-Rodriguez, M.; Martin, C. Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 1998, 125, 3497–3508. [Google Scholar] [PubMed]
- Jakoby, M.J.; Falkenhan, D.; Mader, M.T.; Brininstool, G.; Wischnitzki, E.; Platz, N.; Hudson, A.; Lskamp, M.H.R.; Larkin, J.; Schnittger, A. Transcriptional Profiling of Mature Arabidopsis Trichomes Reveals That NOECK Encodes the MIXTA-Like Transcriptional Regulator MYB106. Plant Physiol. 2008, 148, 1583–1602. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Milliken, O.N.; Pham, H.; Seyit, R.; Napoli, R.; Preston, J.; Koltunow, A.M.; Parisha, R.W. The Arabidopsis MYB5 Transcription Factor Regulates Mucilage Synthesis, Seed Coat Development, and Trichome Morphogenesis. Plant Cell 2009, 21, 72–89. [Google Scholar] [CrossRef] [PubMed]
- Kirik, V.; Lee, M.M.; Wester, K.; Herrmann, U.; Zheng, Z.G.; Oppenheimer, D.; Schiefelbein, J.; Hulskamp, M. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 2005, 132, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Maes, L.; Inze, D.; Goossens, A. Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 2008, 148, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Kasili, R.; Walker, J.D.; Simmons, L.A.; Zhou, J.; De Veylder, L.; Larkin, J.C. SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 2010, 185, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Churchman, M.L.; Brown, M.L.; Kato, N.; Kirik, V.; Hulskamp, M.; Inze, D.; De Veylder, L.; Walker, J.D.; Zheng, Z.; Oppenheimer, D.G.; et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 2006, 18, 3145–3157. [Google Scholar] [CrossRef] [PubMed]
- Oxley, D.; Ktistakis, N.; Farmaki, T. Differential isolation and identification of PI(3)P and PI(3,5)P-2 binding proteins from Arabidopsis thaliana using an agarose-phosphatidylinositol-phosphate affinity chromatography. J. Proteomics 2013, 91, 580–594. [Google Scholar] [CrossRef]
- Wang, X.Y.; Wang, X.L.; Hu, Q.N.; Dai, X.M.; Tian, H.N.; Zheng, K.J.; Wang, X.P.; Mao, T.L.; Chen, J.G.; Wang, S.C. Characterization of an activation-tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis. Plant J. 2015, 83, 300–311. [Google Scholar] [CrossRef]
- Kubo, H.; Peeters, A.J.M.; Aarts, M.G.M.; Pereira, A.; Koornneef, M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 1999, 11, 1217–1226. [Google Scholar] [CrossRef]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2010, 46, 768–779. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Ohl, S.; Hedrick, S.A.; Chory, J.; Lamb, C.J. Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell 1990, 2, 837–848. [Google Scholar] [PubMed]
- Schoenbohm, C.; Martens, S.; Eder, C.; Forkmann, G.; Weisshaar, B. Identification of the Arabidopsis thaliana flavonoid 3 ‘-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol. Chem. 2000, 381, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, M.L.F.; Emiliani, J.; Rodriguez, E.J.; Campos-Bermudez, V.A.; Grotewold, E.; Casati, P. The Identification of Maize and Arabidopsis Type I FLAVONE SYNTHASEs Links Flavones with Hormones and Biotic Interactions. Plant Physiol. 2015, 169, 1090–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preuss, A.; Stracke, R.; Weisshaar, B.; Hillebrecht, A.; Matern, U.; Martens, S. Arabidopsis thaliana expresses a second functional flavonol synthase. Febs Lett. 2009, 583, 1981–1986. [Google Scholar] [CrossRef] [PubMed]
- Shirley, B.W.; Hanley, S.; Goodman, H.M. Effects of Ionizing-Radiation on a Plant Genome - Analysis of 2 Arabidopsis Transparent-Testa Mutations. Plant Cell 1992, 4, 333–347. [Google Scholar] [PubMed]
- Nakajima, J.; Tanaka, Y.; Yamazaki, M.; Saito, K. Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J. Biol. Chem. 2001, 276, 25797–25803. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, J.; Sato, Y.; Hoshino, T.; Yamazaki, M.; Saito, K. Mechanistic study on the oxidation of anthocyanidin synthase by quantum mechanical calculation. J. Biol. Chem. 2006, 281, 21387–21398. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Fukushima, A.; Nakabayashi, R.; Hanada, K.; Matsuda, F.; Sugawara, S.; Inoue, E.; Kuromori, T.; Ito, T.; Shinozaki, K.; et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012, 69, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, J.C.; Reichelt, M.; Luck, K.; Svatos, A.; Gershenzon, J. Identification and characterization of the BAHD acyltransferase malonyl CoA: Anthocyanidin 5-O-glucoside-6’’-O-malonyltransferase (At5MAT) in Arabidopsis thaliana. FEBS Lett 2007, 581, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, H.; Huang, J.R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 2012, 5, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Wiesen, L.B.; Bender, R.L.; Paradis, T.; Larson, A.; Perera, M.; Nikolau, B.J.; Olszewski, N.E.; Carter, C.J. A role for GIBBERELLIN 2-OXIDASE6 and gibberellins in regulating nectar production. Mol. Plant 2016, 9, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Lehotai, N.; Feigl, G.; Koos, A.; Molnar, A.; Ordog, A.; Peto, A.; Erdei, L.; Kolbert, Z. Nitric oxide-cytokinin interplay influences selenite sensitivity in Arabidopsis. Plant Cell Rep. 2016, 35, 2181–2195. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, M.K.; Murrell, J.R.; Shirley, B.W. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis—Further evidence for differential regulation of ‘‘early’’ and ‘‘late’’ genes. Plant Physiol. 1997, 113, 1437–1445. [Google Scholar] [CrossRef] [PubMed]
- Prescott, A.G.; Stamford, N.P.J.; Wheeler, G.; Firmin, J.L. In vitro properties of a recombinant flavonol synthase from Arabidopsis thaliana. Phytochemistry 2002, 60, 589–593. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.H.; Li, D.; Jin, C.Y.; Duan, S.W.; Qi, S.H.; Liu, K.G.; Wang, H.C.; Ma, H.L.; Hai, J.B.; Chen, M.X. Genome-wide identification of GLABRA3 downstream genes for anthocyanin biosynthesis and trichome formation in Arabidopsis. Biochem. Bioph. Res. Co. 2017, 485, 360–365. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Duan, S.W.; Wang, J.J.; Gao, C.H.; Jin, C.Y.; Li, D.; Peng, D.S.; Du, G.M.; Li, Y.Q.; Chen, M.X. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018, 268, 47–53. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
 ) and ttg1-13 (
) and ttg1-13 (  ) plants at 18, 20, and 22 days after germination. The house-keeping gene ACTIN7 was used as the internal control. The expression of each gene was first calculated relative to ACTIN7 and then normalized to its expression level at 18 days after germination in the wild type that was set to 1. Asterisks indicate significant differences in gene expression compared with the wild type control (two-tailed paired Student’s t test, p ≤ 0.05). Values are means ± SD (n = 3). Error bars denote SD.
) plants at 18, 20, and 22 days after germination. The house-keeping gene ACTIN7 was used as the internal control. The expression of each gene was first calculated relative to ACTIN7 and then normalized to its expression level at 18 days after germination in the wild type that was set to 1. Asterisks indicate significant differences in gene expression compared with the wild type control (two-tailed paired Student’s t test, p ≤ 0.05). Values are means ± SD (n = 3). Error bars denote SD.
 ) and ttg1-13 (
) and ttg1-13 (  ) plants at 18, 20, and 22 days after germination. The house-keeping gene ACTIN7 was used as the internal control. The expression of each gene was first calculated relative to ACTIN7 and then normalized to its expression level at 18 days after germination in the wild type that was set to 1. Asterisks indicate significant differences in gene expression compared with the wild type control (two-tailed paired Student’s t test, p ≤ 0.05). Values are means ± SD (n = 3). Error bars denote SD.
) plants at 18, 20, and 22 days after germination. The house-keeping gene ACTIN7 was used as the internal control. The expression of each gene was first calculated relative to ACTIN7 and then normalized to its expression level at 18 days after germination in the wild type that was set to 1. Asterisks indicate significant differences in gene expression compared with the wild type control (two-tailed paired Student’s t test, p ≤ 0.05). Values are means ± SD (n = 3). Error bars denote SD.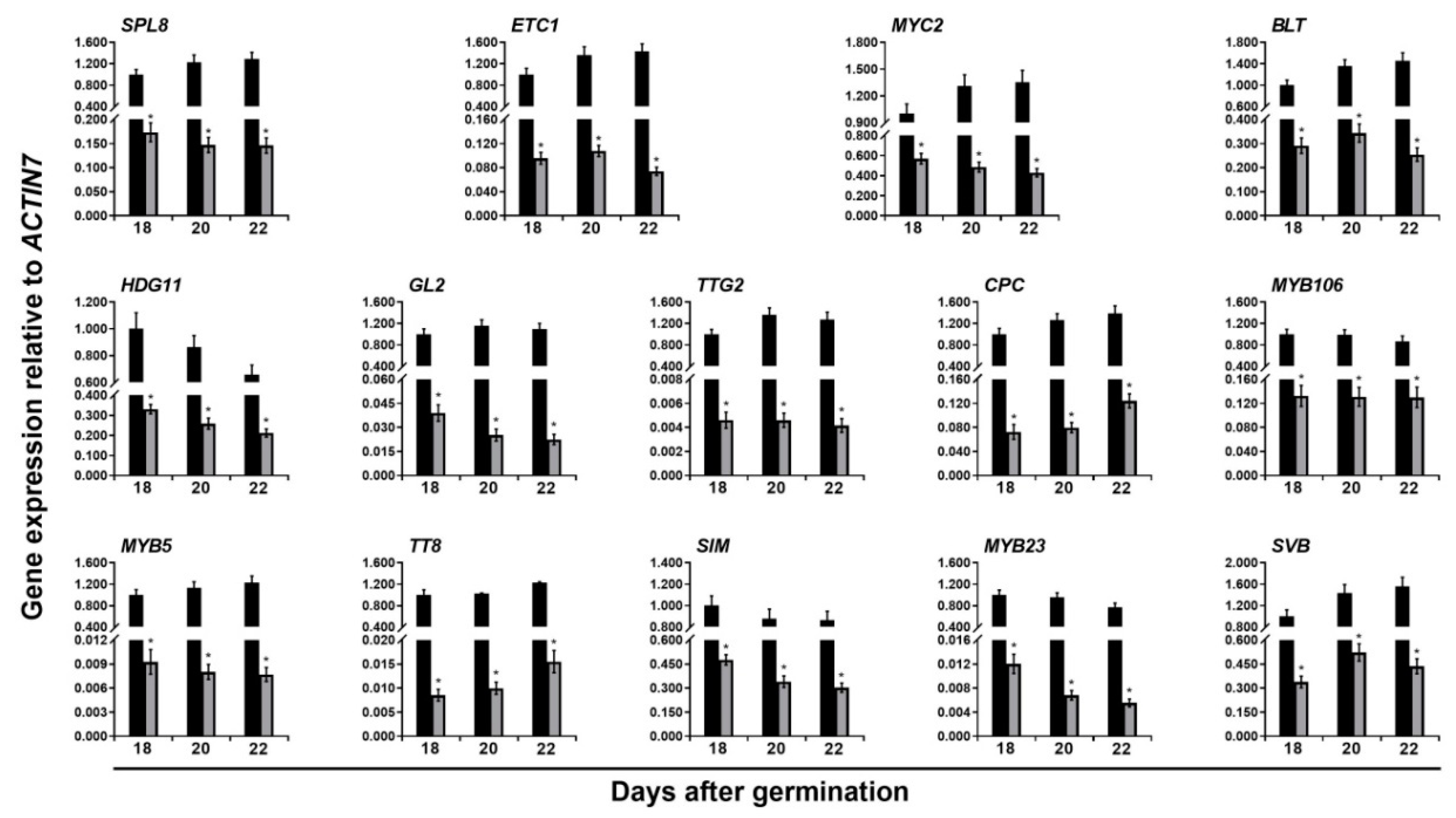
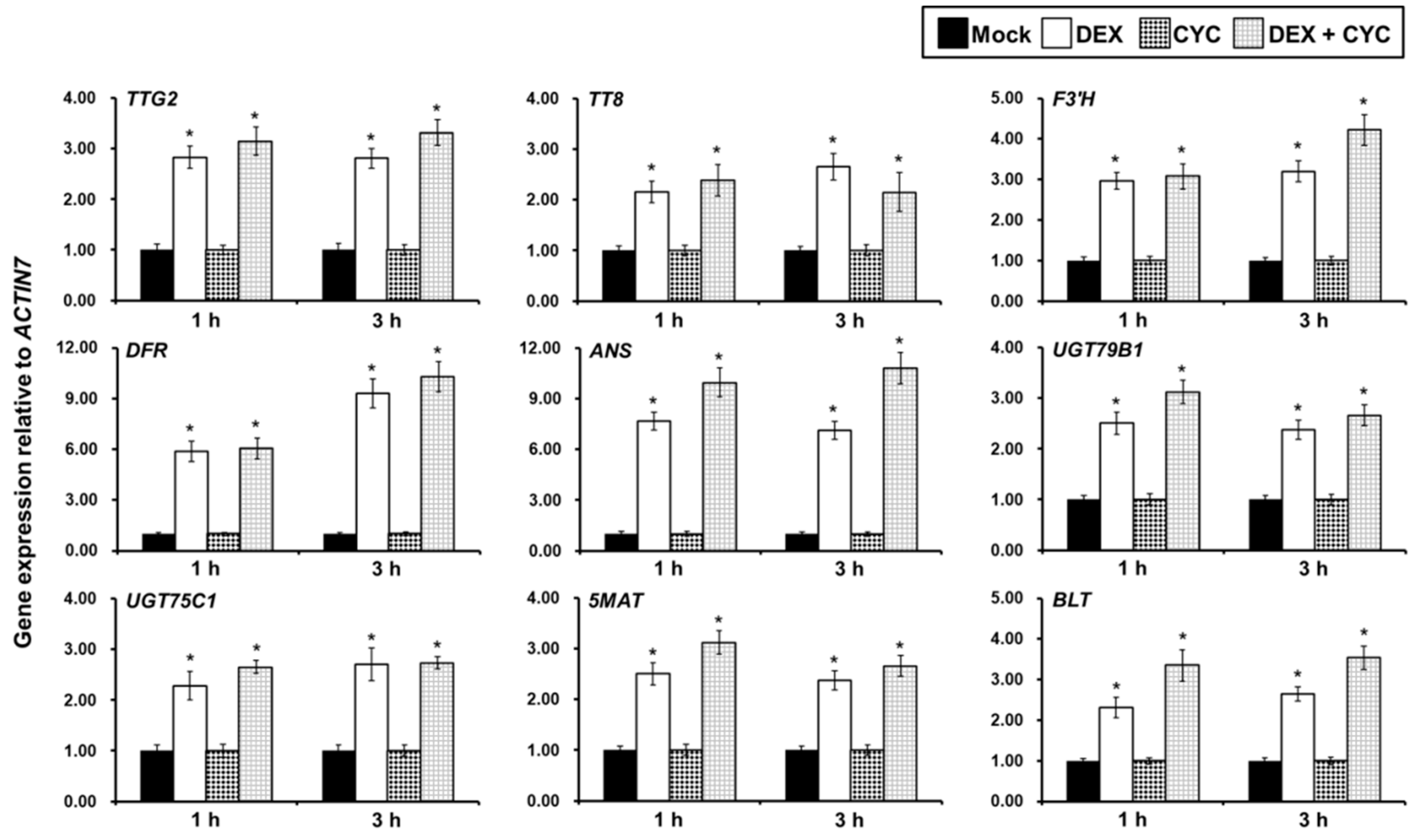
 ) and (
) and (  ) ttg1-13 plants at 18, 20, and 22 days after germination. The house-keeping gene ACTIN7 was used as the internal control. The expression of each gene was first calculated relative to ACTIN7 and then normalized to its expression level at 18 days after germination in the wild type that was set to 1. Asterisks indicate significant differences in gene expression compared with the wild type control (two-tailed paired Student’s t test, p ≤ 0.05). Values are means ± SD (n = 3). Error bars denote SD.
) ttg1-13 plants at 18, 20, and 22 days after germination. The house-keeping gene ACTIN7 was used as the internal control. The expression of each gene was first calculated relative to ACTIN7 and then normalized to its expression level at 18 days after germination in the wild type that was set to 1. Asterisks indicate significant differences in gene expression compared with the wild type control (two-tailed paired Student’s t test, p ≤ 0.05). Values are means ± SD (n = 3). Error bars denote SD.
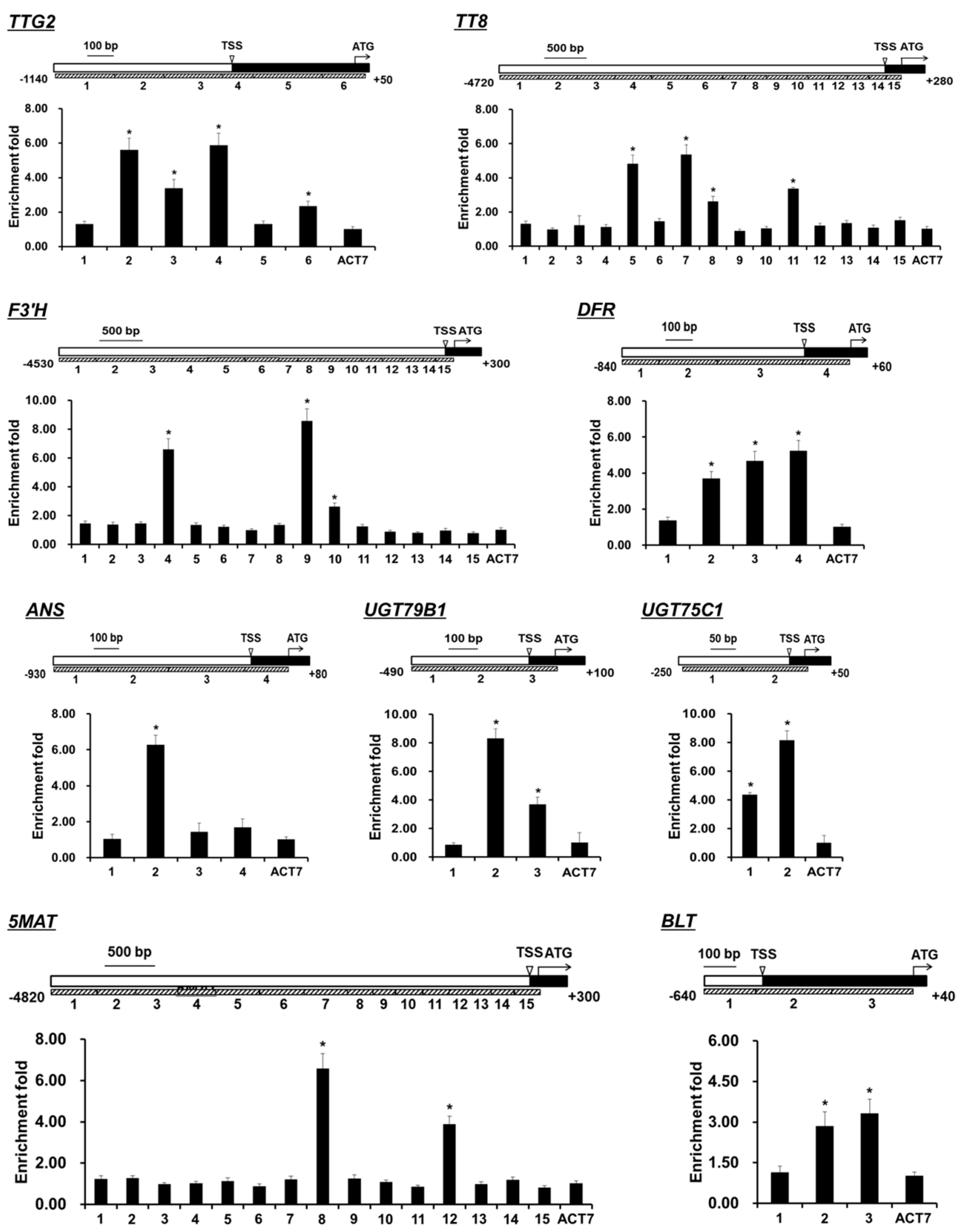
 ) and (
) and (  ) ttg1-13 plants at 18, 20, and 22 days after germination. The house-keeping gene ACTIN7 was used as the internal control. The expression of each gene was first calculated relative to ACTIN7 and then normalized to its expression level at 18 days after germination in the wild type that was set to 1. Asterisks indicate significant differences in gene expression compared with the wild type control (two-tailed paired Student’s t test, p ≤ 0.05). Values are means ± SD (n = 3). Error bars denote SD.
) ttg1-13 plants at 18, 20, and 22 days after germination. The house-keeping gene ACTIN7 was used as the internal control. The expression of each gene was first calculated relative to ACTIN7 and then normalized to its expression level at 18 days after germination in the wild type that was set to 1. Asterisks indicate significant differences in gene expression compared with the wild type control (two-tailed paired Student’s t test, p ≤ 0.05). Values are means ± SD (n = 3). Error bars denote SD.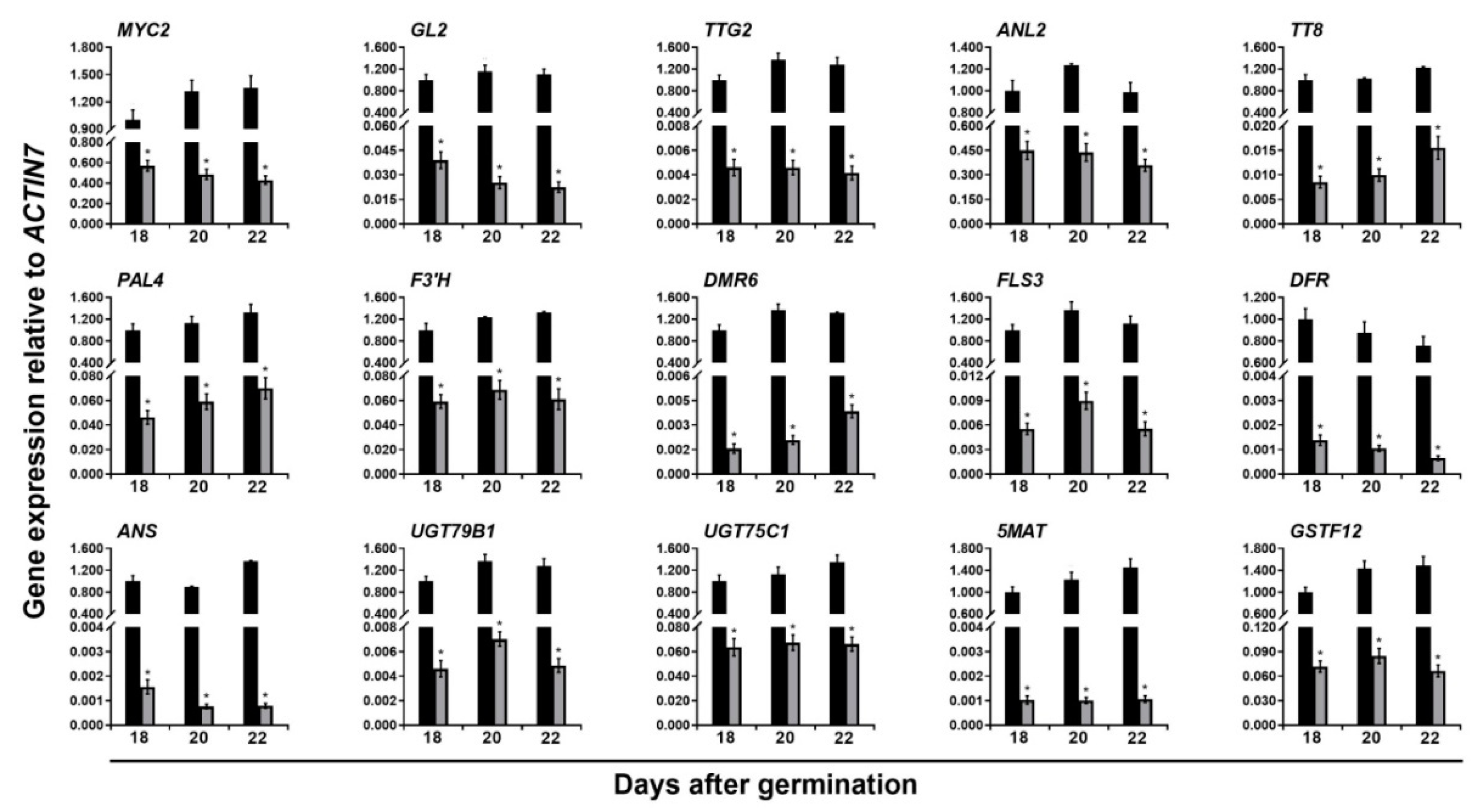
| Differentially Expressed Genes | log2 Ratios | Functions | References |
|---|---|---|---|
| SPL8 (AT1G02065) | −2.36 | Promoting trichome formation | [48] |
| ETC1 (AT1G01380) | −3.73 | Repressing trichome formation | [49] |
| MYC2 (AT1G32640) | −1.27 | Promoting trichome initiation | [50,51,52,53] |
| BLT (AT1G64690) | −2.10 | Promoting trichome branching | [54,55] |
| HDG11 (AT1G73360) | −1.78 | Promoting trichome differentiation | [56] |
| GL2 (AT1G79840) | −6.33 | Promoting trichome differentiation | [22,23,24] |
| TTG2 (AT2G37260) | −7.89 | Promoting trichome formation | [25,57] |
| CPC (AT2G46410) | −4.09 | Repressing trichome formation | [49] |
| MYB106 (AT3G01140) | −2.82 | Promoting trichome differentiation and repressing trichome branching | [58,59] |
| MYB5 (AT3G13540) | −7.07 | Repressing trichome branching | [60,61] |
| TT8 (AT4G09820) | −7.14 | Promoting flavonoid accumulation | [62] |
| SIM (AT5G04470) | −1.94 | Associated with trichome development | [63,64] |
| MYB23 (AT5G40330) | −7.13 | Repressing trichome branching | [60,61] |
| SVB (AT1G56580) | −1.43 | Associated with trichome size and branching | [65] |
| Differentially Expressed Genes | log2 Ratios | Functions | References |
|---|---|---|---|
| MYC2 (AT1G32640) | −1.27 | Promoting flavonoid accumulation | [50,51,52,53] |
| GL2 (AT1G79840) | −6.33 | Inhibiting anthocyanin accumulation | [66] |
| TTG2 (AT2G37260) | −7.89 | Promoting flavonoid accumulation | [25,57] |
| ANL2 (AT4G00730) | −1.06 | Promoting anthocyanin accumulation | [67] |
| TT8 (AT4G09820) | −7.14 | Promoting flavonoid accumulation | [68,69] |
| PAL4 (AT3G10340) | −3.72 | Converting phenylalanine into trans-cinnamic acid | [70,71] |
| F3’H (AT5G07990) | −4.08 | Converting naringenin and dihydrokaempferol into eriodictyol and dihydroquercetin, respectively | [70,72] |
| DMR6 (AT5G24530) | −9.50 | Converting flavanones into flavones | [73] |
| FLS3 (AT5G63590) | −7.55 | Promoting flavonol accumulation | [74] |
| DFR (AT5G42800) | −10.85 | Converting dihydroflavonols into leucoanthocyanidins | [2,75] |
| ANS (AT4G22880) | −10.68 | Converting leucoanthocyanidins into 3-OH-anthocyanins | [76,77] |
| UGT79B1 (AT5G54060) | −8.29 | Involved in the glycosylation of anthocyanins | [70,78] |
| UGT75C1 (AT4G14090) | −4.09 | Involved in the malonylation of anthocyanins | [79] |
| 5MAT (AT3G29590) | −11.05 | Involved in the accumulation of malonylated anthocyanins | [80] |
| GSTF12 (AT5G17220) | −3.73 | Involved in transport and accumulation of both anthocyanins and proanthocyanidins | [81,82] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Z.; Cheng, Y.; Zhou, C.; Li, D.; Gao, X.; Zhang, S.; Chen, M. Genome-Wide Identification of Direct Targets of the TTG1–bHLH–MYB Complex in Regulating Trichome Formation and Flavonoid Accumulation in Arabidopsis Thaliana. Int. J. Mol. Sci. 2019, 20, 5014. https://doi.org/10.3390/ijms20205014
Wei Z, Cheng Y, Zhou C, Li D, Gao X, Zhang S, Chen M. Genome-Wide Identification of Direct Targets of the TTG1–bHLH–MYB Complex in Regulating Trichome Formation and Flavonoid Accumulation in Arabidopsis Thaliana. International Journal of Molecular Sciences. 2019; 20(20):5014. https://doi.org/10.3390/ijms20205014
Chicago/Turabian StyleWei, Zelou, Yalong Cheng, Chenchen Zhou, Dong Li, Xin Gao, Shuoxin Zhang, and Mingxun Chen. 2019. "Genome-Wide Identification of Direct Targets of the TTG1–bHLH–MYB Complex in Regulating Trichome Formation and Flavonoid Accumulation in Arabidopsis Thaliana" International Journal of Molecular Sciences 20, no. 20: 5014. https://doi.org/10.3390/ijms20205014
APA StyleWei, Z., Cheng, Y., Zhou, C., Li, D., Gao, X., Zhang, S., & Chen, M. (2019). Genome-Wide Identification of Direct Targets of the TTG1–bHLH–MYB Complex in Regulating Trichome Formation and Flavonoid Accumulation in Arabidopsis Thaliana. International Journal of Molecular Sciences, 20(20), 5014. https://doi.org/10.3390/ijms20205014





