Comprehensive Analysis and Functional Studies of WRKY Transcription Factors in Nelumbo nucifera
Abstract
:1. Introduction
2. Results
2.1. Identification of WRKY Family Members
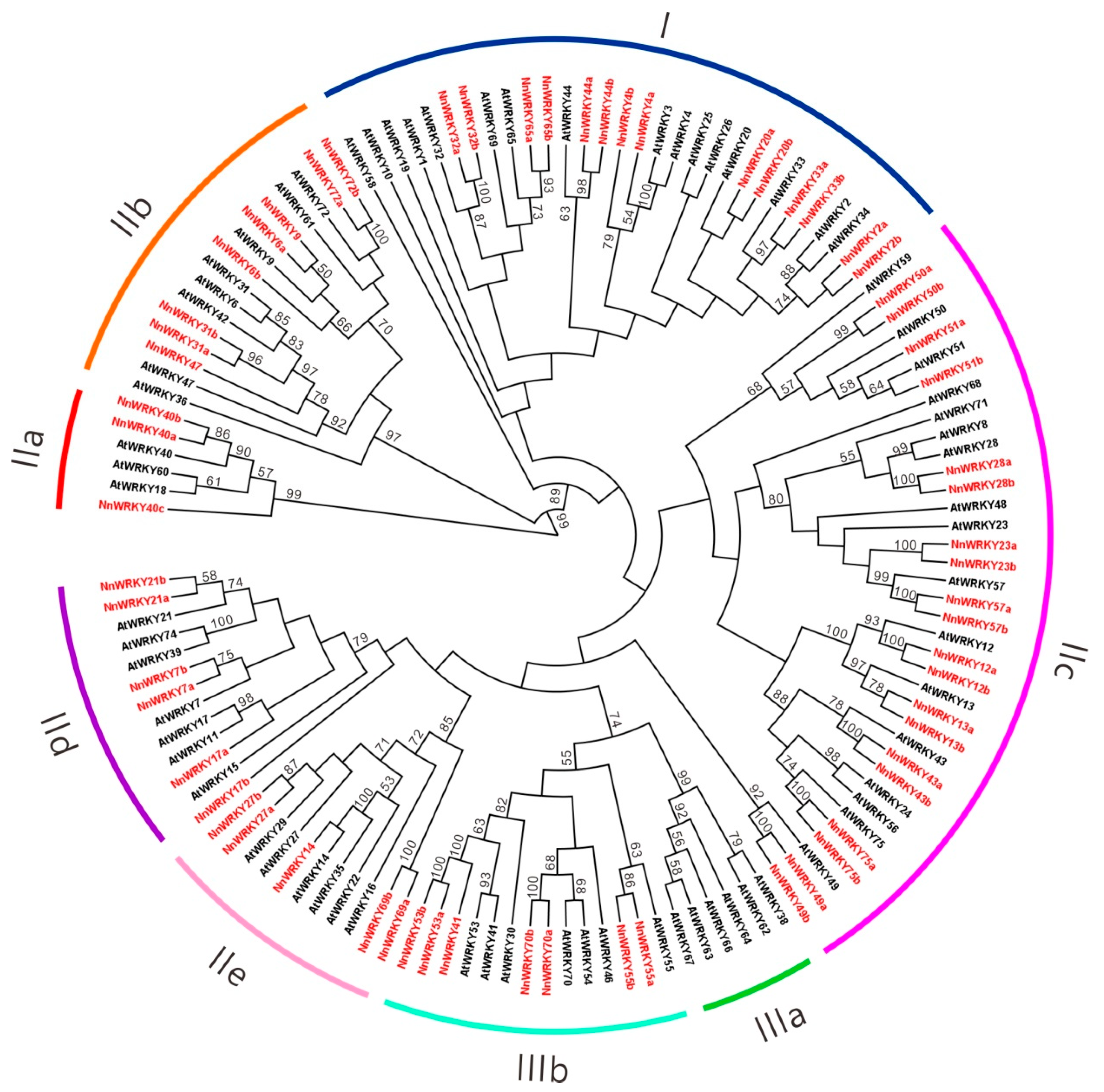
2.2. Phylogenetic Analysis of NnWRKY Family Genes
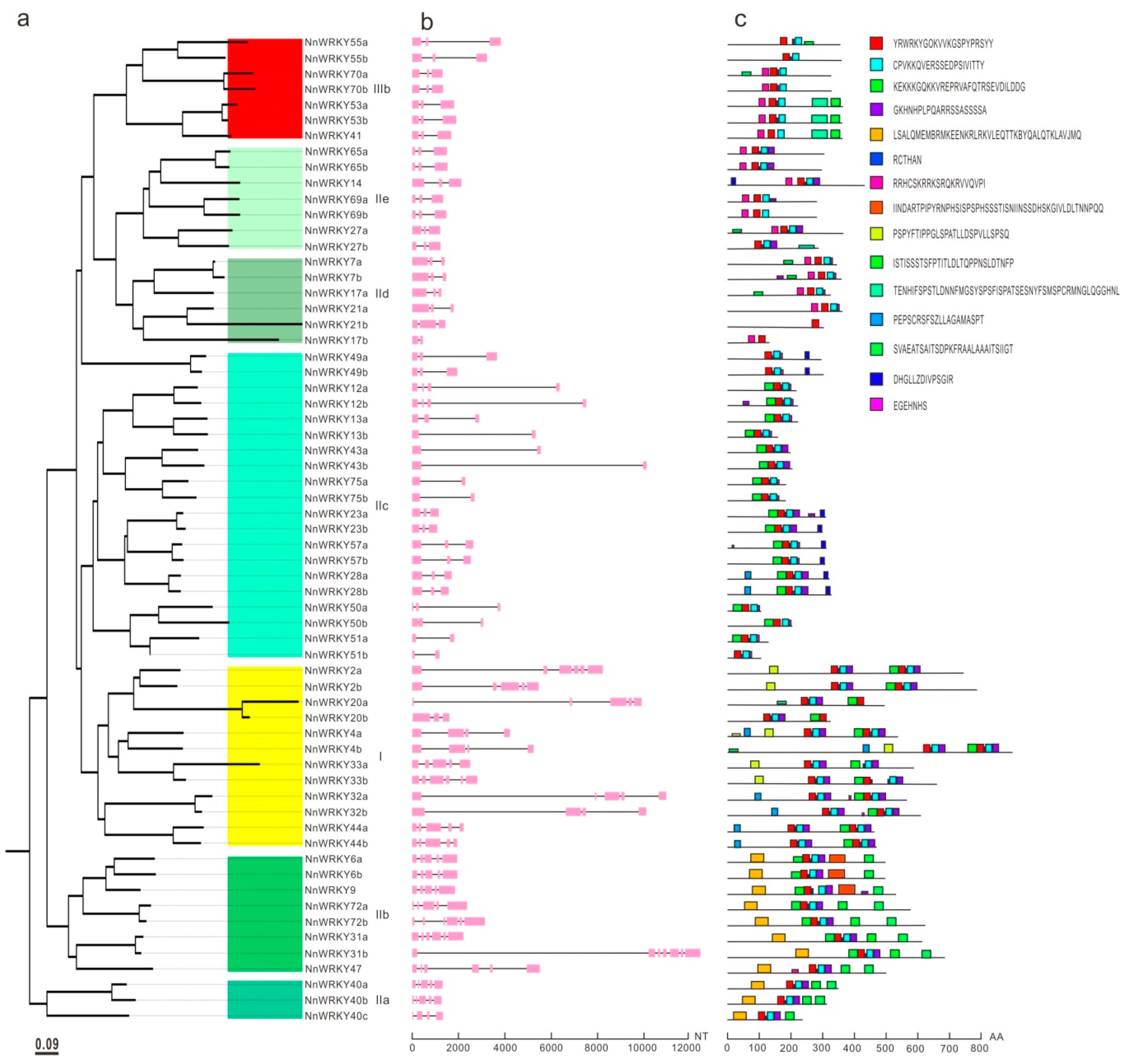
2.3. NnWRKY Gene Structures and Conserved Motifs
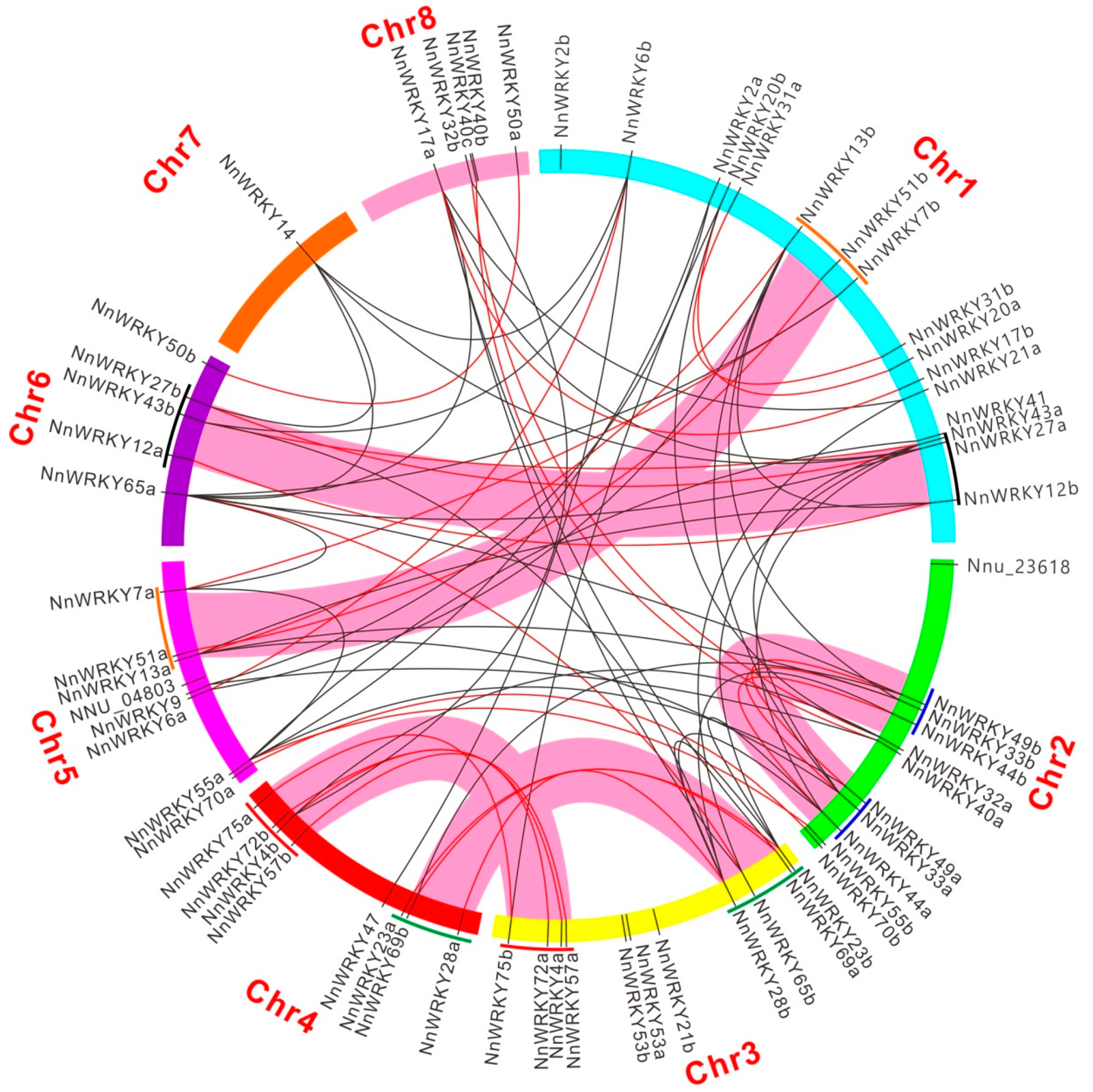
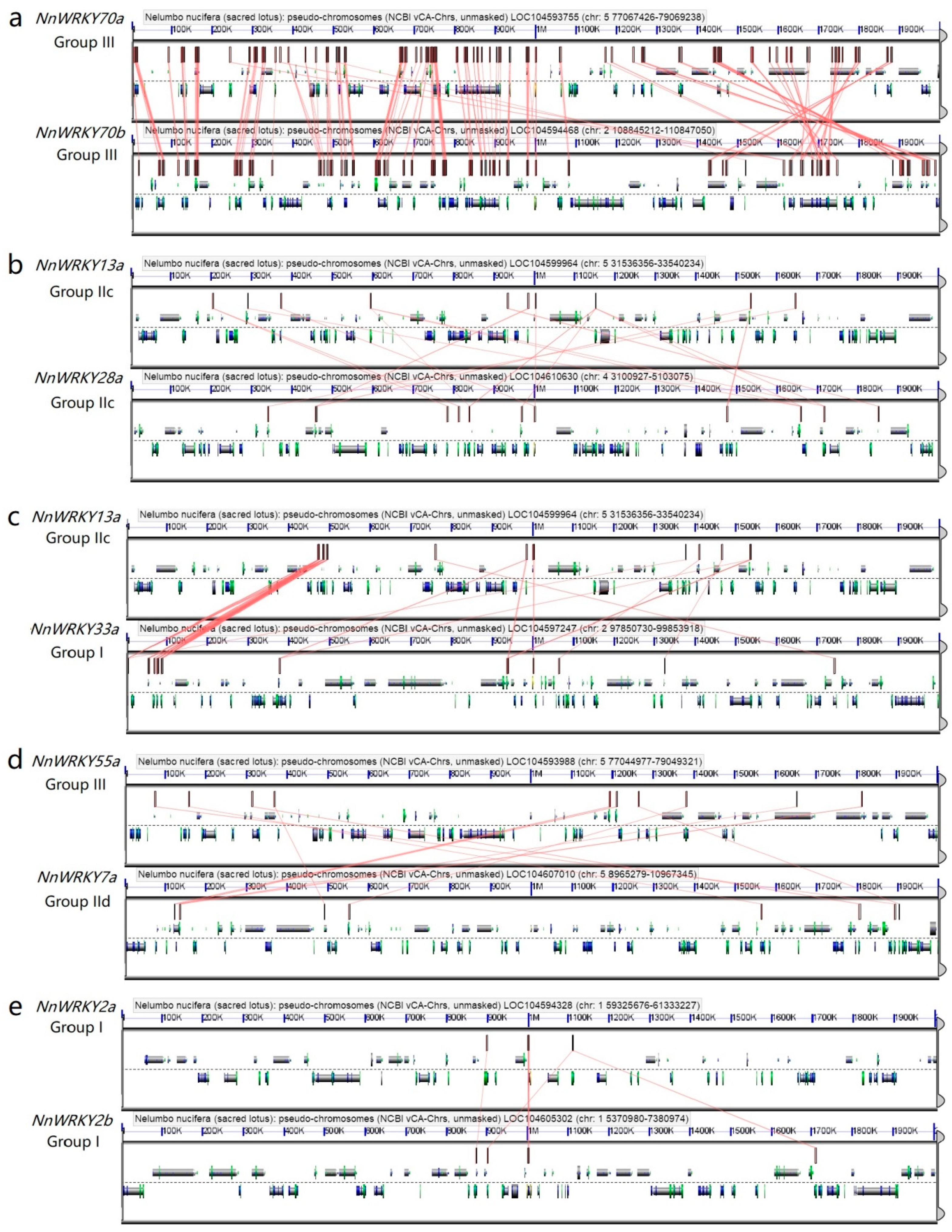
2.4. Synteny Analysis of NnWRKY Genes
2.5. Spatial Expression Patterns of NnWRKYs
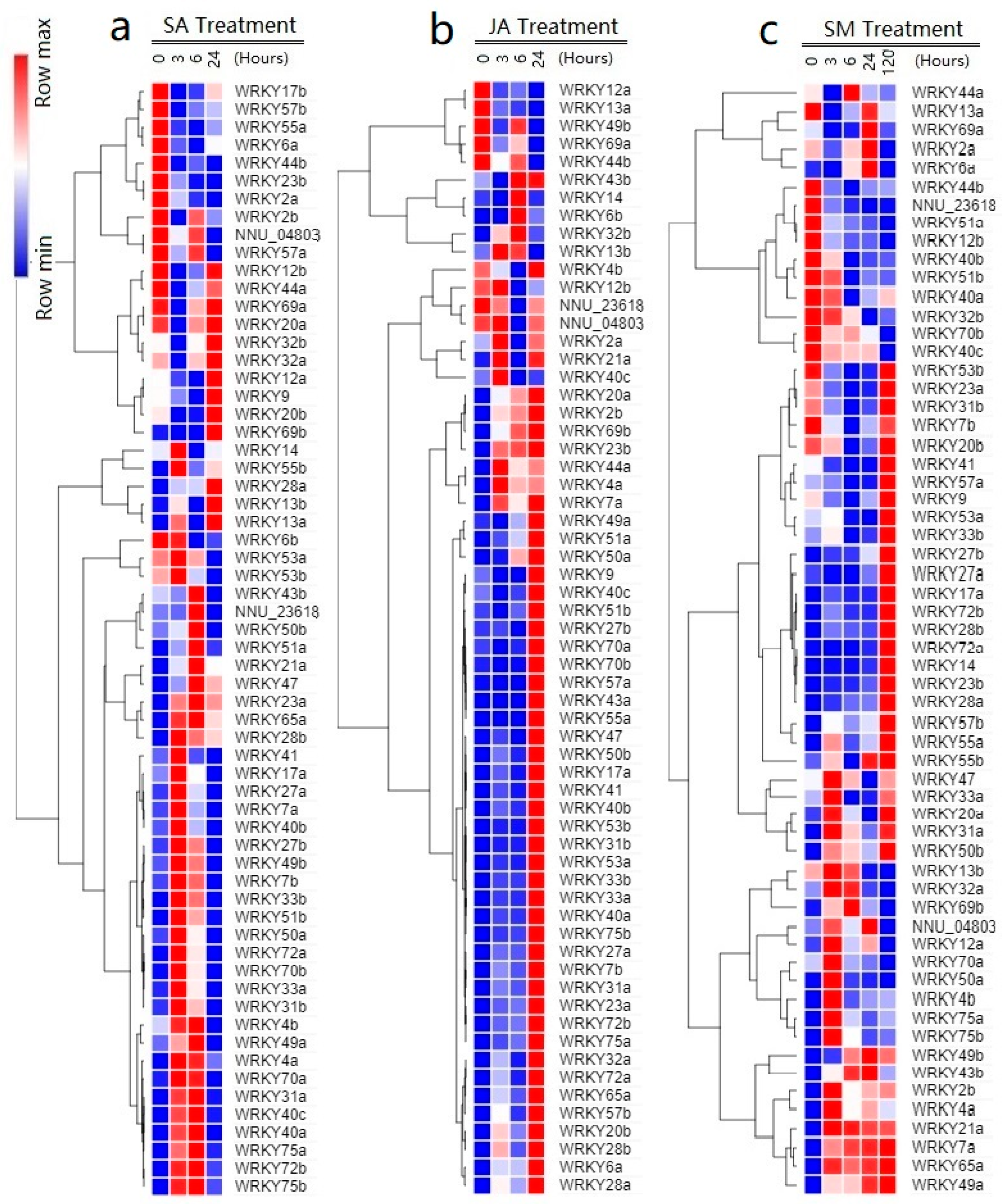
2.6. Responses of NnWRKYs to SA, JA, and Submergence Treatments
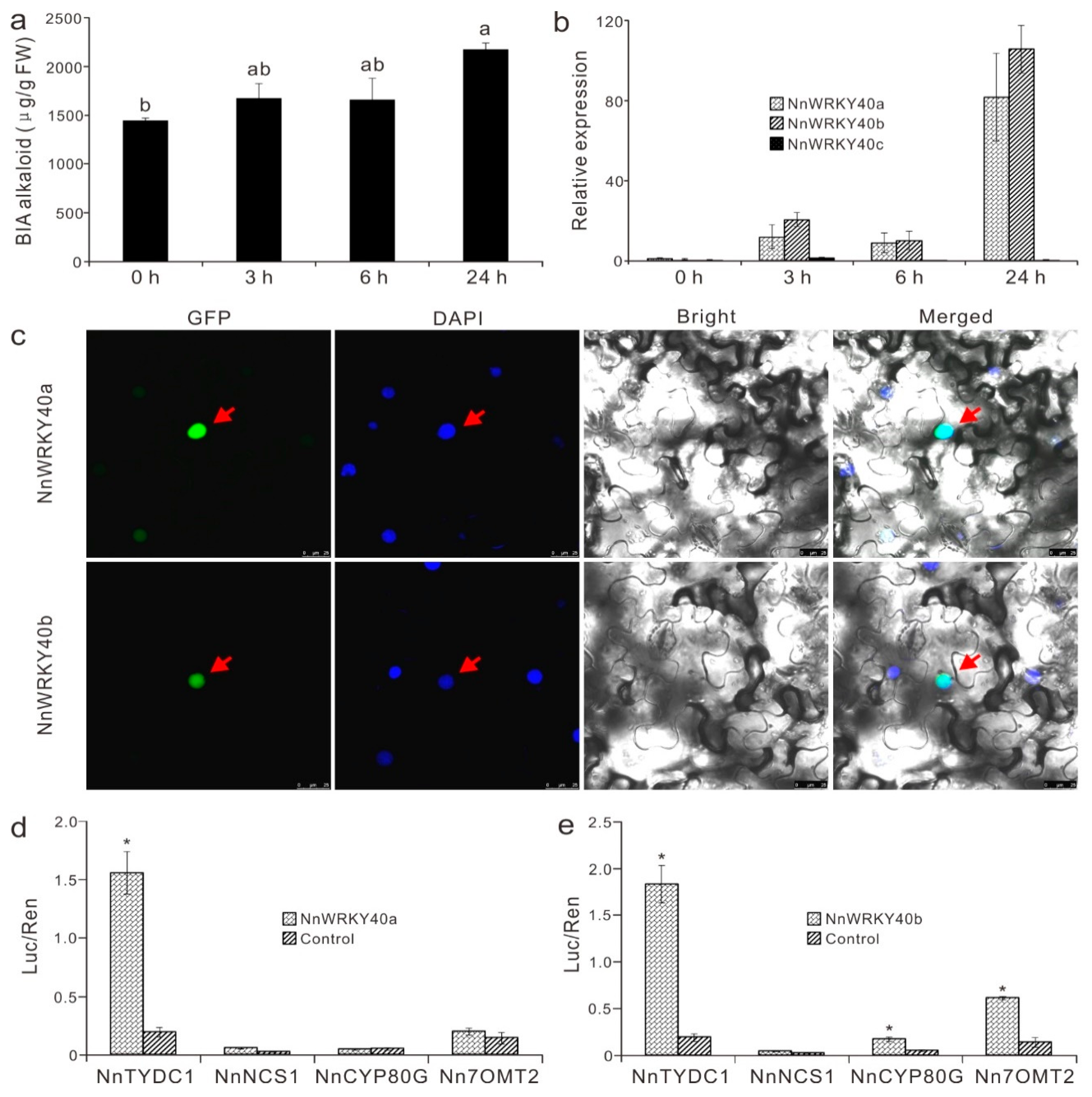
2.7. NnWRKY40a and NnWRKY40b are Involved in Lotus Benzylisoquinoline Alkaloid Biosynthesis
3. Discussion
3.1. Characteristics of Lotus WRKY Gene Family
3.2. The Evolution Pattern of NnWRKY Genes
3.3. Roles of NnWRKYs on Responses to Biotic and Abiotic Stresses
3.4. Role of NnWRKYs in Regulation of Lotus Alkaloid Biosynthesis
4. Materials and Methods
4.1. Plant Materials, Treatments, and Alkaloid Quantification
4.2. Mining of WRKY Genes
4.3. Phylogenetic Analysis
4.4. Analysis of WRKY Gene Structures and Conserved Domains
4.5. Synteny Analysis
4.6. RNA Extraction, Real-Time PCR and RNA Sequencing Analysis
4.7. Dual-Luciferase Report Gene Assay
4.8. Subcellular Localization Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.L.; Guo, Z.J.; Wang, H.H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yue, C.; Chen, D.; Zheng, Y.; Zhang, Q.; Yang, J.; Ye, N. Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes Genom. 2019, 41, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Maeo, K.; Hayashi, S.; Kojima-Suzuki, H.; Morikami, A.; Nakamura, D. Role of conserved residues of the WRKY domain in DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem. 2001, 65, 2428–2436. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1. [Google Scholar]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Agarwal, P.; Reddy, M.P.; Chikara, J. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef]
- Ruston, P.J.; Torresm, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [CrossRef]
- Machens, F.; Becker, M.; Umrath, F.; Hehl, R. Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol. Biol. 2014, 84, 371–385. [Google Scholar] [CrossRef]
- Kanofsky, K.; Bahlmann, A.K.; Hehl, R.; Dong, D.X. Combinatorial requirement of W- and WT-boxes in microbe-associated molecular pattern-responsive synthetic promoters. Plant Cell Rep. 2017, 36, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Van Verk, M.C.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J. A Novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef] [PubMed]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.C.; Cai, H.Y.; Qin, Y.; Mullis, A.; Lin, Z.G.; Zhang, L.S. The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA binding protein, SPF1, that recognizes SP8 sequences in the 5′upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Li, J.; Besseau, S.; Törönen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. N. Phytol. 2013, 200, 457–472. [Google Scholar] [CrossRef]
- Li, J.; Zhong, R.; Palva, E.T. WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS ONE 2017, 12, e0183731. [Google Scholar] [CrossRef]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ahammed, G.J.; Yu, J.; Yao, Z.; Ruan, M.; Ye, Q.; Li, Z.; Wang, R.; Feng, K.; Zhou, G.; et al. Putative WRKYs associated with regulation of fruit ripening revealed by detailed expression analysis of the WRKY gene family in pepper. Sci. Rep. 2016, 7, 43498. [Google Scholar] [CrossRef] [PubMed]
- Kato, N.; Dubouzet, E.; Kokabu, Y.; Yoshida, S.; Taniguchi, Y.; Dubouzet, J.G.; Yazaki, K.; Sato, F. Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol. 2007, 48, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Apuya, N.R.; Park, J.H.; Zhang, L.; Ahyow, M.; Davidow, P.; Van Fleet, J.; Rarang, J.C.; Hippley, M.; Johnson, T.W.; Yoo, H.D.; et al. Enhancement of alkaloid production in opium and California poppy by transactivation using heterologous regulatory factors. Plant Biotechnol. J. 2008, 6, 160–175. [Google Scholar] [CrossRef]
- Miao, Y.; Zentgraf, U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 2007, 19, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Deng, C.; Zhang, Y.; Cheng, Y.; Huo, Q.; Xue, L. The WRKY transcription factor genes in eggplant (Solanum melongena L.) and Turkey Berry (Solanum torvum Sw.). Int. J. Mol. Sci. 2015, 16, 7608–7626. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.X.; Chen, C.H.; Chen, Z.X. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Bencke-Malato, M.; Cabreira, C.; Wiebke-Strohm, B.; Bücker-Neto, L.; Mancini, E.; Osorio, M.B.; Homrich, M.S.; Turchetto-Zolet, A.C.; De Carvalho, M.C.; Stolf, R.; et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 2014, 14, 236. [Google Scholar] [CrossRef]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Dou, L.; Zhang, X.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Yu, S. Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics 2014, 289, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Mukherjee, D.; Maji, A.K.; Rai, S.; Heinrich, M. The sacred lotus (Nelumbo nucifera)–Phytochemical and therapeutic profile. J. Pharm. Pharmacol. 2009, 61, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.R.; Gautam, L.M.S.; Adhikari, D.; Karki, R. A comprehensive review on chemical profiling of Nelumbo nucifera: Potential for drug development. Phytother. Res. 2017, 31, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Zhu, L.; Fang, T.; Vimolmangkang, S.; Yang, D.; Ogutu, C.; Liu, Y.; Han, Y. Analysis of isoquinoline alkaloid composition and wound induced variation in Nelumbo using HPLC-MS/MS. J. Agric. Food Chem. 2016, 64, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Zhao, L.; Fang, T.; Xiong, Y.; Ogutu, C.; Yang, D.; Vimolmangkang, S.; Liu, Y.; Han, Y. Investigation of benzylisoquinoline alkaloid biosynthetic pathway and its transcriptional regulation in lotus. Hortic. Res. 2018, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Gui, S.; Peng, J.; Wang, X.; Wu, Z.; Cao, R.; Salse, J.; Zhang, H.; Zhu, Z.; Xia, Q.; Quan, Z.; et al. Improving Nelumbo nucifera genome assemblies using high-resolution genetic maps and BioNano genome mapping reveals ancient chromosome rearrangements. Plant J. 2018, 94, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Lyons, E.; Freeling, M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008, 53, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 2011, 12, 471. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genomics 2018, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Guo, R.; Xu, X.; Gao, M.; Li, X.; Song, J.; Zheng, Y.; Wang, X. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.; Li, Y.; Bai, Y.; Li, M.; Cheng, L. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 2016, 103, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Mangelsen, E.; Kilian, J.; Berendzen, K.W.; Kolukisaoglu, U.H.; Harter, K.; Jansson, C.; Wanke, D. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 2008, 9, 194–211. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef]
- Ming, R.; VanBuren, R.; Liu, Y.; Yang, M.; Han, Y.; Li, L.T.; Zhang, Q.; Kim, M.J.; Schatz, M.C.; Campbell, M.; et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 2013, 14, R41. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, G.; Liu, Y.; Sun, F.; Shi, C.; Liu, X.; Peng, J.; Chen, W.; Huang, X.; Cheng, S.; et al. The sacred lotus genome provides insights into the evolution of flowering plants. Plant J. 2013, 76, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wofe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef] [PubMed]
- De Smet, R.; Adams, K.L.; Vandepoele, K.; Van Montagu, M.C.; Maere, S.; Van de Peer, Y. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. USA 2013, 110, 2898–2903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Xie, D. Jasmonate in plant defence: Sentinel or double agent? Plant Biotechnol. J. 2015, 13, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, C.M.J.; van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Van Wees, S.C.M.; De Swart, E.A.M.; van Pelt, J.A.; van Loon, L.C.; Pieterse, C.M.J. Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 8711–8716. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.J.; Kenton, P.; Atzorn, R.; Miersch, O.; Wasternack, C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 2006, 140, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef]
- Mishra, S.; Triptahi, V.; Singh, S.; Phukan, U.J.; Gupta, M.M.; Shanker, K.; Shukla, R.K. Wound induced transcriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS ONE 2013, 8, e52784. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W. Noble WS MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]

| No | Lotus WRKY Pairs | NA Identity | AA Identity |
|---|---|---|---|
| 1 | NnWRKY2a and NnWRKY2b | 54.8% | 41.6% |
| 2 | NnWRKY4a and NnWRKY4b * | 47.7% | 44.8% |
| 3 | NnWRKY6a and NnWRKY6b * | 73.0% | 60.1% |
| 4 | NnWRKY7a and NnWRKY7b * | 80.4% | 79.5% |
| 5 | NnWRKY12a and NnWRKY12b * | 81.6% | 78.5% |
| 6 | NnWRKY13a and NnWRKY13b * | 60.4% | 53.9% |
| 7 | NnWRKY17a and NnWRKY17b * | 56.9% | 60.1% |
| 8 | NnWRKY20a and NnWRKY20b * | 55.3% | 50.9% |
| 9 | NnWRKY21a and NnWRKY21b | 67.4% | 57.5% |
| 10 | NnWRKY23a and NnWRKY23b * | 78.7% | 76.3% |
| 11 | NnWRKY27a and NnWRKY27b * | 65.4% | 62.4% |
| 12 | NnWRKY28a and NnWRKY28b * | 80.2% | 77.2% |
| 13 | NnWRKY31a and NnWRKY31b * | 76.2% | 76.6% |
| 14 | NnWRKY32a and NnWRKY32b * | 73.9% | 70.0% |
| 15 | NnWRKY33a and NnWRKY33b * | 69.8% | 64.3% |
| 16 | NnWRKY43a and NnWRKY43b * | 78.6% | 75.2% |
| 17 | NnWRKY44a and NnWRKY44b * | 82.0% | 75.6% |
| 18 | NnWRKY49a and NnWRKY49b * | 80.0% | 74.8% |
| 19 | NnWRKY50a and NnWRKY50b * | 63.6% | 64.2% |
| 20 | NnWRKY51a and NnWRKY51b * | 70.5% | 72.5% |
| 21 | NnWRKY53a and NnWRKY53b | 90.4% | 86.0% |
| 22 | NnWRKY55a and NnWRKY55b * | 54.4% | 43.2% |
| 23 | NnWRKY57a and NnWRKY57b * | 79.4% | 78.0% |
| 24 | NnWRKY65a and NnWRKY65b * | 78.8% | 72.1% |
| 25 | NnWRKY69a and NnWRKY69b * | 77.6% | 66.9% |
| 26 | NnWRKY70a and NnWRKY70b * | 70.8% | 63.7% |
| 27 | NnWRKY72a and NnWRKY72b * | 77.3% | 73.0% |
| 28 | NnWRKY75a and NnWRKY75b * | 81.9% | 75.4% |
| 29 | NnWRKY40a and NnWRKY40b/c * | 75.6%/45.0% | 73.8%/42.0% |
| 30 | NnWRKY9 and NNU_24697 * | 52.2% | 57.7% |
| Treatments | Positively Regulated WRKYs | Negatively Regulated WRKYs |
|---|---|---|
| SA | 32 NnWRKYs: NnWRKY4aNnWRKY7a NnWRKY7b NnWRKY17a NnWRKY23a NnWRKY27a NnWRKY28a NnWRKY28b NnWRKY31a NnWRKY31b WRKY33a NnWRKY33b NnWRKY40a NnWRKY40b NnWRKY41 NnWRKY43b NnWRKY47 NnWRKY49a NnWRKY49b NnWRKY50a NnWRKY50b NnWRKY51a NnWRKY51b NnWRKY55b NnWRKY65a NnWRKY70a NnWRKY70b NnWRKY72a NnWRKY72b NnWRKY75a NnWRKY75b Nnu_23618 | 6 NnWRKYs: NnWRKY6a NnWRKY12b NnWRKY23b NnWRKY44a NnWRKY55a NnWRKY69a |
| JA | 34 NnWRKYs: NnWRKY2b NnWRKY4a NnWRKY6a NnWRKY7a NnWRKY7b NnWRKY17a NnWRKY23a NnWRKY23b NnWRKY27a NnWRKY27b NnWRKY28a NnWRKY28b NnWRKY31a NnWRKY31b NnWRKY33a NnWRKY33b NnWRKY40a NnWRKY40b NnWRKY41 NnWRKY47 NnWRKY50a NnWRKY50b NnWRKY51a NnWRKY51b NnWRKY53a NnWRKY53b NnWRKY57a NnWRKY57b NnWRKY65a NnWRKY70a NnWRKY70b NnWRKY72a NnWRKY75a NnWRKY75b | 4 NnWRKYs: NnWRKY12a NnWRKY40c NnWRKY49b NnWRKY69a |
| SM | 28 NnWRKYs: NnWRKY4a NnWRKY4b NnWRKY6a NnWRKY7a NnWRKY12a NnWRKY17a NnWRKY21a NnWRKY23b NnWRKY27a NnWRKY27b NnWRKY28a NnWRKY28b NnWRKY31a NnWRKY33a NnWRKY43b NnWRKY49a NnWRKY49b NnWRKY50a NnWRKY50b NnWRKY55a NnWRKY55b NnWRKY57b NnWRKY65a NnWRKY69b NnWRKY72a NnWRKY72b NnWRKY75a NnWRKY75b | 12 NnWRKYs: NnWRKY9 NnWRKY12b NnWRKY23a NnWRKY40a NnWRKY40b NnWRKY41 NnWRKY44b NnWRKY51a NnWRKY51b NnWRKY53a NnWRKY53b Nnu_23618 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Xiong, Y.; Li, Y.; Ye, S.; Yin, Q.; Gao, S.; Yang, D.; Yang, M.; Palva, E.T.; Deng, X. Comprehensive Analysis and Functional Studies of WRKY Transcription Factors in Nelumbo nucifera. Int. J. Mol. Sci. 2019, 20, 5006. https://doi.org/10.3390/ijms20205006
Li J, Xiong Y, Li Y, Ye S, Yin Q, Gao S, Yang D, Yang M, Palva ET, Deng X. Comprehensive Analysis and Functional Studies of WRKY Transcription Factors in Nelumbo nucifera. International Journal of Molecular Sciences. 2019; 20(20):5006. https://doi.org/10.3390/ijms20205006
Chicago/Turabian StyleLi, Jing, Yacen Xiong, Yi Li, Shiqi Ye, Qi Yin, Siqi Gao, Dong Yang, Mei Yang, E. Tapio Palva, and Xianbao Deng. 2019. "Comprehensive Analysis and Functional Studies of WRKY Transcription Factors in Nelumbo nucifera" International Journal of Molecular Sciences 20, no. 20: 5006. https://doi.org/10.3390/ijms20205006





