Targeting of LRRC59 to the Endoplasmic Reticulum and the Inner Nuclear Membrane
Abstract
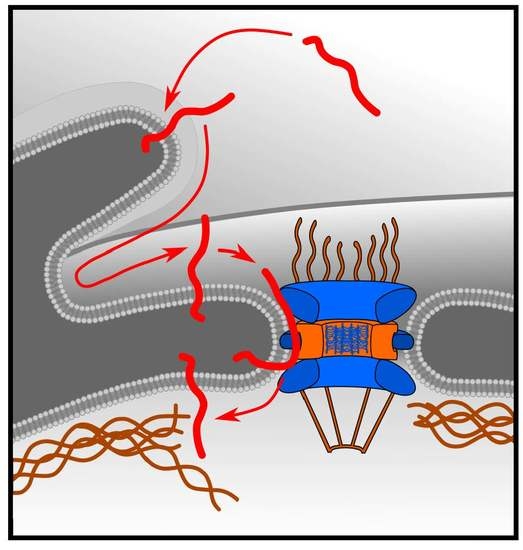
:1. Introduction
2. Results
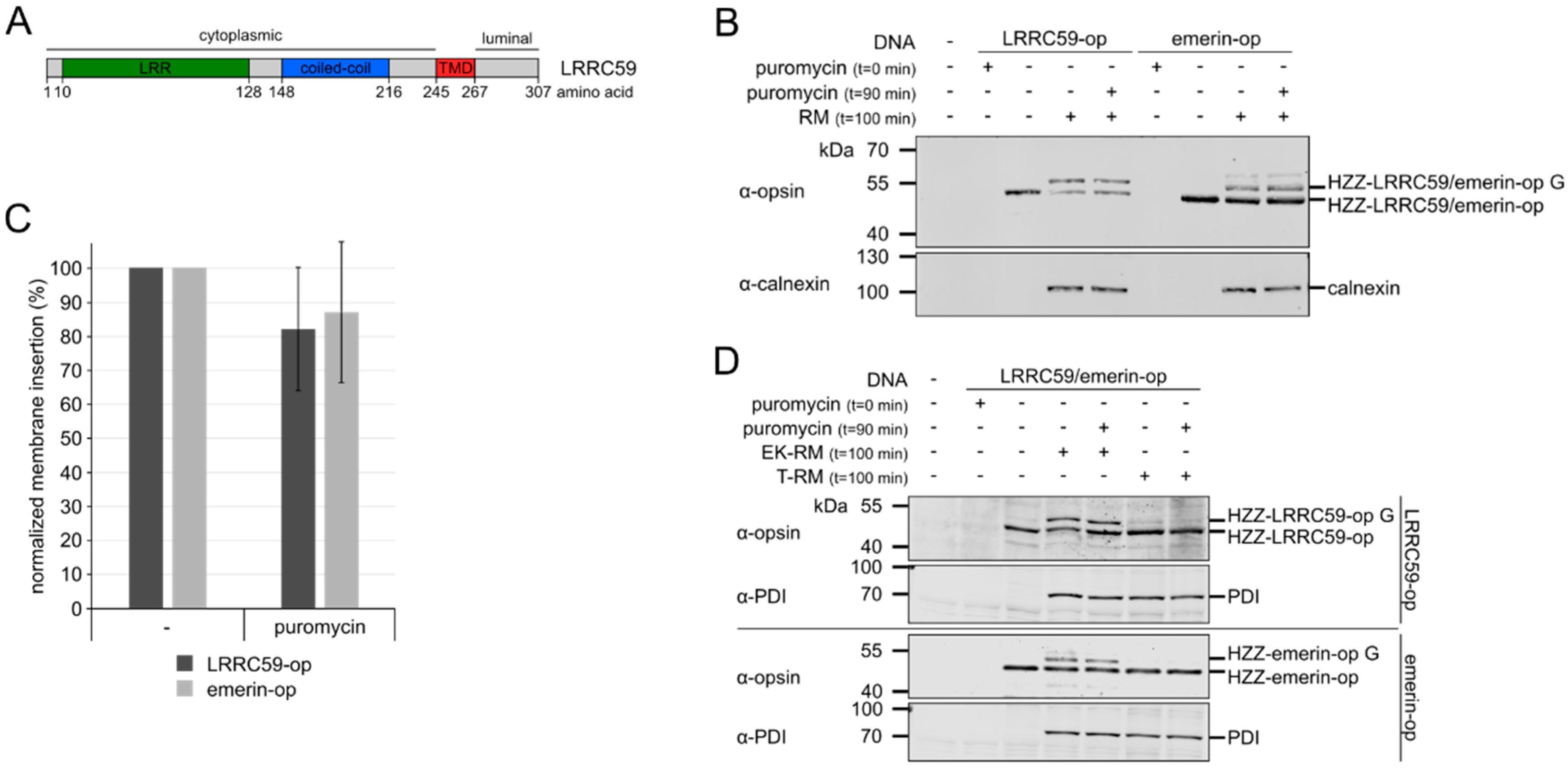
2.1. LRRC59 can be Post-Translationally Inserted into Microsomal Membranes
2.2. Membrane Insertion of LRRC59 Does Not Require the TRC40 System
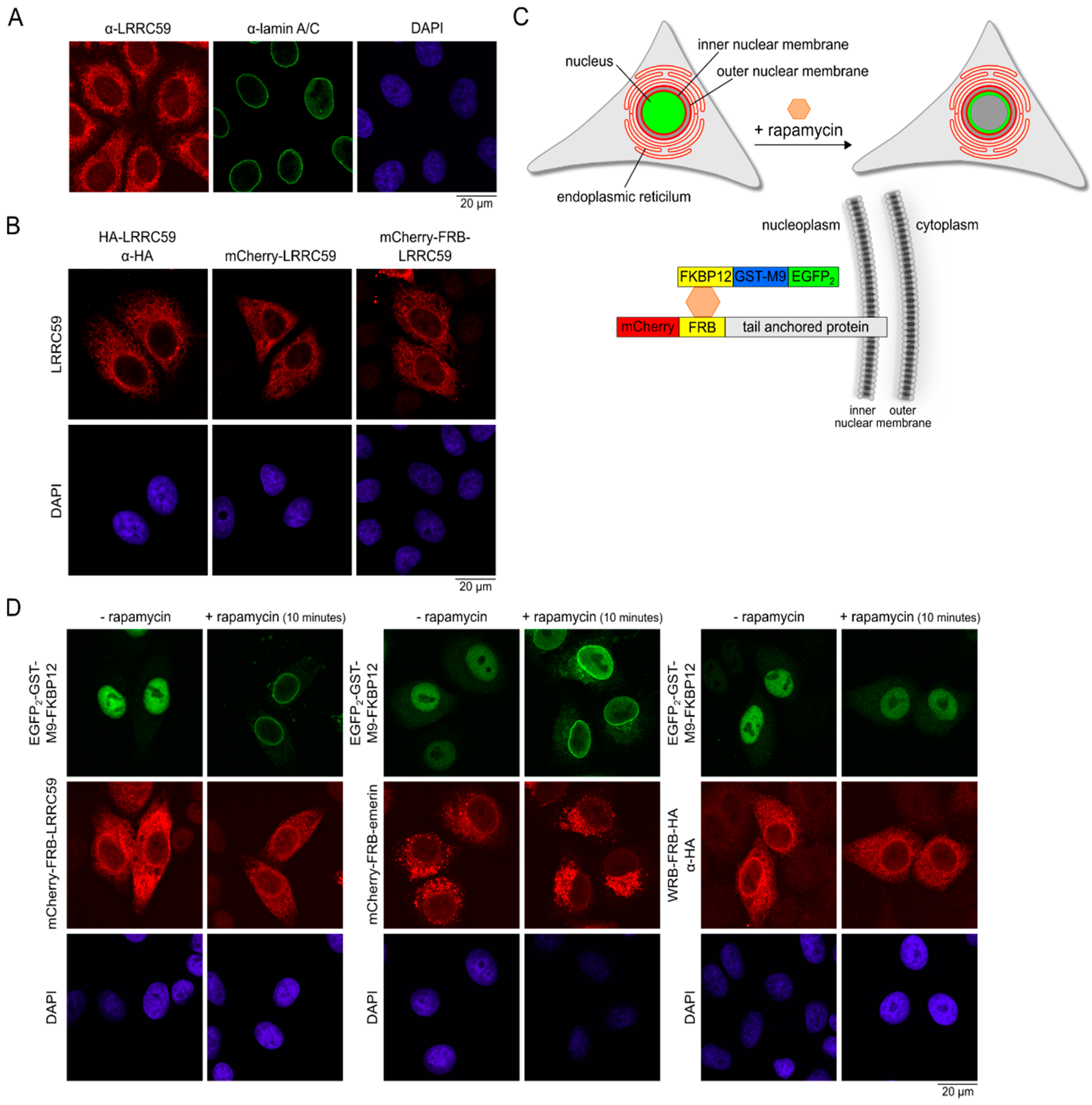
2.3. LRRC59 Localizes to the INM
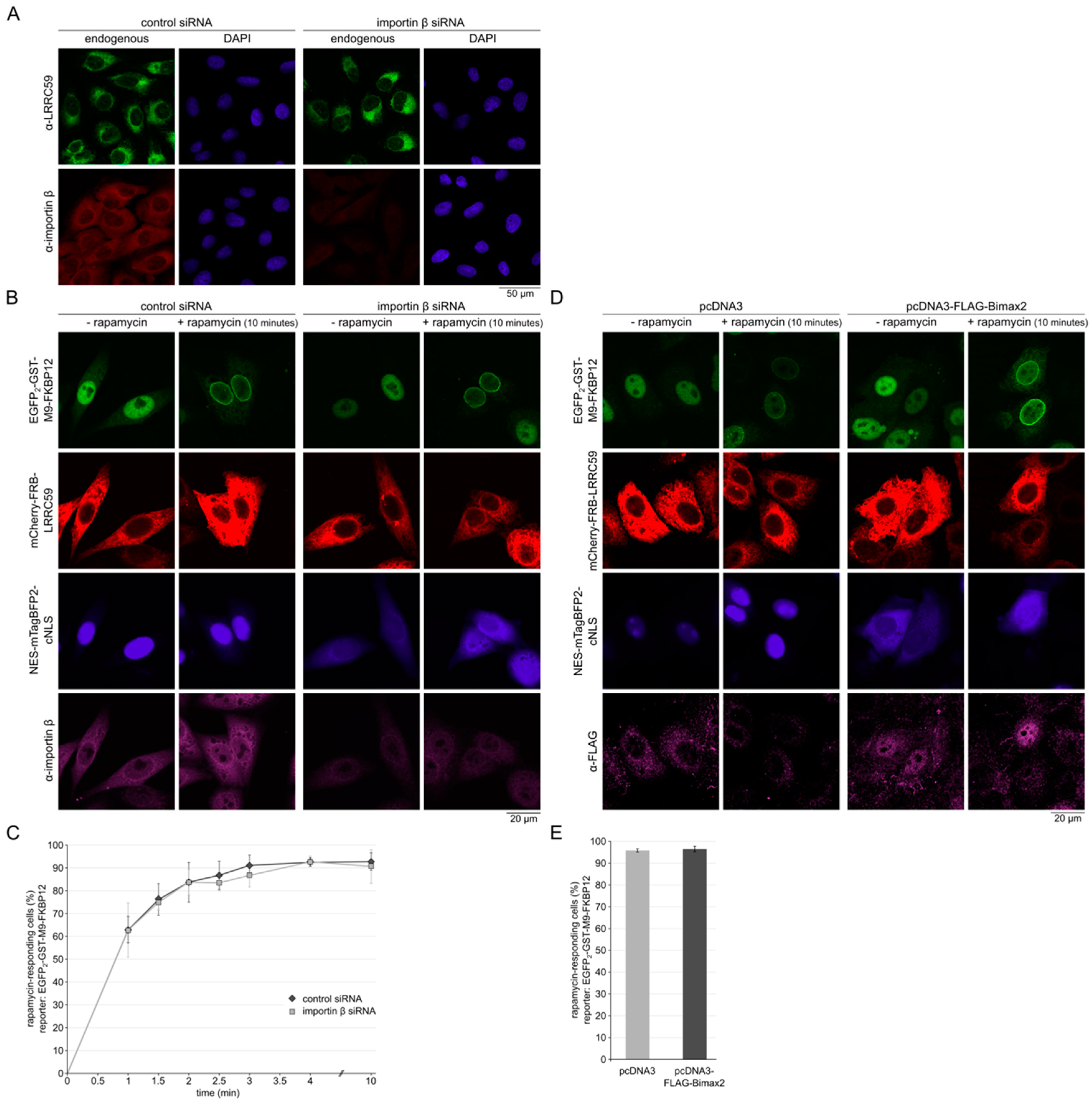
2.4. Importin β Is Not Required for Localization of LRRC59 at the INM
2.5. Nuclear Accumulation of LRRC59 Lacking the TMD
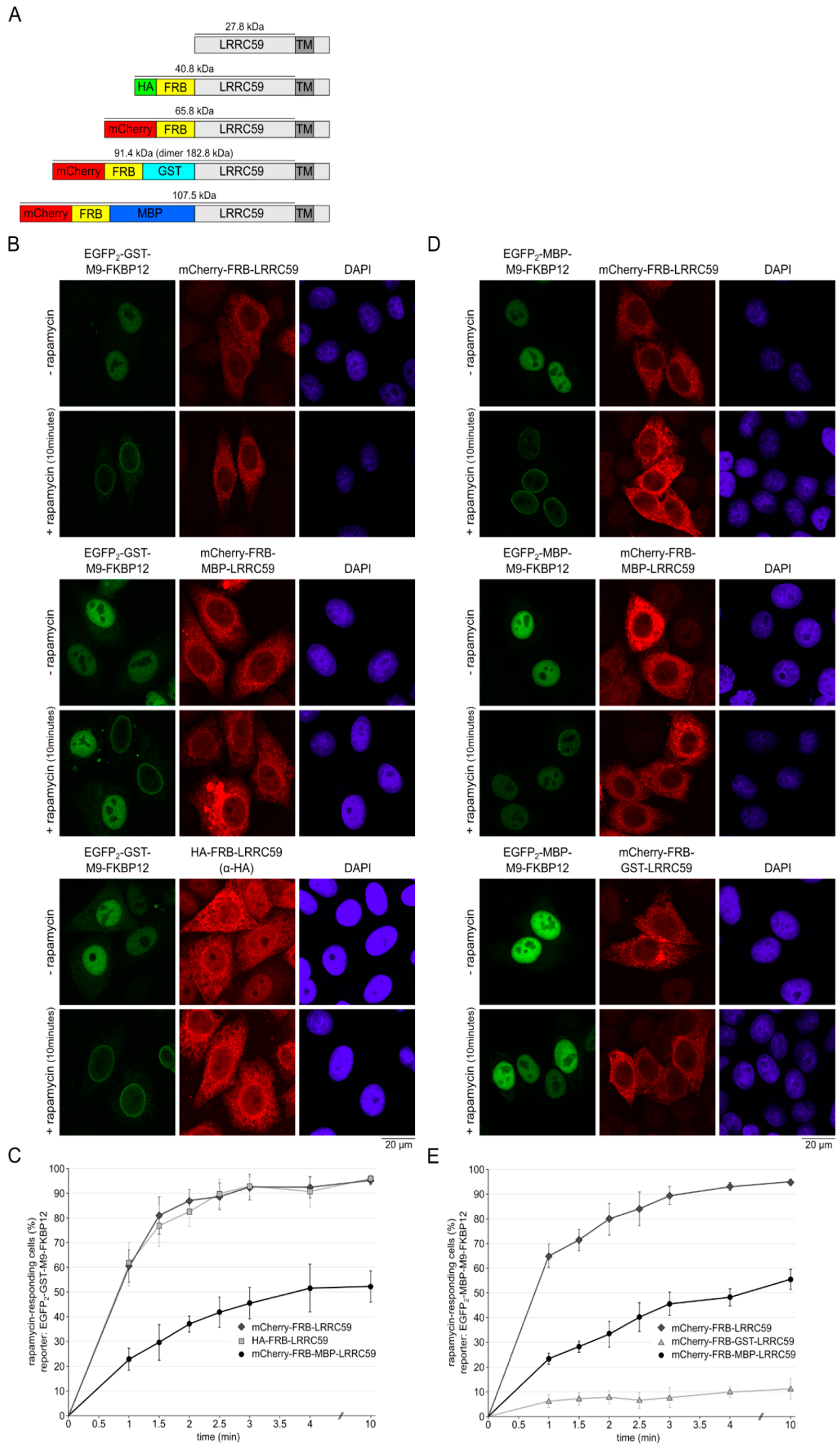
2.6. The Size of the Cytoplasmic Domain Controls INM Localization of LRRC59
3. Discussion
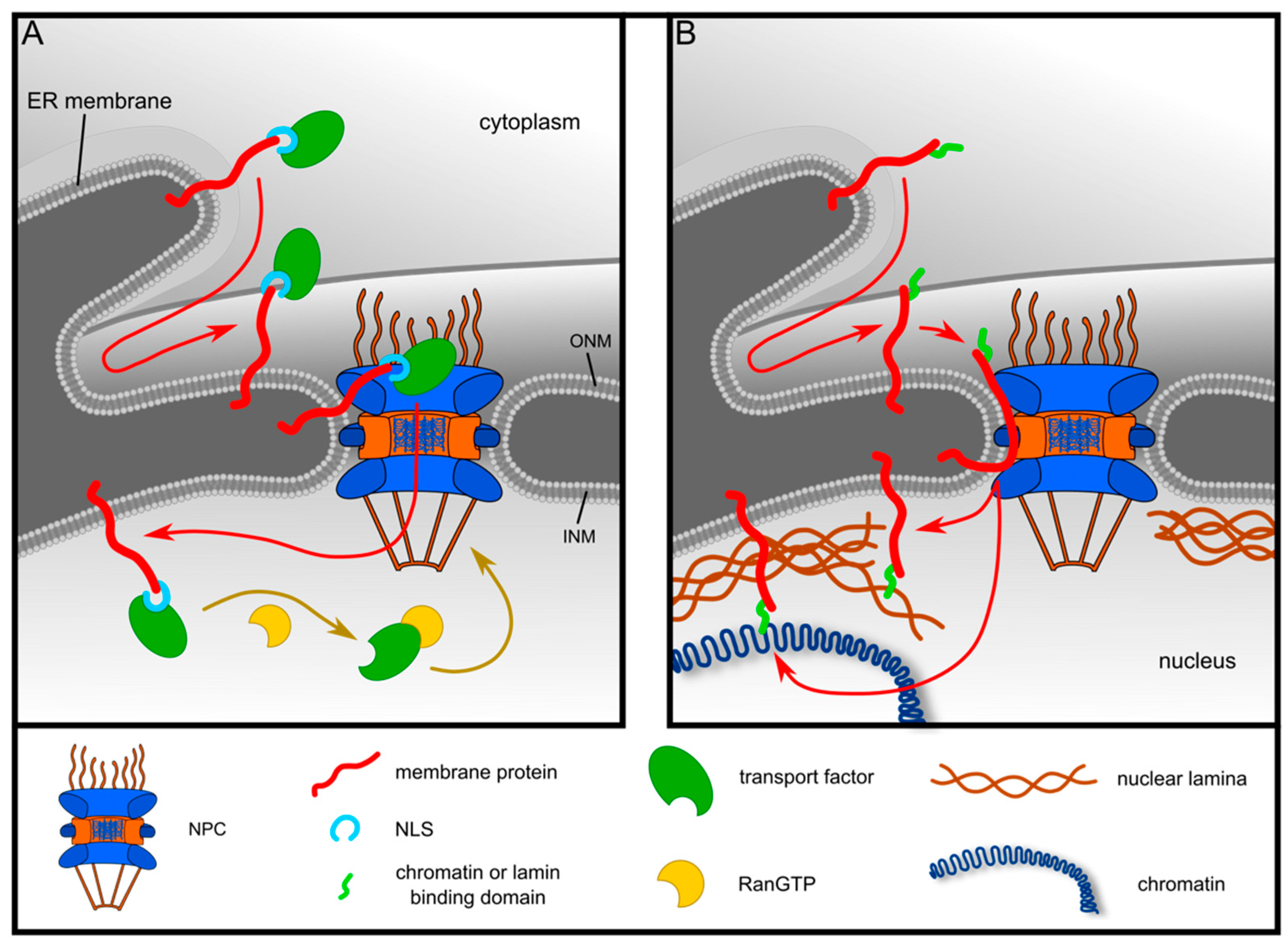
3.1. Post-Translational Targeting of LRRC59 to ER-Membranes
3.2. Targeting of LRRC59 to the INM
4. Materials and Methods
4.1. Plasmids and Constructs
4.2. Cell Culture, Plasmid- and siRNA- Transfection and Immunofluorescence Microscopy
4.3. SDS-PAGE, Ponceau S Staining and Western Blotting
4.4. Microsome Integration Assay
4.5. Rapamycin Induced Dimerization Assay
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| CAML | Ca2+-modulating cyclophilin ligand |
| CIP2A | cancerous inhibitor of PP2A |
| cNLS | classical nuclear localization signal |
| ER | endoplasmic reticulum |
| EK-RM | EDAT and high salt treated rough microsomes |
| FKBP12 | 12 kDa FK506 binding protein |
| FG repeats | phenylalanine-glycine repeats |
| FRB | FKBP rapamycin binding domain |
| INM | inner nuclear membrane |
| LBR | Lamin B receptor |
| LRRC59 | leucine-rich repeat-containing protein 59 |
| NES | nuclear export sequence |
| NLS | nuclear localization signal |
| NPC | nuclear pore complex |
| ONM | outer nuclear membrane |
| PNGase F | Peptide-N-glycosidase F |
| RM | rough microsomes |
| T-RM | trypsin-treated rough microsomes |
| TA | tail-anchored |
| TMD | Transmembrane domain |
| TRC40 | ATPase ASNA1 |
| WRB | tryptophan-rich basic protein |
References
- Korfali, N.; Wilkie, G.S.; Swanson, S.K.; Srsen, V.; de las Heras, J.; Batrakou, D.G.; Malik, P.; Zuleger, N.; Kerr, A.R.W.; Florens, L.; et al. The nuclear envelope proteome differs notably between tissues. Nucleus 2012, 3, 552–564. [Google Scholar] [CrossRef] [Green Version]
- Schirmer, E.C.; Florens, L.; Guan, T.; Yates, J.R., 3rd; Gerace, L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 2003, 301, 1380–1382. [Google Scholar] [CrossRef]
- Burns, L.T.; Wente, S.R. Trafficking to uncharted territory of the nuclear envelope. Curr. Opin. Cell Biol. 2012, 24, 341–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laba, J.K.; Steen, A.; Veenhoff, L.M. Traffic to the inner membrane of the nuclear envelope. Curr. Opin. Cell Biol. 2014, 28, 36–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ungricht, R.; Kutay, U. Establishment of NE asymmetry-targeting of membrane proteins to the inner nuclear membrane. Curr. Opin. Cell Biol. 2015, 34, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Zuleger, N.; Kerr, A.R.; Schirmer, E.C. Many mechanisms, one entrance: Membrane protein translocation into the nucleus. Cell. Mol. Life Sci. 2012, 69, 2205–2216. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Nath, A.; Hauber, J.; Kehlenbach, R.H. Multiple importins function as nuclear transport receptors for the Rev protein of human immunodeficiency virus type 1. J. Biol. Chem. 2006, 281, 20883–20890. [Google Scholar] [CrossRef]
- Lam, M.H.; Briggs, L.J.; Hu, W.; Martin, T.J.; Gillespie, M.T.; Jans, D.A. Importin beta recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin alpha. J. Biol. Chem. 1999, 274, 7391–7398. [Google Scholar] [CrossRef]
- Moore, J.D.; Yang, J.; Truant, R.; Kornbluth, S. Nuclear import of Cdk/cyclin complexes: Identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J. Cell Biol. 1999, 144, 213–224. [Google Scholar] [CrossRef]
- Fried, H.; Kutay, U. Nucleocytoplasmic transport: Taking an inventory. Cell. Mol. Life Sci. 2003, 60, 1659–1688. [Google Scholar] [CrossRef]
- Terry, L.J.; Wente, S.R. Flexible gates: Dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell 2009, 8, 1814–1827. [Google Scholar] [CrossRef] [PubMed]
- Lusk, C.P.; Blobel, G.; King, M.C. Highway to the inner nuclear membrane: Rules for the road. Nat. Rev. Mol. Cell Biol. 2007, 8, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Östlund, C.; Ellenberg, J.; Hallberg, E.; Lippincott-Schwartz, J.; Worman, H.J. Intracellular trafficking of emerin, the Emery-Dreifuss muscular dystrophy protein. J. Cell Sci. 1999, 112, 1709–1719. [Google Scholar] [PubMed]
- Soullam, B.; Worman, H.J. The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J. Cell Biol. 1993, 120, 1093–1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchiya, Y.; Hase, A.; Ogawa, M.; Yorifuji, H.; Arahata, K. Distinct regions specify the nuclear membrane targeting of emerin, the responsible protein for Emery-Dreifuss muscular dystrophy. Eur. J. Biochem. 1999, 259, 859–865. [Google Scholar] [CrossRef] [PubMed]
- King, M.C.; Lusk, C.P.; Blobel, G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 2006, 442, 1003–1007. [Google Scholar] [CrossRef]
- Meinema, A.C.; Laba, J.K.; Hapsari, R.A.; Otten, R.; Mulder, F.A.; Kralt, A.; van den Bogaart, G.; Lusk, C.P.; Poolman, B.; Veenhoff, L.M. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science 2011, 333, 90–93. [Google Scholar] [CrossRef]
- Kralt, A.; Jagalur, N.B.; van den Boom, V.; Lokareddy, R.K.; Steen, A.; Cingolani, G.; Fornerod, M.; Veenhoff, L.M. Conservation of inner nuclear membrane targeting sequences in mammalian Pom121 and yeast Heh2 membrane proteins. Mol. Biol. Cell 2015, 26, 3301–3312. [Google Scholar] [CrossRef] [Green Version]
- Boni, A.; Politi, A.Z.; Strnad, P.; Xiang, W.; Hossain, M.J.; Ellenberg, J. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J. Cell Biol. 2015, 209, 705–720. [Google Scholar] [CrossRef] [Green Version]
- Ungricht, R.; Klann, M.; Horvath, P.; Kutay, U. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J. Cell Biol. 2015, 209, 687–704. [Google Scholar] [CrossRef] [Green Version]
- Soullam, B.; Worman, H.J. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 1995, 130, 15–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maimon, T.; Elad, N.; Dahan, I.; Medalia, O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 2012, 20, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Zwerger, M.; Eibauer, M.; Medalia, O. Insights into the gate of the nuclear pore complex. Nucleus 2016, 7, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohba, T.; Schirmer, E.C.; Nishimoto, T.; Gerace, L. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 2004, 167, 1051–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaughan, A.; Alvarez-Reyes, M.; Bridger, J.M.; Broers, J.L.; Ramaekers, F.C.; Wehnert, M.; Morris, G.E.; Whitfield, W.G.F.; Hutchison, C.J. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J. Cell Sci. 2001, 114, 2577–2590. [Google Scholar] [PubMed]
- Turgay, Y.; Ungricht, R.; Rothballer, A.; Kiss, A.; Csucs, G.; Horvath, P.; Kutay, U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010, 29, 2262–2275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Lin, F.; Worman, H.J. Intracellular trafficking of MAN1, an integral protein of the nuclear envelope inner membrane. J. Cell Sci. 2002, 115, 1361–1371. [Google Scholar] [PubMed]
- Laba, J.K.; Steen, A.; Popken, P.; Chernova, A.; Poolman, B.; Veenhoff, L.M. Active Nuclear Import of Membrane Proteins Revisited. Cells 2015, 4, 653–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katta, S.S.; Smoyer, C.J.; Jaspersen, S.L. Destination: Inner nuclear membrane. Trends Cell Biol. 2014, 24, 221–229. [Google Scholar] [CrossRef]
- Skjerpen, C.S.; Wesche, J.; Olsnes, S. Identification of ribosome-binding protein p34 as an intracellular protein that binds acidic fibroblast growth factor. J. Biol. Chem. 2002, 277, 23864–23871. [Google Scholar] [CrossRef]
- Zhen, Y.; Sorensen, V.; Skjerpen, C.S.; Haugsten, E.M.; Jin, Y.; Walchli, S.; Olsnes, S.; Wiedlocha, A. Nuclear import of exogenous FGF1 requires the ER-protein LRRC59 and the importins Kpnalpha1 and Kpnbeta1. Traffic 2012, 13, 650–664. [Google Scholar] [CrossRef] [PubMed]
- Pallai, R.; Bhaskar, A.; Barnett-Bernodat, N.; Gallo-Ebert, C.; Pusey, M.; Nickels, J.T., Jr.; Rice, L.M. Leucine-rich repeat-containing protein 59 mediates nuclear import of cancerous inhibitor of PP2A in prostate cancer cells. Tumour Biol. 2015, 36, 6383–6390. [Google Scholar] [CrossRef] [PubMed]
- Borgese, N.; Colombo, S.; Pedrazzini, E. The tale of tail-anchored proteins: Coming from the cytosol and looking for a membrane. J. Cell Biol. 2003, 161, 1013–1019. [Google Scholar] [CrossRef]
- Kutay, U.; Hartmann, E.; Rapoport, T.A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993, 3, 72–75. [Google Scholar] [CrossRef]
- Denks, K.; Vogt, A.; Sachelaru, I.; Petriman, N.A.; Kudva, R.; Koch, H.G. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 2014, 31, 58–84. [Google Scholar] [CrossRef] [PubMed]
- Hegde, R.S.; Keenan, R.J. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2011, 12, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.; Powis, K.; High, S. Post-translational translocation into the endoplasmic reticulum. Biochim. Biophys. Acta 2013, 1833, 2403–2409. [Google Scholar] [CrossRef] [Green Version]
- Favaloro, V.; Spasic, M.; Schwappach, B.; Dobberstein, B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci. 2008, 121, 1832–1840. [Google Scholar] [CrossRef] [Green Version]
- Stefanovic, S.; Hegde, R.S. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 2007, 128, 1147–1159. [Google Scholar] [CrossRef]
- Pfaff, J.; Rivera Monroy, J.; Jamieson, C.; Rajanala, K.; Vilardi, F.; Schwappach, B.; Kehlenbach, R.H. Emery-Dreifuss muscular dystrophy mutations impair TRC40-mediated targeting of emerin to the inner nuclear membrane. J. Cell Sci. 2016, 129, 502–516. [Google Scholar] [CrossRef]
- Vilardi, F.; Lorenz, H.; Dobberstein, B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J. Cell Sci. 2011, 124, 1301–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, Y.; Sakisaka, T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol. Cell 2012, 48, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chan, C.; Weir, N.R.; Denic, V. The Get1/2 transmembrane complex is an endoplasmic-reticulum membrane protein insertase. Nature 2014, 512, 441–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abell, B.M.; Rabu, C.; Leznicki, P.; Young, J.C.; High, S. Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J. Cell Sci. 2007, 120, 1743–1751. [Google Scholar] [CrossRef] [Green Version]
- Favaloro, V.; Vilardi, F.; Schlecht, R.; Mayer, M.P.; Dobberstein, B. Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell Sci. 2010, 123, 1522–1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casson, J.; McKenna, M.; Hassdenteufel, S.; Aviram, N.; Zimmerman, R.; High, S. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017, 130, 3851–3861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilardi, F.; Stephan, M.; Clancy, A.; Janshoff, A.; Schwappach, B. WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS ONE 2014, 9, e85033. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, X.F.; Brown, E.J.; Schreiber, S.L. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 1995, 92, 4947–4951. [Google Scholar] [CrossRef] [PubMed]
- Pollard, V.W.; Michael, W.M.; Nakielny, S.; Siomi, M.C.; Wang, F.; Dreyfuss, G. A novel receptor-mediated nuclear protein import pathway. Cell 1996, 86, 985–994. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Entani, T.; Takayama, S.; Tomita, M.; Yanagawa, H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem. Biol. 2008, 15, 940–949. [Google Scholar] [CrossRef]
- Bell, M.R.; Engleka, M.J.; Malik, A.; Strickler, J.E. To fuse or not to fuse: What is your purpose? Protein Sci. 2013, 22, 1466–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abell, B.M.; Pool, M.R.; Schlenker, O.; Sinning, I.; High, S. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 2004, 23, 2755–2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.C.; Chuartzman, S.G.; Jan, C.H.; Hassdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guna, A.; Volkmar, N.; Christianson, J.C.; Hegde, R.S. The ER membrane protein complex is a transmembrane domain insertase. Science 2018, 359, 470–473. [Google Scholar] [CrossRef]
- Hassdenteufel, S.; Sicking, M.; Schorr, S.; Aviram, N.; Fecher-Trost, C.; Schuldiner, M.; Jung, M.; Zimmermann, R.; Lang, S. hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett. 2017, 591, 3211–3224. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Itzhak, D.N.; Hussmann, J.A.; Schirle Oakdale, N.T.; Costa, E.A.; Jonikas, M.; Weibezahn, J.; Popova, K.D.; Jan, C.H.; Sinitcyn, P.; et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife 2018, 7, e37018. [Google Scholar] [CrossRef] [PubMed]
- Zuleger, N.; Kelly, D.A.; Richardson, A.C.; Kerr, A.R.; Goldberg, M.W.; Goryachev, A.B.; Schirmer, E.C. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J. Cell Biol. 2011, 193, 109–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamus, G.; Zam, Z.S.; Arendt, A.; Palczewski, K.; McDowell, J.H.; Hargrave, P.A. Anti-rhodopsin monoclonal antibodies of defined specificity: Characterization and application. Vis. Res. 1991, 31, 17–31. [Google Scholar] [CrossRef]
- Charneau, P.; Mirambeau, G.; Roux, P.; Paulous, S.; Buc, H.; Clavel, F. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 1994, 241, 651–662. [Google Scholar] [CrossRef]
- Chen, C.; Okayama, H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987, 7, 2745–2752. [Google Scholar] [CrossRef]
- Frohnert, C.; Hutten, S.; Wälde, S.; Nath, A.; Kehlenbach, R.H. Importin 7 and Nup358 promote nuclear import of the protein component of human telomerase. PLoS ONE 2014, 9, e88887. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Blobel, G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983, 96, 84–93. [Google Scholar] [PubMed]
- Leznicki, P.; Clancy, A.; Schwappach, B.; High, S. Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 2010, 123, 2170–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blenski, M.; Kehlenbach, R.H. Targeting of LRRC59 to the Endoplasmic Reticulum and the Inner Nuclear Membrane. Int. J. Mol. Sci. 2019, 20, 334. https://doi.org/10.3390/ijms20020334
Blenski M, Kehlenbach RH. Targeting of LRRC59 to the Endoplasmic Reticulum and the Inner Nuclear Membrane. International Journal of Molecular Sciences. 2019; 20(2):334. https://doi.org/10.3390/ijms20020334
Chicago/Turabian StyleBlenski, Marina, and Ralph H. Kehlenbach. 2019. "Targeting of LRRC59 to the Endoplasmic Reticulum and the Inner Nuclear Membrane" International Journal of Molecular Sciences 20, no. 2: 334. https://doi.org/10.3390/ijms20020334
APA StyleBlenski, M., & Kehlenbach, R. H. (2019). Targeting of LRRC59 to the Endoplasmic Reticulum and the Inner Nuclear Membrane. International Journal of Molecular Sciences, 20(2), 334. https://doi.org/10.3390/ijms20020334