Translesion DNA Synthesis Across Lesions Induced by Oxidative Products of Pyrimidines: An Insight into the Mechanism by Microscale Thermophoresis
Abstract
:1. Introduction
2. Results and Discussion
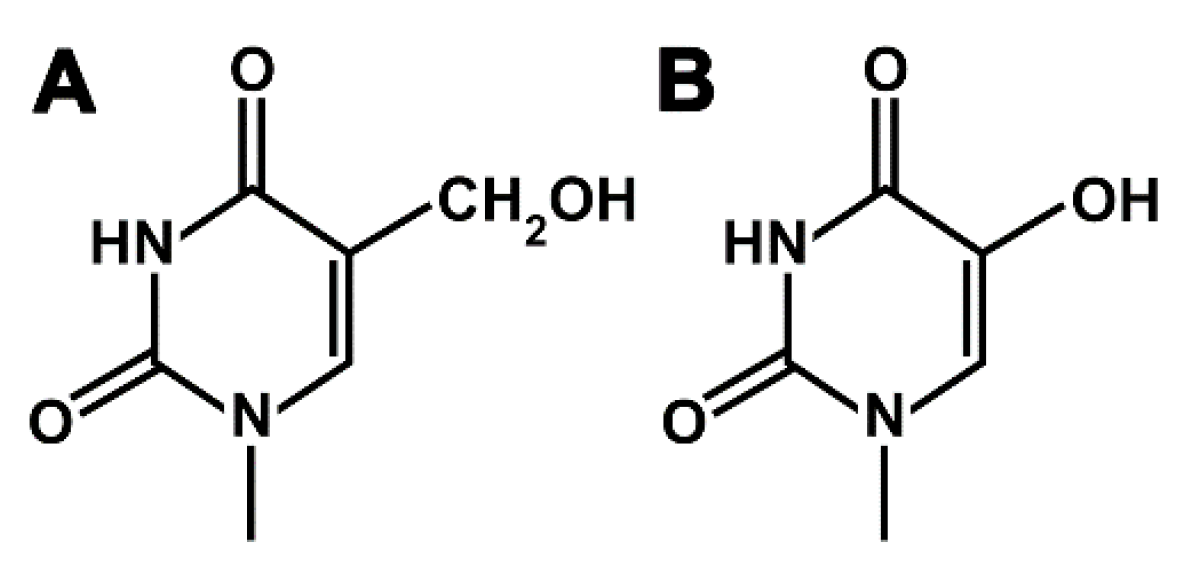
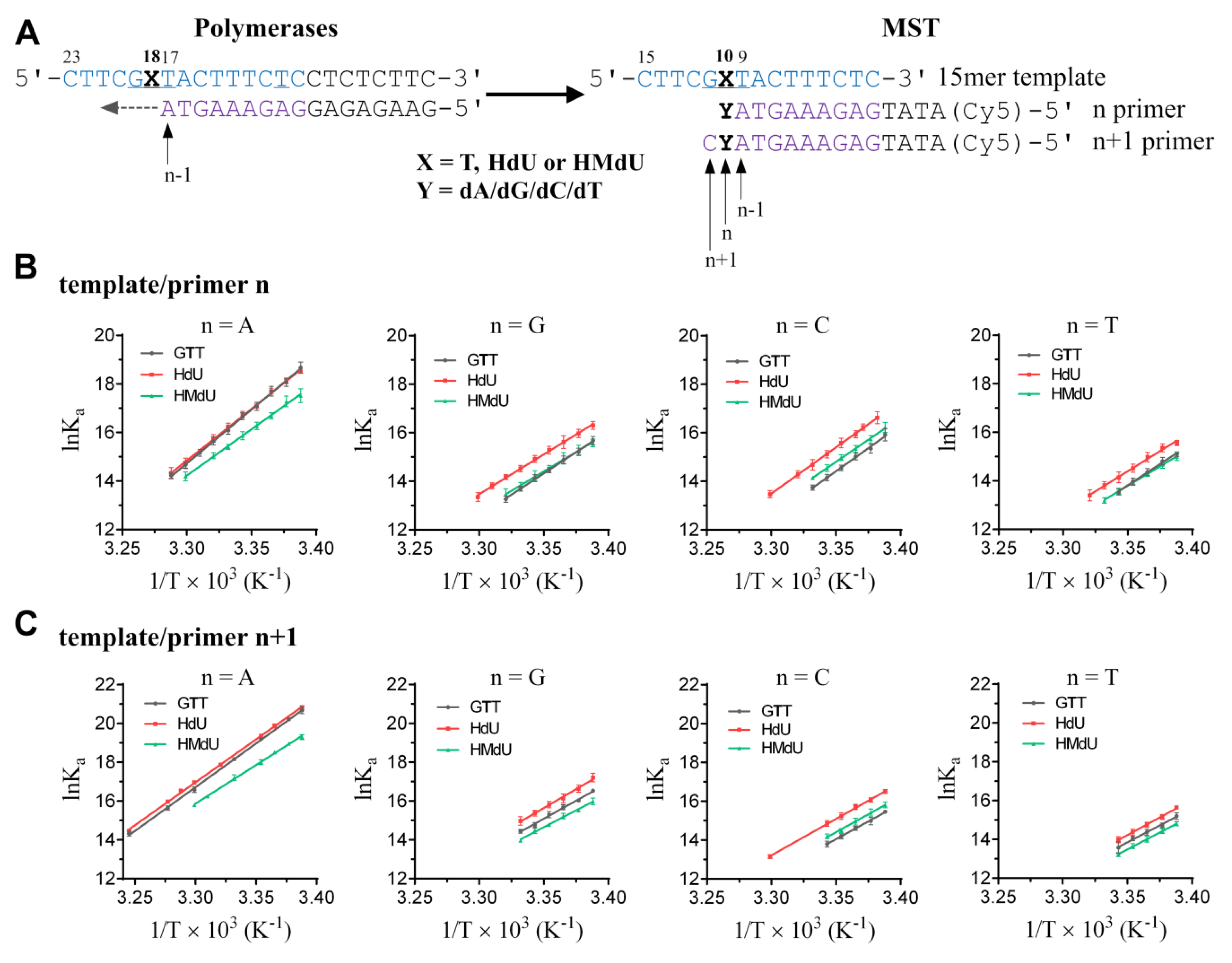
2.1. Determination of Thermodynamic Parameters of Duplex DNA Constructs with Nucleotide Misincorporation Opposite HdU and HMdU Lesions by Microscale Thermophoresis
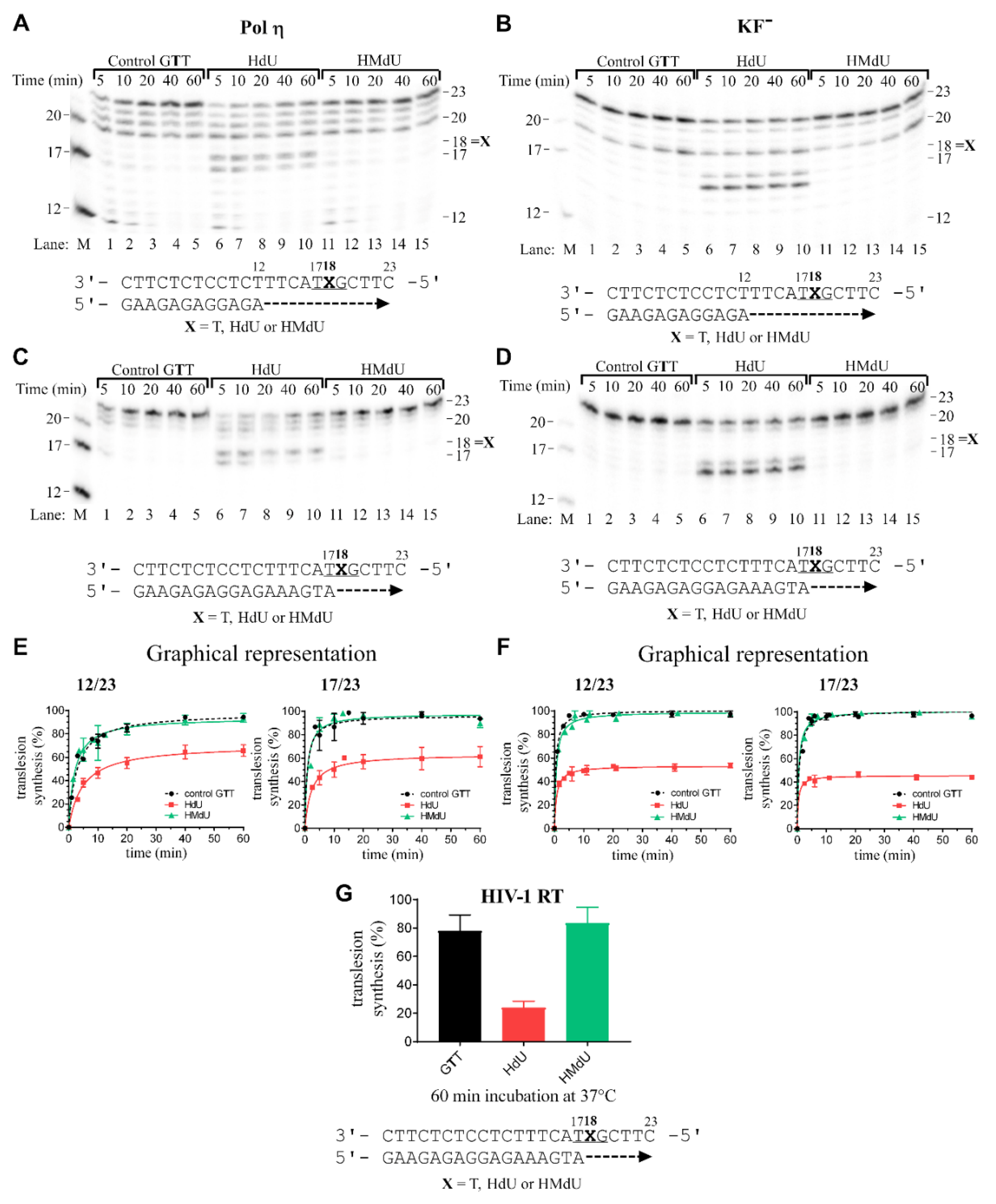
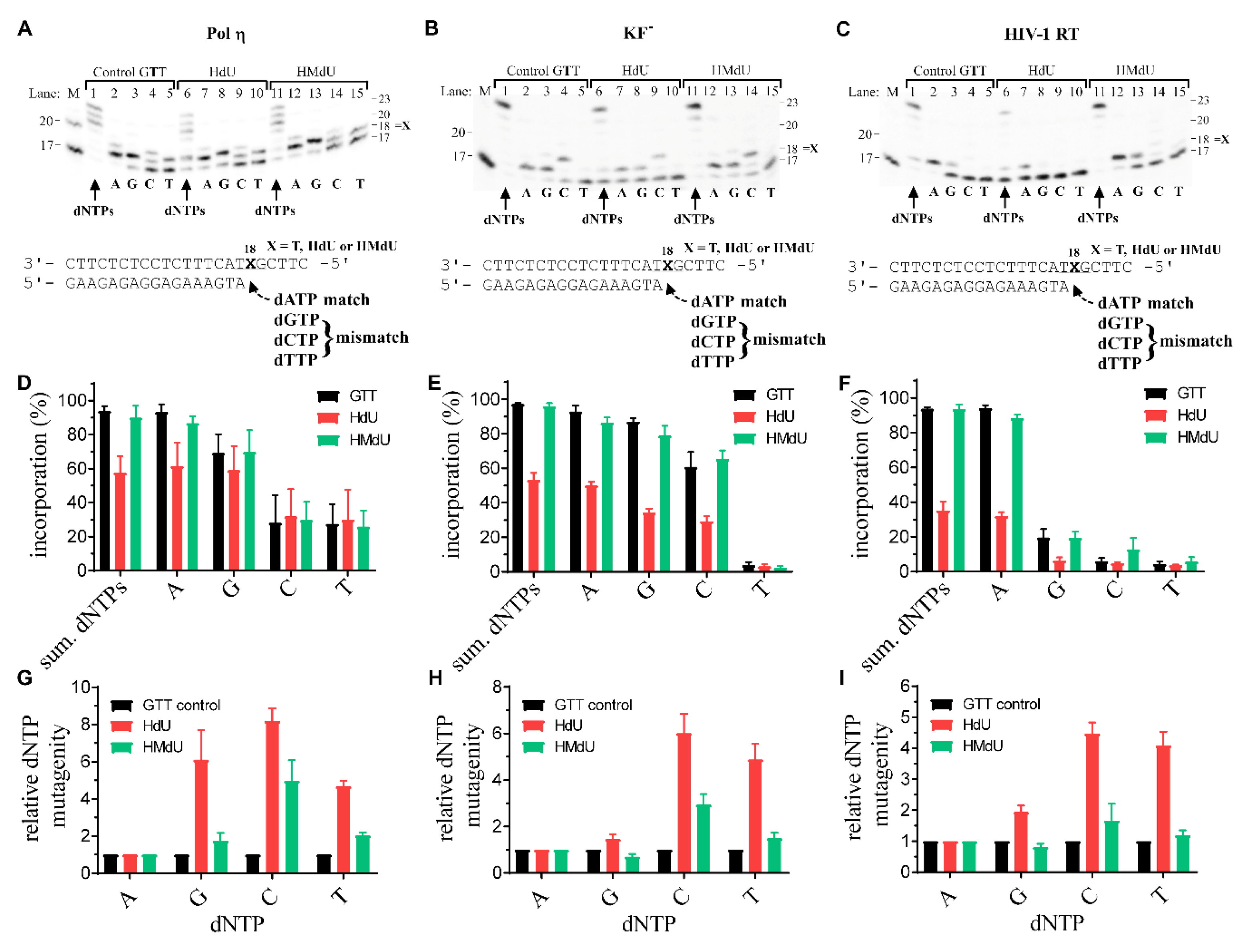
2.2. DNA Synthesis by DNA Polymerases across Pyrimidine Oxidative Products HdU and HMdU
3. Conclusions
4. Materials and Methods
4.1. Chemicals
4.2. Purification of Oligonucleotides
4.3. Microscale Thermophoresis (MST) Determination of Thermodynamic Parameters of DNA Constructs with Nucleotide Misincorporation Opposite Pyrimidine Derivatives
4.4. Primer Extension Activity of DNA Polymerases
4.5. Analysis of Nucleotide Misincorporation Opposite Pyrimidine Derivatives
4.6. Other Physical Methods
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HdU | 2’-deoxyribo-5-hydroxyuridin |
| HMdU | 2’-deoxyribo-5-hydroxymethyluridin |
| TLS | translesion DNA synthesis |
| polη | DNA polymerase η |
| KF– | Klenow fragment of DNA polymerase I (the exonuclease deficient) |
| HIV-1 RT | reverse transcriptase from human immunodeficiency virus type 1 |
| dT | thymidine |
| dNTP | 2’-deoxyribonucleotide-5‘-triphosphate |
| PAA | polyacrylamide |
| MST | microscale thermophoresis |
| DSC | differential scanning calorimetry |
| DTT | dithiothreitol |
| BSA | bovine serum albumin |
| TET | ten eleven translocation |
References
- Douki, T.; Delatour, T.; Paganon, F.; Cadet, J. Measurement of oxidative damage at pyrimidine bases in gamma-irradiated DNA. Chem. Res. Toxicol. 1996, 9, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Purmal, A.A.; Kow, Y.W.; Wallace, S.S. Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in-vitro. Nucleic Acids Res. 1994, 22, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Thiviyanathan, V.; Somasunderam, A.; Volk, D.E.; Gorenstein, D.G. 5-Hydroxyuracil can form stable base pairs with all four bases in a DNA duplex. Chem. Commun. 2005, 3, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell. Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, F.; Cuesta, S.M.; Beraldi, D.; Mahtey, A.; Hardisty, R.E.; Carrington, M.; Balasubramanian, S. Sequencing 5-hydroxymethyluracil at single-base resolution. Angew. Chem. Int. Ed. 2018, 57, 9694–9696. [Google Scholar] [CrossRef]
- Kreutzer, D.A.; Essigmann, J.M. Oxidized, deaminated cytosines are a source of C- > T transitions in vivo. Proc. Natl. Acade Sci. USA 1998, 95, 3578–3582. [Google Scholar] [CrossRef] [PubMed]
- Purmal, A.A.; Lampman, G.W.; Bond, J.P.; Hatahet, Z.; Wallace, S.S. Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J. Biol. Chem. 1998, 273, 10026–10035. [Google Scholar] [CrossRef]
- Thiviyanathan, V.; Somasunderam, A.; Volk, D.E.; Hazra, T.K.; Mitra, S.; Gorenstein, D.G. Base-pairing properties of the oxidized cytosine derivative, 5-hydroxy uracil. Biochem. Biophys. Res. Commun. 2008, 366, 752–757. [Google Scholar] [CrossRef] [Green Version]
- Greim, H.; Albertini, R.J. The Cellular Response to the Genotoxic Insult. The Question of Treshold for Genootoxic Carcinogens; The Royal Society of Chemistry: Cambridge, UK, 2012. [Google Scholar]
- Mellac, S.; Fazakerley, G.V.; Sowers, L.C. Structures of base-pairs with 5-(hydroxymethyl)-2′-deoxyuridine in DNA determined by NMR-spectroscopy. Biochemistry 1993, 32, 7779–7786. [Google Scholar] [CrossRef]
- Carson, S.; Wilson, J.; Aksimentiev, A.; Weigele, P.R.; Wanunu, M. Hydroxymethyluracil modifications enhance the flexibility and hydrophilicity of double-stranded DNA. Nucleic Acids Res. 2016, 44, 2085–2092. [Google Scholar] [CrossRef]
- Greene, J.R.; Morrissey, L.M.; Foster, L.M.; Geiduschek, E.P. DNA-binding by the bacteriophage-SPO1-encoded type-II DNA-binding protein, transcription factor-1 formation of nested complexes at a selective binding-site. J. Biol. Chem. 1986, 261, 2820–2827. [Google Scholar]
- Pfaffeneder, T.; Spada, F.; Wagner, M.; Brandmayr, C.; Laube, S.K.; Eisen, D.; Truss, M.; Steinbacher, J.; Hackner, B.; Kotljarova, O. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014, 10, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Herrala, A.M.; Vilpo, J.A. Template-primer activity of 5-(hydroxymethyl)uracil-containing DNA for prokaryotic and eukaryotic DNA and RNA polymerases. Biochemistry 1989, 28, 8274–8277. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.D.; Teebor, G.W. Site directed substitution of 5-hydroxymethyluracil for thymine in replicating øX-M4am3 DNA via synthesis of 5-hydroxymethyl-2′-deoxyuridine-5′-triphosphate. Nucleic Acids Res. 1991, 19, 3337–3343. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, F.; Murat, P.; Li, Z.; Santner, T.; Balasubramanian, S. Synthesis and biophysical analysis of modified thymine-containing DNA oligonucleotides. Chem. Commun. 2017, 53, 1389–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaluca, A.; Wagner, J.R.; Saragovi, H.U.; Ramotar, D. UNG-1 and APN-1 are the major enzymes to efficiently repair 5-hydroxymethyluracil DNA lesions in C. elegans. Sci. Rep. 2018, 8, 6860. [Google Scholar] [CrossRef]
- Jacobs, A.L.; Schar, P. DNA glycosylases: In DNA repair and beyond. Chromosoma 2012, 121, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Boorstein, R.J.; Teebor, G.W. Mutagenicity of 5-hydroxymethyl-2′-deoxyuridine to Chinese Hamster cells. Cancer Res. 1988, 48, 5466–5470. [Google Scholar] [PubMed]
- Boorstein, R.J.; Teebor, G.W. Effects of 5-hydroxymethyluracil and 3-aminobenzamide on the repair and toxicity of 5-hydroxymethyl-2′-deoxyuridine in mammalian cells. Cancer Res. 1989, 49, 1509–1514. [Google Scholar] [PubMed]
- Shiau, G.T.; Schinazi, R.F.; Chen, M.S.; Prusoff, W.H. Synthesis and biological activities of 5-(hydroxymethyl, azidomethyl, or aminomethyl)-2′-deoxyuridine and related 5′-substituted analogs. J. Med. Chem. 1980, 23, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Bassett, E.; Vaisman, A.; Havener, J.M.; Masutani, C.; Hanaoka, F.; Chaney, S.G. Efficiency of extension of mismatched primer termini across from cisplatin and oxaliplatin adducts by human DNA polymerases beta and eta in vitro. Biochemistry 2003, 42, 14197–14206. [Google Scholar] [CrossRef] [PubMed]
- Arana, M.E.; Song, L.; Le Gac, N.T.; Parris, D.S.; Villani, G.; Boehmer, P.E.B. On the role of proofreading exonuclease in bypass of a 1,2 d(GpG) cisplatin adduct by the herpes simplex virus-1 DNA polymerase. DNA Repair 2004, 3, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Wickramaratne, S.; Boldry, E.J.; Buehler, C.; Wang, Y.-C.; Distefano, M.D.; Tretyakova, N.Y. Error-prone translesion synthesis past DNA-peptide cross-links conjugated to the major groove of DNA via C5 of thymidine. J. Biol. Chem. 2015, 290, 775–787. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, D.K.; Guengerich, F.P.; Egli, M.; Wilds, C.J. Backbone flexibility influences nucleotide incorporation by human translesion DNA polymerase η opposite intrastrand cross-linked DNA. Biochemistry 2015, 54, 7449–7456. [Google Scholar] [CrossRef] [PubMed]
- Villani, G.; Hubscher, U.; Gironis, N.; Parkkinen, S.; Pospiech, H.; Shevelev, I.; di Cicco, G.; Markkanen, E.; Syvaoja, J.E.; Tanguy Le Gac, N. In vitro gap-directed translesion DNA synthesis of an abasic site involving human DNA polymerases epsilon, lambda, and beta. J. Biol. Chem. 2011, 286, 32094–32104. [Google Scholar] [CrossRef]
- Ho, T.V.; Guainazzi, A.; Derkunt, S.B.; Enoiu, M.; Schärer, O.D. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 2011, 39, 7455–7464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beard, W.A.; Wilson, S.H. Structural insights into the origins of DNA polymerase fidelity. Structure 2003, 11, 489–496. [Google Scholar] [CrossRef]
- Novakova, O.; Farrell, N.P.; Brabec, V. Translesion DNA synthesis across double-base lesions derived from cross-links of an antitumor trinuclear platinum compound: Primer extension, conformational and thermodynamic studies. Metallomics 2018, 10, 132–144. [Google Scholar] [CrossRef]
- Kasparkova, J.; Suchankova, T.; Halamikova, A.; Zerzankova, L.; Vrana, O.; Margiotta, N.; Natile, G.; Brabec, V. Cytotoxicity, cellular uptake, glutathione and DNA interactions of an antitumor large-ring PtII chelate complex incorporating the cis-1,4-diaminocyclohexane carrier ligand. Biochem. Pharmacol. 2010, 79, 552–564. [Google Scholar] [CrossRef] [Green Version]
- Minetti, C.; Remeta, D.P.; Miller, H.; Gelfand, C.A.; Plum, G.E.; Grollman, A.P.; Breslauer, K.J. The thermodynamics of template-directed DNA synthesis: Base insertion and extension enthalpies. Proc. Natl. Acad. Sci. USA 2003, 100, 14719–14724. [Google Scholar] [CrossRef] [Green Version]
- Kieft, R.; Bullard, W.; Sabatini, R. A method for the efficient and selective identification of 5-hydroxymethyluracil in genomic DNA. Biol. Methods Protoc. 2017, 2, 1–10. [Google Scholar]
- Liang, F.; Cho, B.P. Probing the thermodynamics of aminofluorene-induced translesion DNA synthesis by differential scanning calorimetry. J. Am. Chem. Soc. 2007, 129, 12108–12109. [Google Scholar] [CrossRef] [PubMed]
- Novakova, O.; Malina, J.; Kasparkova, J.; Halamikova, A.; Bernard, V.; Intini, F.; Natile, G.; Brabec, V. Energetics, conformation, and recognition of DNA duplexes modified by methylated analogues of [PtCl(dien)]+. Chem. Eur. J. 2009, 15, 6211–6221. [Google Scholar] [CrossRef] [PubMed]
- Novakova, O.; Malina, J.; Suchankova, T.; Kasparkova, J.; Bugarcic, T.; Sadler, P.J.; Brabec, V. Energetics, conformation, and recognition of DNA duplexes modified by monodentate RuII complexes containing terphenyl arenes. Chem. Eur. J. 2010, 16, 5744–5754. [Google Scholar] [CrossRef] [PubMed]
- Florian, J.; Brabec, V. Thermodynamics of translesion synthesis across a major DNA adduct of antitumor oxaliplatin: Differential scanning calorimetric study. Chem. Eur. J. 2012, 18, 1634–1639. [Google Scholar] [CrossRef] [PubMed]
- Malina, J.; Novakova, O.; Natile, G.; Brabec, V. The thermodynamics of translesion DNA synthesis past major adducts of enantiomeric analogues of antitumor cisplatin. Chem. Asian J. 2012, 7, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Malina, J.; Brabec, V. Thermodynamic impact of abasic sites on simulated translesion DNA synthesis. Chem. Eur. J. 2014, 20, 7566–7570. [Google Scholar] [CrossRef] [PubMed]
- Minetti, C.; Remeta, D.P.; Iden, C.R.; Johnson, F.; Grollman, A.P.; Breslauer, K.J. Impact of thymine glycol damage on DNA duplex energetics: Correlations with lesion-induced biochemical and structural consequences. Biopolymers 2015, 103, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Cai, A.; Wilson, K.A.; Patnaik, S.; Wetmore, S.D.; Cho, B.P. DNA base sequence effects on bulky lesion-induced conformational heterogeneity during DNA replication. Nucleic Acids Res. 2018, 46, 6356–6370. [Google Scholar] [CrossRef] [PubMed]
- Malina, J.; Brabec, V. Probing the thermodynamics of incorporation of N6-methyl-dATP opposite an abasic site, dCMP, and dTMP during simulated DNA synthesis by differential scanning calorimetry. ChemSelect 2018, 3, 13076–13080. [Google Scholar]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef] [Green Version]
- Prakash, S.; Johnson, R.E.; Prakash, L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005, 74, 317–353. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.M.; Grindley, N.D.F.; Joyce, C.M. Interaction of DNA polymerase I (Klenow fragment) with the single-stranded template beyond the site of synthesis. Biochemistry 2003, 42, 2373–2385. [Google Scholar] [CrossRef] [PubMed]
- Lam, W.C.; Van der Schans, E.J.C.; Sowers, L.C.; Millar, D.P. Interaction of DNA polymerase I (Klenow fragment) with DNA substrates containing extrahelical bases: Implications for proofreading of frameshift errors during DNA synthesis. Biochemistry 1999, 38, 2661–2668. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.H.; Suzuki, M.; Adman, E.; Shinkai, A.; Loeb, L.A. Prokaryotic DNA polymerase I: Evolution, structure, and "base flipping" mechanism for nucleotide selection. J. Mol. Biol. 2001, 308, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Villani, G.; Le Gac, N.T.; Wasungu, L.; Burnouf, D.; Fuchs, R.P.; Boehmer, P.E. Effect of manganese on in vitro replication of damaged DNA catalyzed by the herpes simplex virus type-1 DNA polymerase. Nucl. Acids. Res. 2002, 30, 3323–3332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, K.A. Conformational coupling in DNA-polymerase fidelity. Annu. Rev. Biochem. 1993, 62, 685–713. [Google Scholar] [CrossRef]
- Brabec, V.; Reedijk, J.; Leng, M. Sequence-dependent distortions induced in DNA by monofunctional platinum(II) binding. Biochemistry 1992, 31, 12397–12402. [Google Scholar] [CrossRef]
- Asmari, M.; Ratih, R.; Alhazmi, H.A.; El Deeb, S. Thermophoresis for characterizing biomolecular interaction. Methods 2018, 146, 107–119. [Google Scholar] [CrossRef]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef]
- Seidel, S.A.I.; Dijkman, P.M.; Lea, W.A.; van den Bogaart, G.; Jerabek-Willemsen, M.; Lazic, A.; Joseph, J.S.; Srinivasan, P.; Baaske, P.; Simeonov, A.; et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2013, 59, 301–3015. [Google Scholar] [CrossRef] [PubMed]
- Kasparkova, J.; Novakova, O.; Marini, V.; Najajreh, Y.; Gibson, D.; Perez, J.-M.; Brabec, V. Activation of trans geometry in bifunctional mononuclear platinum complexes by a piperidine ligand: Mechanistic studies on antitumor action. J. Biol. Chem. 2003, 278, 47516–47525. [Google Scholar] [CrossRef] [PubMed]
- Novakova, O.; Chen, H.; Vrana, O.; Rodger, A.; Sadler, P.J.; Brabec, V. DNA interactions of monofunctional organometallic ruthenium(II) antitumor complexes in cell-free media. Biochemistry 2003, 42, 11544–11554. [Google Scholar] [CrossRef] [PubMed]
| Control Duplexes | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GTT(15) n = A | 367 | 1.090 | 29.4 | 11.38 µM | 38.9 nM |
| GTT(15) n+1 n = A | 377 | 1.104 | 34.3 | 1.66 µM | 4.6 nM |
| Duplexes containing HdU lesions | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GXT(15) n = A | 354 (–13) | 1.046 (–0.044) | 29.9 (0.5) | 9.12 µM | 37.8 nM |
| GXT(15) n+1 n = A | 362 (–15) | 1.052 (–0.052) | 35.3 (1.0) | 1.15 µM | 3.9 nM |
| Duplexes containing HMdU lesions | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GXT(15) n = A | 314 (–53) | 0.917 (–0.173) | 29.3 (–0.1) | 11.44 µM | 86.8 nM |
| GXT(15) n+1 n = A | 330 (–47) | 0.955 (–0.149) | 33.2 (–1.1) | 2.53 µM | 14.9 nM |
| Control Duplexes | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GTT(15) n = G | 290 | 0.851 | 25.6 | 48.22 µM | 528 nM |
| GTT(15) n+1 n = G | 308 | 0.905 | 26.9 | 29.54 µM | 231 nM |
| Duplexes containing HdU lesions | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GXT(15) n = G | 271 (–19) | 0.782 (–0.069) | 28.4 (2.8) | 16.64 µM | 233 nM |
| GXT(15) n+1 n = G | 322 (14) | 0.950 (0.045) | 27.7 (0.8) | 21.74 µM | 143 nM |
| Duplexes containing HMdU lesions | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GXT(15) n = G | 269 (–21) | 0.780 (–0.071) | 26.5 (0.9) | 34.03 µM | 522 nM |
| GXT(15) n+1 n = G | 272 (–36) | 0.790 (–0.115) | 27.3 (0.4) | 25.65 µM | 378 nM |
| Control Duplexes | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GTT(15) n = C | 311 | 0.923 | 25.0 | 60.69 µM | 484 nM |
| GTT(15) n+1 n = C | 303 | 0.899 | 24.4 | 78.29 µM | 687 nM |
| Duplexes containing HdU lesions | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GXT(15) n = C | 314 (3) | 0.925 (0.002) | 27.4 (2.4) | 24.22 µM | 173 nM |
| GXT(15) n+1 n = C | 311 (8) | 0.916 (0.017) | 26.8 (2.4) | 30.82 µM | 244 nM |
| Duplexes containing HMdU lesions | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GXT(15) n = C | 299 (–12) | 0.880 (–0.043) | 26.5 (1.5) | 34.45 µM | 313 nM |
| GXT(15) n+1 n = C | 304 (1) | 0.899 (0) | 25.3 (0.9) | 55.54 µM | 509 nM |
| Control Duplexes | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GTT(15) n = T | 290 | 0.858 | 24.3 | 81.14 µM | 851 nM |
| GTT(15) n+1 n = T | 293 | 0.865 | 24.2 | 82.57 µM | 798 nM |
| Duplexes containing HdU lesions | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GXT(15) n = T | 276 (–14) | 0.806 (–0.052) | 26.3 (2.0) | 36.83 µM | 498 nM |
| GXT(15) n+1 n = T | 303 (10) | 0.897 (0.032) | 24.8 (0.6) | 65.54 µM | 579 nM |
| Duplexes containing HMdU lesions | ΔH (kJ mol−1)2 | ΔS (kJ K−1 mol−1)2 | ΔG0310 (kJ mol−1)2 | Kd[310]3 | Kd[298]3 |
| GXT(15) n = T | 264 (–26) | 0.770 (–0.088) | 25.3 (1.0) | 55.42 µM | 873 nM |
| GXT(15) n+1 n = T | 289 (–4) | 0.855 (–0.010) | 23.5 (–0.7) | 109.91 µM | 1.18 µM |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hrabina, O.; Brabec, V.; Novakova, O. Translesion DNA Synthesis Across Lesions Induced by Oxidative Products of Pyrimidines: An Insight into the Mechanism by Microscale Thermophoresis. Int. J. Mol. Sci. 2019, 20, 5012. https://doi.org/10.3390/ijms20205012
Hrabina O, Brabec V, Novakova O. Translesion DNA Synthesis Across Lesions Induced by Oxidative Products of Pyrimidines: An Insight into the Mechanism by Microscale Thermophoresis. International Journal of Molecular Sciences. 2019; 20(20):5012. https://doi.org/10.3390/ijms20205012
Chicago/Turabian StyleHrabina, Ondrej, Viktor Brabec, and Olga Novakova. 2019. "Translesion DNA Synthesis Across Lesions Induced by Oxidative Products of Pyrimidines: An Insight into the Mechanism by Microscale Thermophoresis" International Journal of Molecular Sciences 20, no. 20: 5012. https://doi.org/10.3390/ijms20205012
APA StyleHrabina, O., Brabec, V., & Novakova, O. (2019). Translesion DNA Synthesis Across Lesions Induced by Oxidative Products of Pyrimidines: An Insight into the Mechanism by Microscale Thermophoresis. International Journal of Molecular Sciences, 20(20), 5012. https://doi.org/10.3390/ijms20205012







