Effects of CD4 Binding on Conformational Dynamics, Molecular Motions, and Thermodynamics of HIV-1 gp120
Abstract
1. Introduction
2. Results
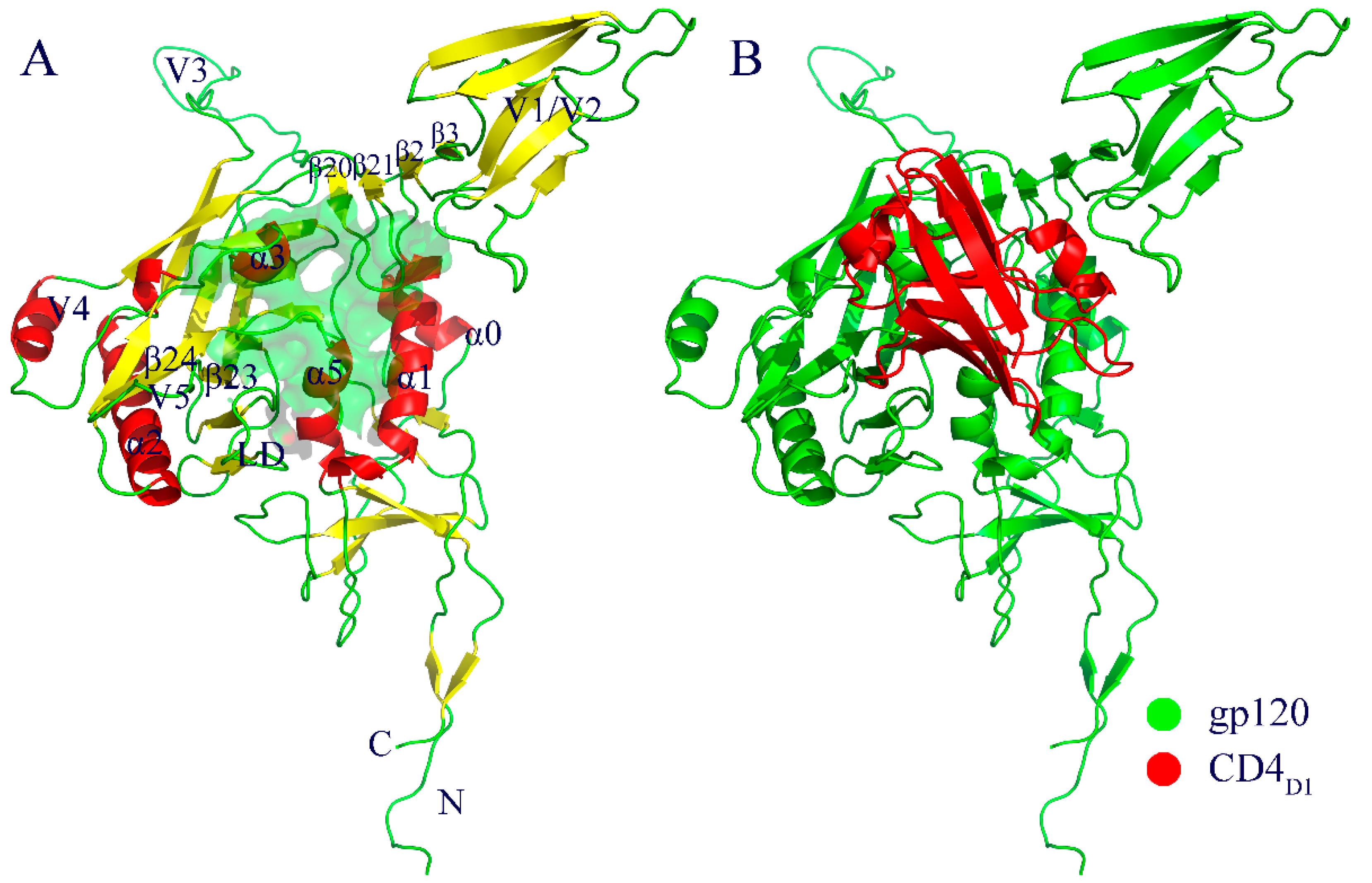
2.1. Molecular Architecture of gp120
2.2. Conformational Dynamics
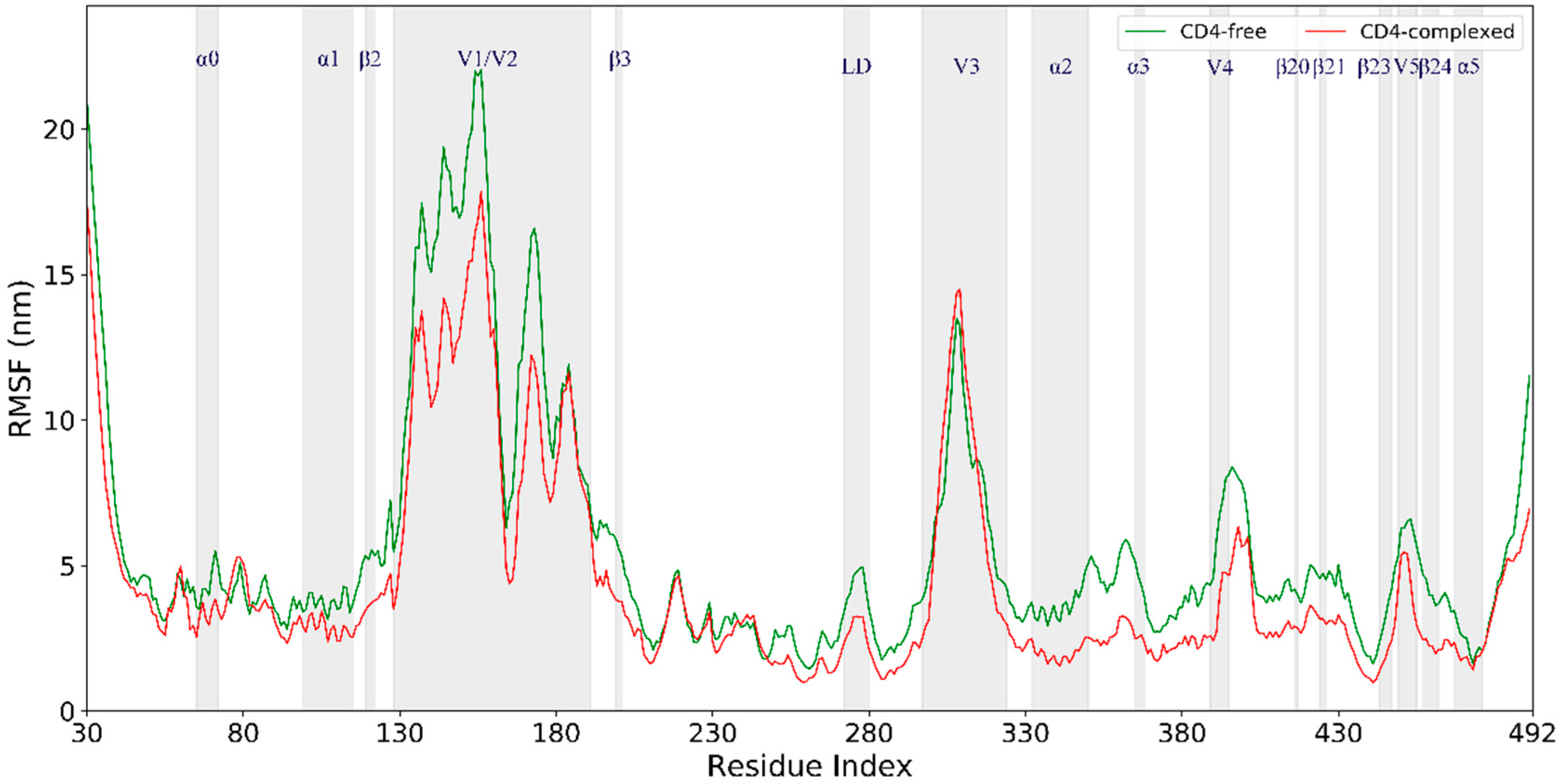
2.3. Conformational Flexibility
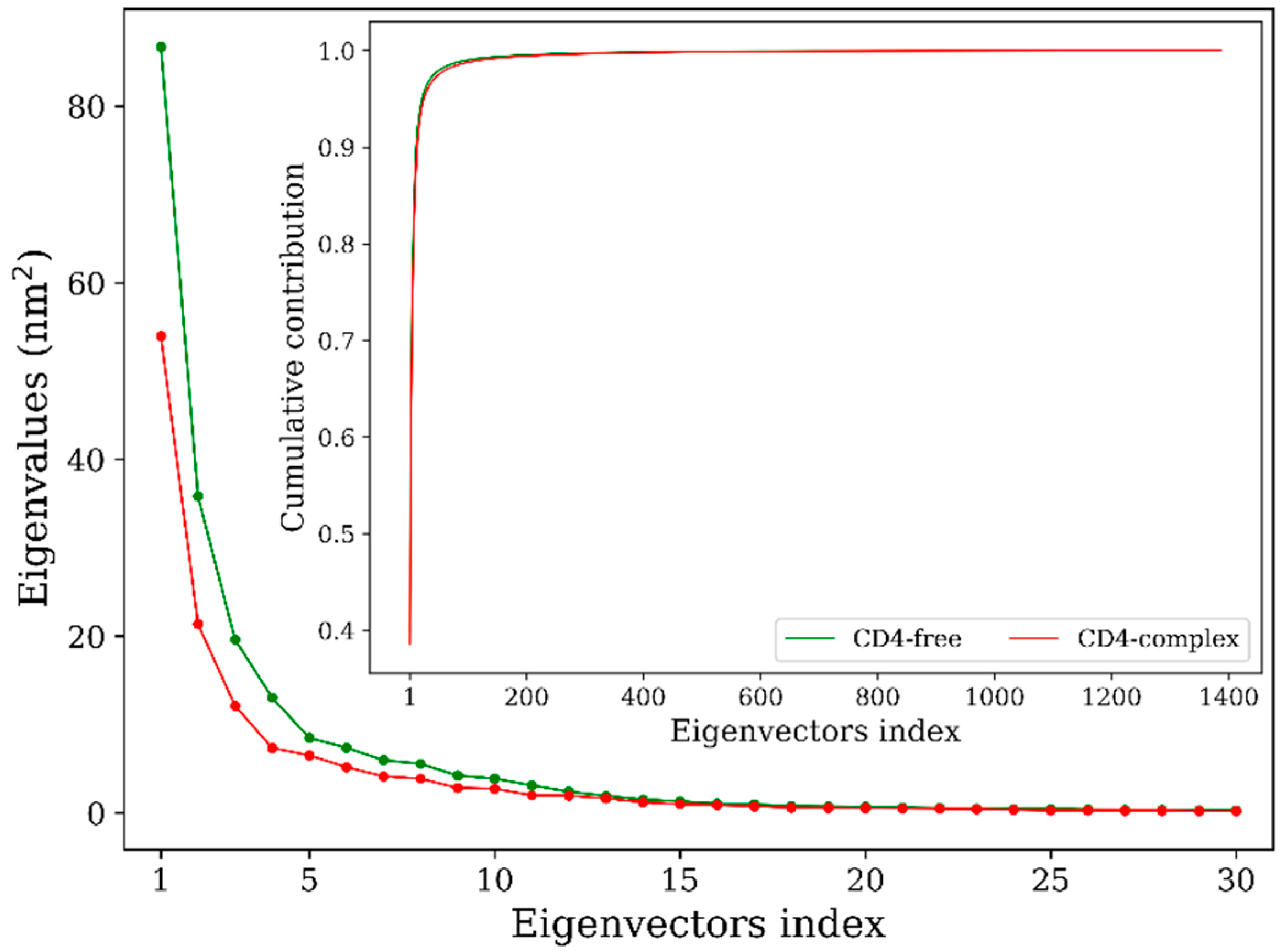
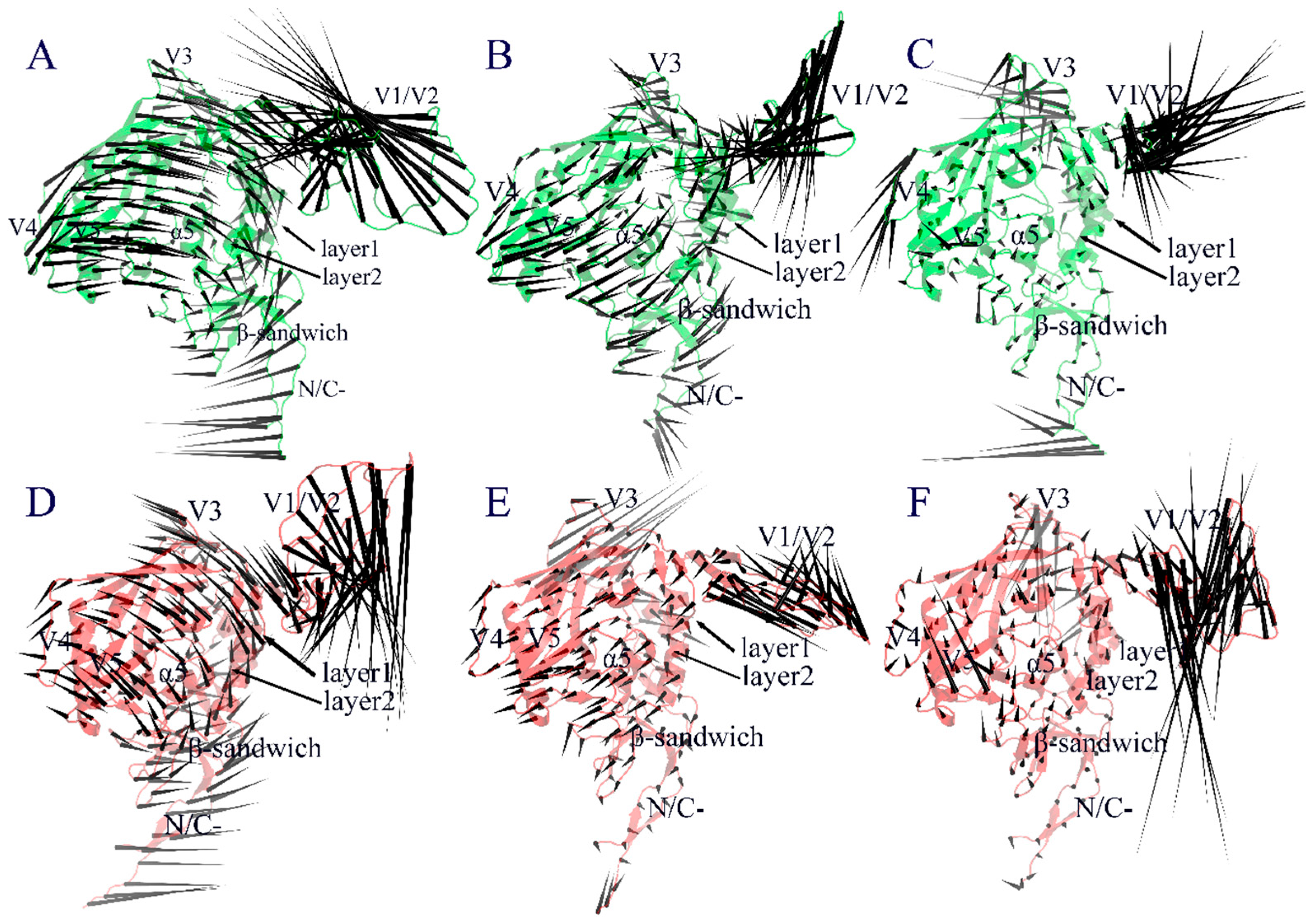
2.4. Collective Motions
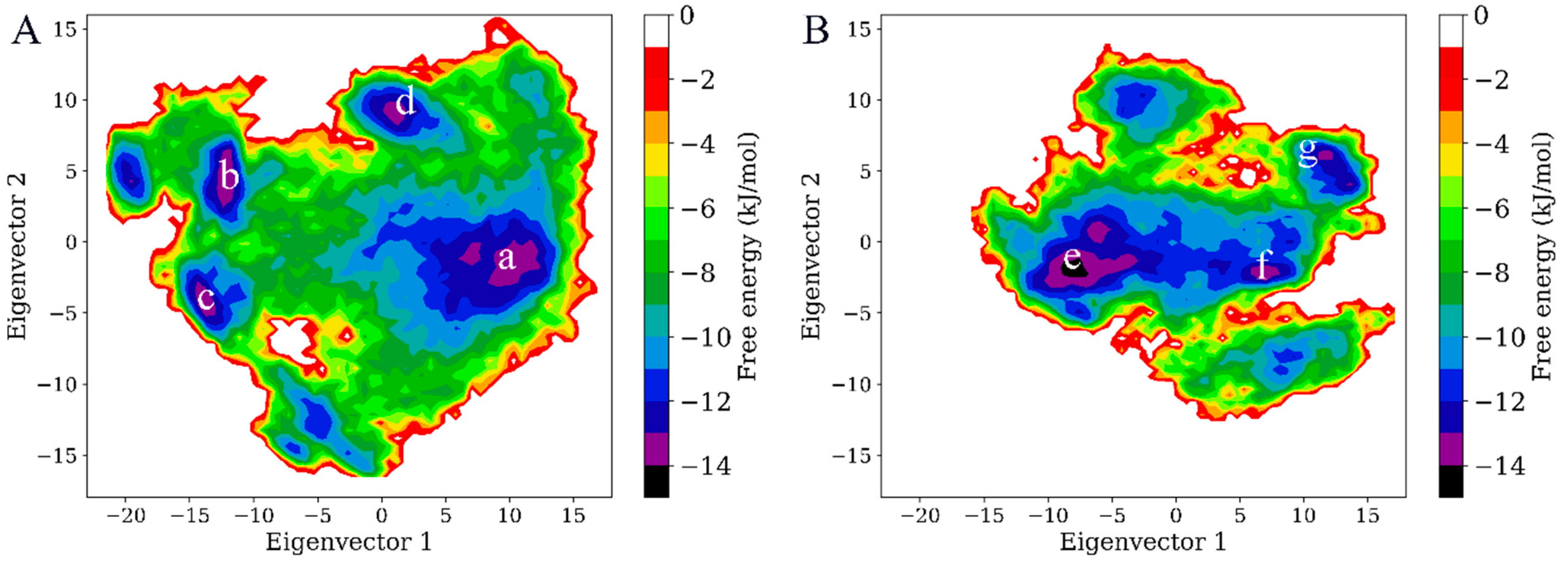
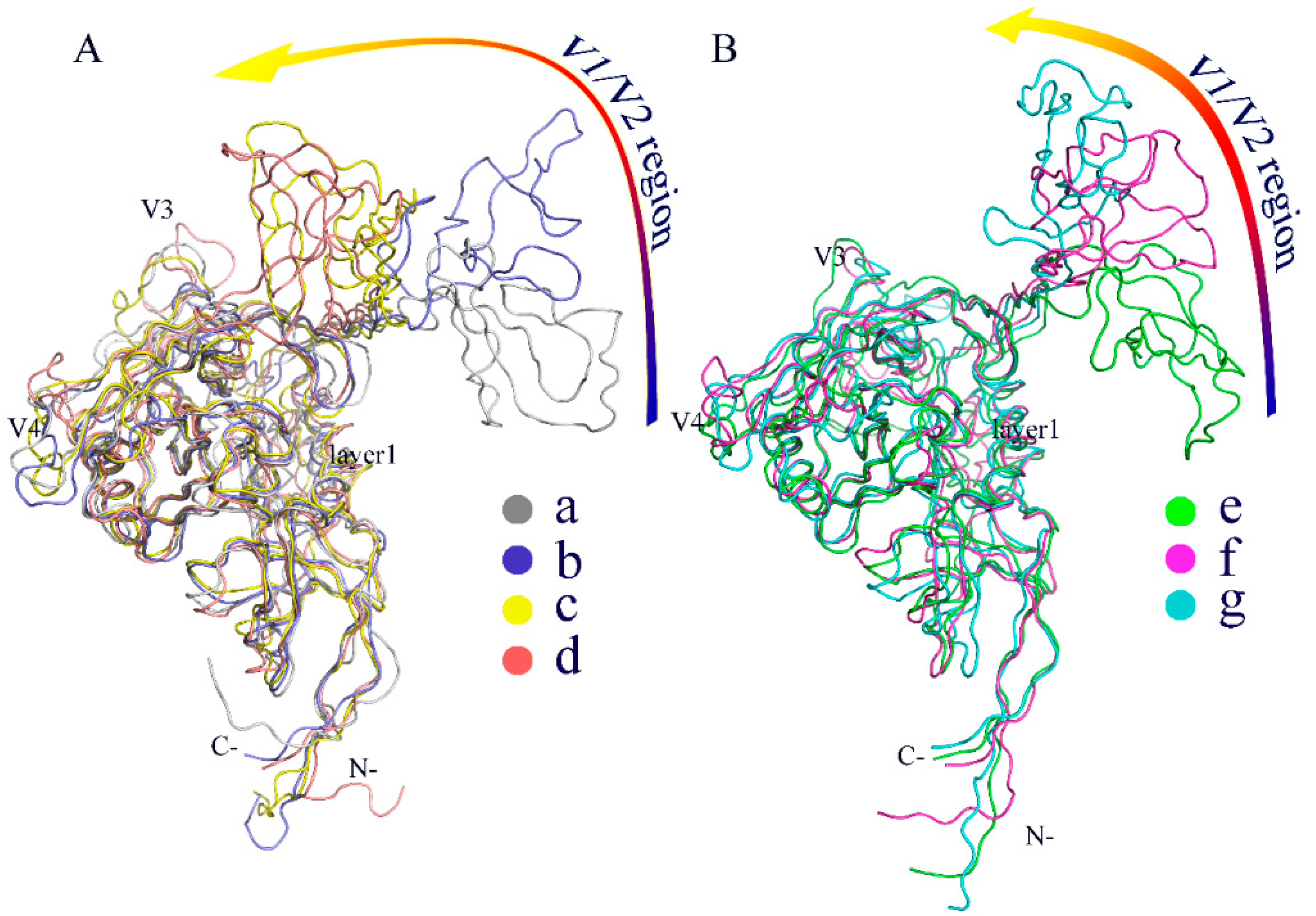
2.5. Free Energy Landscape (FEL) and Representative Structures
3. Discussion
4. Materials and Methods
4.1. Simulation System Preparation
4.2. MD Simulations
4.3. Analysis Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Liu, J.; Bartesaghi, A.; Borgnia, M.J.; Sapiro, G.; Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 2008, 455, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Pancera, M.; Majeed, S.; Ban, Y.E.A.; Chen, L.; Huang, C.C.; Kong, L.; Kwon, Y.D.; Stuckey, J.; Zhou, T.; Robinson, J.E.; et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. USA 2009, 107, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, R.; Sodroski, J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science 1998, 280, 1884–1888. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.D.; Finzi, A.; Wu, X.; Dogo-Isonagie, C.; Lee, L.K.; Moore, L.R.; Schmidt, S.D.; Stuckey, J.; Yang, Y.; Zhou, T.; et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. USA 2012, 109, 5663–5668. [Google Scholar] [CrossRef] [PubMed]
- Raja, A.; Venturi, M.; Kwong, P.; Sodroski, J. CD4 binding site antibodies inhibit human immunodeficiency virus gp120 envelope glycoprotein interaction with CCR5. J. Virol. 2003, 77, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, P.; Gruell, H.; Nogueira, L.; Pai, J.A.; Butler, A.L.; Millard, K.; Lehmann, C.; Suárez, I.; Oliveira, T.Y.; Lorenzi, J.C.C.; et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 2018, 561, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Kwong, P.D.; Wyatt, R.; Robinson, J.; Sweet, R.W.; Sodroski, J.; Hendrickson, W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998, 393, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Kwong, P.D.; Doyle, M.L.; Casper, D.J.; Cicala, C.; Leavitt, S.A.; Majeed, S.; Steenbeke, T.D.; Venturi, M.; Chaiken, I.; Fung, M. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 2002, 420, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Julien, J.P.; Cupo, A.; Sok, D.; Stanfield, R.L.; Lyumkis, D.; Deller, M.C.; Klasse, P.J.; Burton, D.R.; Sanders, R.W.; Moore, J.P.; et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 2013, 342, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, M.; Bettadapura, R.; Bajaj, C. Computational refinement and validation protocol for proteins with large variable regions applied to model HIV Env spike in CD4 and 17b bound state. Structure 2015, 23, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Ozorowski, G.; Pallesen, J.; de Val, N.; Lyumkis, D.; Cottrell, C.A.; Torres, J.L.; Copps, J.; Stanfield, R.L.; Cupo, A.; Pugach, P.; et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 2017, 547, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cohen, A.A.; Galimidi, R.P.; Gristick, H.B.; Jensen, G.J.; Bjorkman, P.J. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc. Natl. Acad. Sci. USA 2016, 113, E7151–E7158. [Google Scholar] [CrossRef] [PubMed]
- Myszka, D.G.; Sweet, R.W.; Hensley, P.; Brigham-Burke, M.; Kwong, P.D.; Hendrickson, W.A.; Wyatt, R.; Sodroski, J.; Doyle, M.L. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 2000, 97, 9026–9031. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Garcia, N.K.; Cupo, A.; Matsui, T.; Julien, J.-P.; Sanders, R.W.; Wilson, I.A.; Moore, J.P.; Lee, K.K. CD4-Induced activation in a soluble HIV-1 Env trimer. Structure 2014, 22, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Munro, J.B.; Gorman, J.; Ma, X.; Zhou, Z.; Arthos, J.; Burton, D.R.; Koff, W.C.; Courter, J.R.; Smith, A.B., 3rd; Kwong, P.D.; et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 2014, 346, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Scharf, L.; Wang, H.; Gao, H.; Chen, S.; McDowall, A.W.; Bjorkman, P.J. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell 2015, 162, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ozorowski, G.; Ward, A.B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 2016, 351, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Stewart-Jones, G.B.; Soto, C.; Lemmin, T.; Chuang, G.-Y.; Druz, A.; Kong, R.; Thomas, P.V.; Wagh, K.; Zhou, T.; Behrens, A.-J.; et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 2016, 165, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Caves, L.S.; Evanseck, J.D.; Karplus, M. Locally accessible conformations of proteins: Multiple molecular dynamics simulations of crambin. Protein Sci. 1998, 7, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Herschhorn, A.; Gu, C.; Moraca, F.; Ma, X.; Farrell, M.; Smith, A.B., 3rd; Pancera, M.; Kwong, P.D.; Schon, A.; Freire, E.; et al. The β20-β21 of gp120 is a regulatory switch for HIV-1 Env conformational transitions. Nat. Commun. 2017, 8, 1049. [Google Scholar] [CrossRef] [PubMed]
- Sang, P.; Yang, Q.; Du, X.; Yang, N.; Yang, L.Q.; Ji, X.L.; Fu, Y.X.; Meng, Z.H.; Liu, S.Q. Effect of the solvent temperatures on dynamics of serine protease proteinase K. Int. J. Mol. Sci. 2016, 17, 254. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, M.; Poon, B.K.; Wang, Q.; Ma, J. Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement. Acta Crystallogr. D Biol. Crystallogr. 2010, 65, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Vogan, E.M.; Gong, H.; Skehel, J.J.; Wiley, D.C.; Harrison, S.C. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 2005, 433, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into protein-ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Tang, M.; Zhang, M.Y.; Majeed, S.; Montabana, E.; Stanfield, R.L.; Dimitrov, D.S.; Korber, B.; Sodroski, J.; Wilson, I.A.; et al. Structure of a V3-containing HIV-1 gp120 core. Science 2005, 310, 1025–1028. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Fan, S.; Sun, Z. Structural and functional characterization of the human CCR5 receptor in complex with HIV gp120 envelope glycoprotein and CD4 receptor by molecular modeling studies. J. Mol. Model. 2003, 9, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Langley, D.R.; Kimura, S.R.; Sivaprakasam, P.; Zhou, N.; Dicker, I.; Mcauliffe, B.; Wang, T.; Kadow, J.F.; Meanwell, N.A.; Krystal, M. Homology models of the HIV-1 attachment inhibitor BMS-626529 bound to gp120 suggest a unique mechanism of action. Proteins 2015, 83, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Pinter, A.; Honnen, W.J.; He, Y.; Gorny, M.K.; Zolla-Pazner, S.; Kayman, S.C. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 2004, 78, 5205–5215. [Google Scholar] [CrossRef] [PubMed]
- Rusert, P.; Krarup, A.; Magnus, C.; Brandenberg, O.F.; Weber, J.; Ehlert, A.K.; Regoes, R.R.; Günthard, H.F.; Trkola, A. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J. Exp. Med. 2011, 208, 1419–1433. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, L.; Ai, S.M.; Sang, P.; Yang, J.; Xia, Y.L.; Zhang, Z.B.; Fu, Y.X.; Liu, S.Q. Insights into the molecular mechanism underlying CD4-dependency and neutralization sensitivity of HIV-1: A comparative molecular dynamics study on gp120s from isolates with different phenotypes. RSC Adv. 2018, 8, 14355–14368. [Google Scholar] [CrossRef]
- Truneh, A.; Buck, D.; Cassatt, D.R.; Juszczak, R.; Kassis, S.; Ryu, S.E.; Healey, D.; Sweet, R.; Sattentau, Q. A region in domain 1 of CD4 distinct from the primary gp120 binding site is involved in HIV infection and virus-mediated fusion. J. Biol. Chem. 1991, 266, 5942–5948. [Google Scholar] [PubMed]
- Esser, U.; Speck, R.F.; Deen, K.C.; Atchison, R.E.; Sweet, R.; Goldsmith, M.A. Molecular function of the CD4 D1 domain in coreceptor-mediated entry by HIV type 1. AIDS Res. Hum. Retroviruses 2000, 16, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Price, D.J.; Brooks, C.L., 3rd. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Pall, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Aliev, A.E.; Kulke, M.; Khaneja, H.S.; Chudasama, V.; Sheppard, T.D.; Lanigan, R.M. Motional timescale predictions by molecular dynamics simulations: Case study using proline and hydroxyproline sidechain dynamics. Proteins 2014, 82, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Q.; Meng, Z.H.; Fu, Y.X.; Zhang, K.Q. Insights derived from molecular dynamics simulation into the molecular motions of serine protease proteinase K. J. Mol. Model. 2010, 16, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comp. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An, N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Deng, L.; Yang, L.-Q.; Sang, P.; Liu, S.-Q. Effects of CD4 Binding on Conformational Dynamics, Molecular Motions, and Thermodynamics of HIV-1 gp120. Int. J. Mol. Sci. 2019, 20, 260. https://doi.org/10.3390/ijms20020260
Li Y, Deng L, Yang L-Q, Sang P, Liu S-Q. Effects of CD4 Binding on Conformational Dynamics, Molecular Motions, and Thermodynamics of HIV-1 gp120. International Journal of Molecular Sciences. 2019; 20(2):260. https://doi.org/10.3390/ijms20020260
Chicago/Turabian StyleLi, Yi, Lei Deng, Li-Quan Yang, Peng Sang, and Shu-Qun Liu. 2019. "Effects of CD4 Binding on Conformational Dynamics, Molecular Motions, and Thermodynamics of HIV-1 gp120" International Journal of Molecular Sciences 20, no. 2: 260. https://doi.org/10.3390/ijms20020260
APA StyleLi, Y., Deng, L., Yang, L.-Q., Sang, P., & Liu, S.-Q. (2019). Effects of CD4 Binding on Conformational Dynamics, Molecular Motions, and Thermodynamics of HIV-1 gp120. International Journal of Molecular Sciences, 20(2), 260. https://doi.org/10.3390/ijms20020260





