Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants
Abstract
:1. Introduction
2. ABA Core Pathways for the Regulation of Leaf Senescence
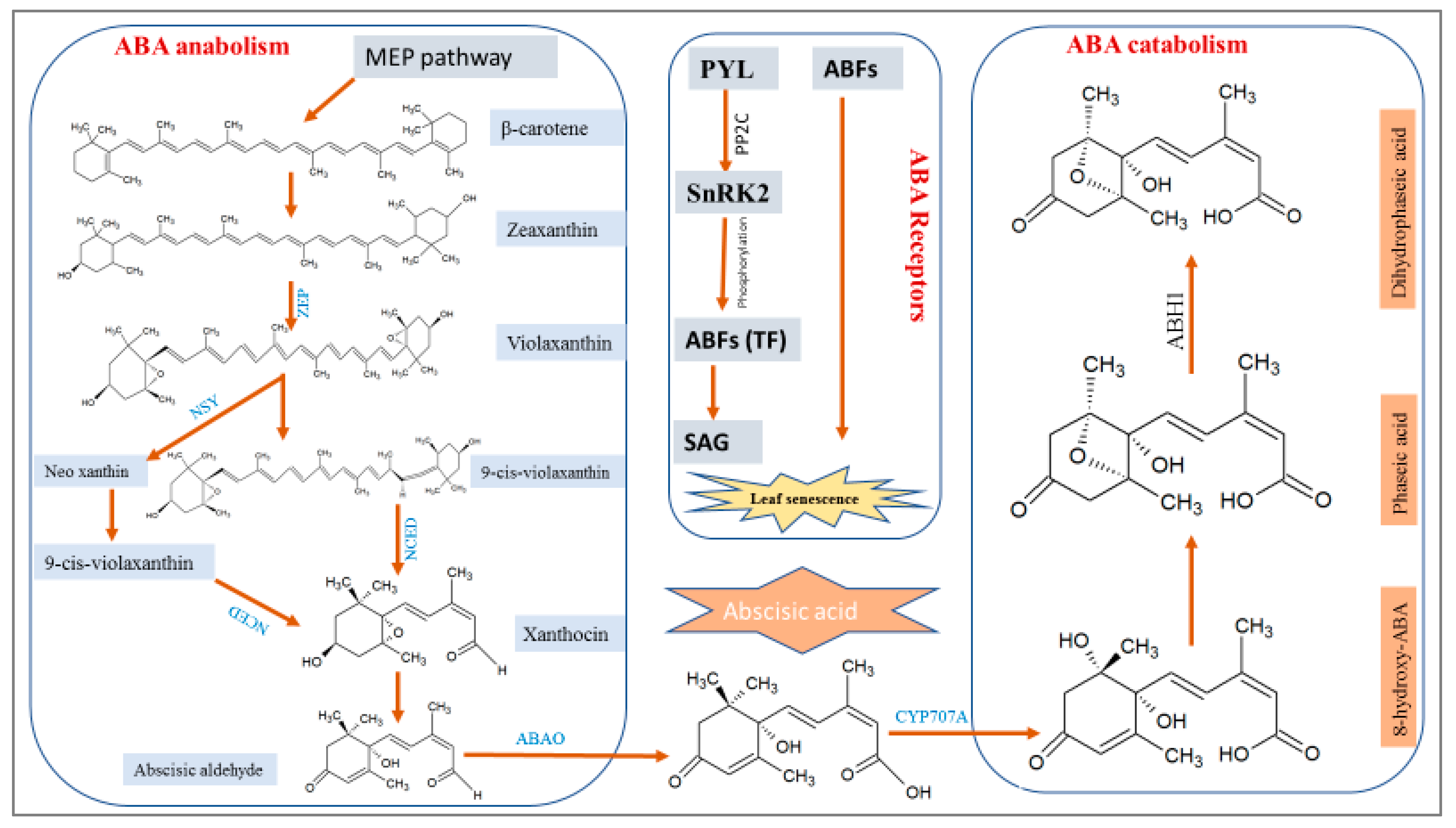
2.1. ABA Biosynthesis and Catabolism
2.2. ABA Transport
2.3. ABA Signaling Receptors
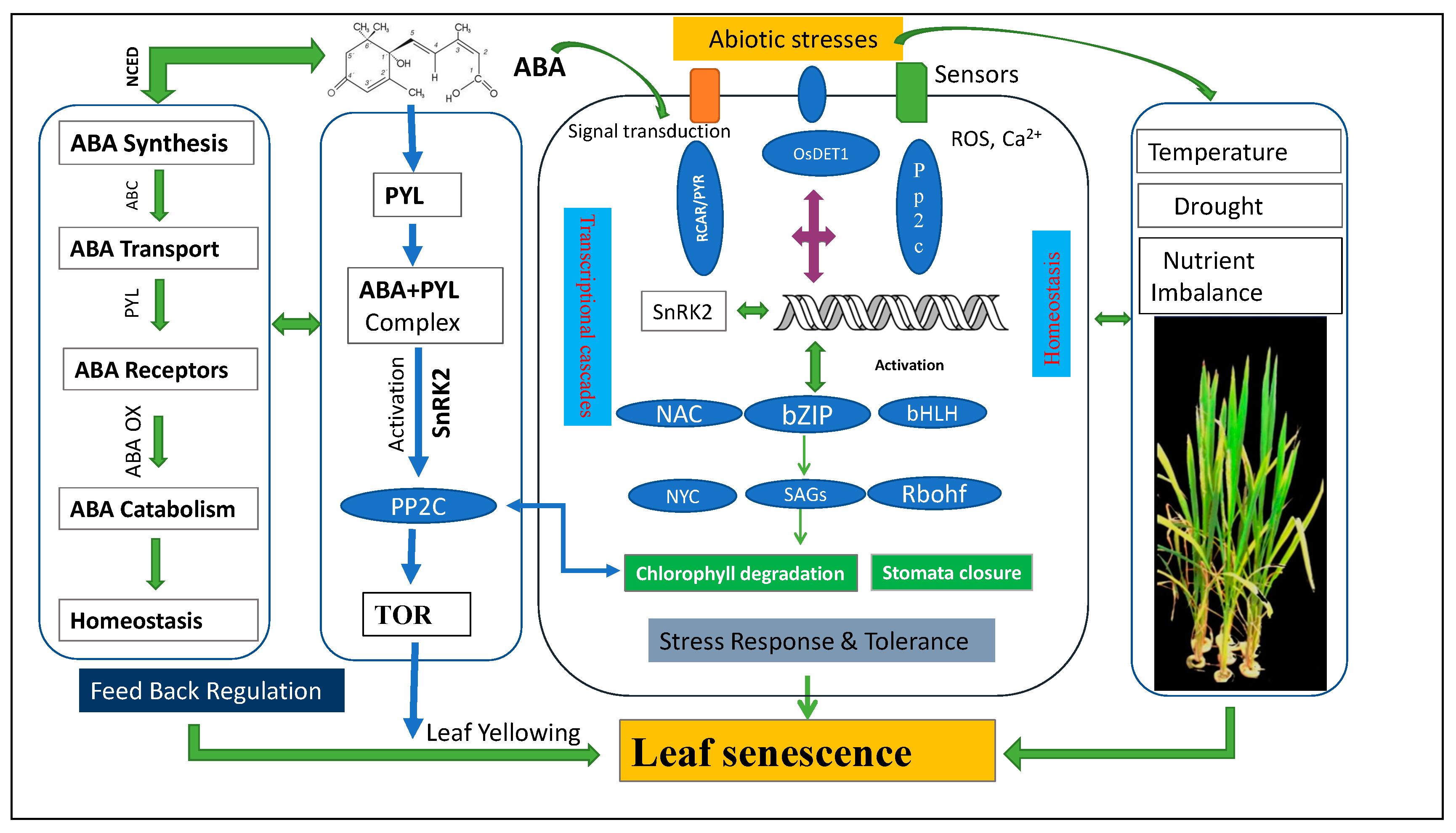
3. Integrated Mechanism of ABA-Induced Leaf Senescence
3.1. Relationship of ABA-Induced Leaf Senescence with Chloroplast Degradation and Photosynthesis Decline
3.2. Relationship of ABA-Induced Leaf Senescence with ROS Generation and Oxidative Stress
3.3. ABA Modulates Leaf Senescence by Activating Regulating Kinase Protein
3.4. Involvement of Secondary Messenger Action Ca2+ in ABA Signaling and Leaf Senescence Regulation
4. ABA Regulates TFs to Induce SAG Expressions
4.1. Induction of NAC TFs
4.2. Modulating OsNAC2 TF Expression
4.3. Regulation of bZIP TF
5. ABA Regulation of Senescence-Related Membrane-Associated Protein to Transduce Leaf Senescence
ABA mediates the Target of Rapamycin (TOR) to Induce Leaf Senescence
6. Gene Mutation as the Key Step to Identify ABA Regulation for Senescence
7. Integrated Crosstalk that Initiates the Onset of Leaf Senescence
7.1. Interaction of Sugar Signaling with ABA during Leaf Senescence
7.2. Interacted Crosstalk between Ethylene and ABA
8. Conclusions and Future Recommendations
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| PYL | Pyrabactin resistance 1-like |
| SnRK2 | Sucrose nonfermenting 1-related protein kinase 2 |
| ABFs | ABA-responsive element-binding factors |
| SAPK | Stress-activated protein kinase |
| PPH | Pheophytinase |
| SGR | Stay green |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| MDA | Malondialdehyde |
| PAO | Polyamine oxidases |
| NYC1 | Non yellow coloring 1 |
| SASP | Senescence associated secretory phenotype |
| PP2C | Protein phosphatase 2C |
| AAO3 | Arabidopsis aldehyde oxidase 3 |
| OST1 | Open stomata 1 |
| BZR1 | Brassinazole-resistant 1 |
| ABA2 | Abscisic acid deficient 2 |
| SRE1 | Salt resistance 1 |
| GIN1 | Glucose insensitivity 1 |
| T6P | Trehalose-6-phosphate |
| HXK1 | Hexokinase 1 |
| SUC | Sucrose |
| FRU | Fructose |
| GLC | Glucose |
| YFP | Yellow fluorescent protein |
| SAGs | Senescence-associated genes |
| EIN | Ethylene resistant mutant |
| LUC | Luciferase |
| AVG | Aminoethoxyvinylglycine |
| DET1 | DE-ETIOLATED1 |
References
- Park, D.-Y.; Shim, Y.; Gi, E.; Lee, B.-D.; An, G.; Kang, K.; Paek, N.-C. The MYB-related transcription factor RADIALIS-LIKE3 (OsRL3) functions in ABA-induced leaf senescence and salt sensitivity in rice. Environ. Exp. Bot. 2018, 156, 86–95. [Google Scholar] [CrossRef]
- Lim, P.O.; Woo, H.R.; Nam, H.G. Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 2003, 8, 272–278. [Google Scholar] [CrossRef]
- Xu, X.; Guo, K.; Liang, W.; Chen, Q.; Shi, J.; Shen, B. Quantitative proteomics analysis of proteins involved in leaf senescence of rice (Oryza sativa L.). Plant Growth Regul. 2018, 84, 341–349. [Google Scholar] [CrossRef]
- Wang, F.; Liu, J.; Zhou, L.; Pan, G.; Li, Z.; Zaidi, S.-H.-R.; Cheng, F. Senescence-specific change in ROS scavenging enzyme activities and regulation of various SOD isozymes to ROS levels in psf mutant rice leaves. Plant Physiol. Biochem. 2016, 109, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Gao, X.; Guo, Y. Initiation, Progression, and Genetic Manipulation of Leaf Senescence. In Plant Senescence: Methods and Protocols; Guo, Y., Ed.; Springer: New York, NY, USA, 2018; pp. 9–31. [Google Scholar]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Bai, Q.; Xu, P.; Wu, T.; Guo, D.; Peng, Y.; Zhang, H.; Deng, X.; Chen, X.; Luo, M.; et al. Mutation in Rice Abscisic Acid2 Results in Cell Death, Enhanced Disease-Resistance, Altered Seed Dormancy and Development. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, J.; Chen, M.; Zhou, L.; Li, Z.; Zhao, Q.; Pan, G.; Zaidi, S.-H.-R.; Cheng, F. Involvement of Abscisic Acid in PSII Photodamage and D1 Protein Turnover for Light-Induced Premature Senescence of Rice Flag Leaves. PLoS ONE 2016, 11, e0161203. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Wu, Y.; Xie, Q. Ubiquitin–Proteasome System in ABA Signaling: From Perception to Action. Mol. Plant 2016, 9, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.; Davies, W.J. Drought, ozone, ABA and ethylene: New insights from cell to plant to community. Plantcell Environ. 2010, 33, 510–525. [Google Scholar] [CrossRef]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.-C.; Joseph, L.M.; Deng, W.-T.; Liu, L.; Li, Q.-B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, G.K.; Tripathy, B.C. Modulation of biosynthesis of photosynthetic pigments and light-harvesting complex in wild-type and gun5 mutant of Arabidopsis thaliana during impaired chloroplast development. Protoplasma 2016, 253, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Pudelski, B.; Soll, J.; Philippar, K. A search for factors influencing etioplast–chloroplast transition. Proc. Natl. Acad. Sci. USA 2009, 106, 12201–12206. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Balazadeh, S.; Tanaka, R.; Mueller-Roeber, B.; Tanaka, A. Overproduction of Chl b Retards Senescence through Transcriptional Reprogramming in Arabidopsis. Plant Cell Physiol. 2012, 53, 505–517. [Google Scholar] [CrossRef]
- Lin, Y.-P.; Lee, T.-Y.; Tanaka, A.; Charng, Y.-Y. Analysis of an Arabidopsis heat-sensitive mutant reveals that chlorophyll synthase is involved in reutilization of chlorophyllide during chlorophyll turnover. Plant J. 2014, 80, 14–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Q.; Liang, Y.; Zhang, J.; Zheng, H.; Dong, G.; Qian, Q.; Zuo, J. Involvement of a Putative Bipartite Transit Peptide in Targeting Rice Pheophorbide a Oxygenase into Chloroplasts for Chlorophyll Degradation during Leaf Senescence. J. Genet. Genom. 2016, 43, 145–154. [Google Scholar] [CrossRef]
- Wang, D.; Liu, H.; Zhai, G.; Wang, L.; Shao, J.; Tao, Y. OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. J. Genet. Genom. 2016, 43, 601–608. [Google Scholar] [CrossRef]
- Quiles, M.J.; García, C.; Cuello, J. Differential effects of abscisic acid and methyl jasmonate on endoproteinases in senescing barley leaves. Plant Growth Regul. 1995, 16, 197–204. [Google Scholar] [CrossRef]
- Li, Z.; Wang, F.; Zhao, Q.; Liu, J.; Cheng, F. Involvement of NADPH oxidase isoforms in the production of O2− manipulated by ABA in the senescing leaves of early-senescence-leaf (esl) mutant rice (Oryza sativa). PLoS ONE 2018, 13, e0190161. [Google Scholar] [CrossRef]
- Akhter, D.; Qin, R.; Nath, U.; Alamin, M.; Jin, X.; Shi, C. The Brown Midrib Leaf (bml) Mutation in Rice (Oryza sativa L.) Causes Premature Leaf Senescence and the Induction of Defense Responses. Genes 2018, 9, 203. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [Green Version]
- Ye, N.; Jia, L.; Zhang, J. ABA signal in rice under stress conditions. Rice 2012, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Coumou, D.; Rahmstorf, S. A decade of weather extremes. Nat. Clim. Chang. 2012, 2, 491. [Google Scholar] [CrossRef]
- Kollist, H.; Zandalinas, S.I.; Sengupta, S.; Nuhkat, M.; Kangasjärvi, J.; Mittler, R. Rapid Responses to Abiotic Stress: Priming the Landscape for the Signal Transduction Network. Trends Plant Sci. 2018. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, J.-K. Regulation of Abscisic Acid Biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Zhang, S.J.; Song, G.Q.; Li, Y.L.; Gao, J.; Liu, J.J.; Fan, Q.Q.; Huang, C.Y.; Sui, X.X.; Chu, X.S.; Guo, D.; et al. Cloning of 9-cis-epoxycarotenoid dioxygenase gene (TaNCED1) from wheat and its heterologous expression in tobacco. Biol. Plant. 2014, 58, 89–98. [Google Scholar] [CrossRef]
- North, H.M.; Almeida, A.D.; Boutin, J.-P.; Frey, A.; To, A.; Botran, L.; Sotta, B.; Marion-Poll, A. The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 2007, 50, 810–824. [Google Scholar] [CrossRef] [Green Version]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [Green Version]
- Riahi, L.; Zoghlami, N.; Dereeper, A.; Laucou, V.; Mliki, A.; This, P. Molecular characterization and evolutionary pattern of the 9-cis-epoxycarotenoid dioxygenase NCED1 gene in grapevine. Mol. Breed. 2013, 32, 253–266. [Google Scholar] [CrossRef]
- González-Guzmán, M.; Apostolova, N.; Bellés, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodríguez, P.L. The Short-Chain Alcohol Dehydrogenase ABA2 Catalyzes the Conversion of Xanthoxin to Abscisic Aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef] [Green Version]
- Endo, A.; Nelson, K.M.; Thoms, K.; Abrams, S.R.; Nambara, E.; Sato, Y. Functional characterization of xanthoxin dehydrogenase in rice. J. Plant Physiol. 2014, 171, 1231–1240. [Google Scholar] [CrossRef]
- Seo, M.; Peeters, A.J.M.; Koiwai, H.; Oritani, T.; Marion-Poll, A.; Zeevaart, J.A.D.; Koornneef, M.; Kamiya, Y.; Koshiba, T. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 2000, 97, 12908–12913. [Google Scholar] [CrossRef]
- Rötter, R.P.; Hoffmann, M.P.; Koch, M.; Müller, C. Progress in modelling agricultural impacts of and adaptations to climate change. Curr. Opin. Plant Biol. 2018, 45, 255–261. [Google Scholar] [CrossRef]
- Babacar, F.; Heidi, W.; Jesse, B.N.; Dilys, S.M.; Myriam, A.; Frank, E.; John, P.A.L.; Carl-Friedrich, S.; Alex, R.; Ursula, G.; et al. Impacts of 1.5 versus 2.0 °C on cereal yields in the West African Sudan Savanna. Environ. Res. Lett. 2018, 13, 034014. [Google Scholar]
- Dinneny, J.R.; Long, T.A.; Wang, J.Y.; Jung, J.W.; Mace, D.; Pointer, S.; Barron, C.; Brady, S.M.; Schiefelbein, J.; Benfey, P.N. Cell Identity Mediates the Response of Arabidopsis Roots to Abiotic Stress. Science 2008, 320, 942–945. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8’-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As Encode (+)-Abscisic Acid 8′-Hydroxylase, a Key Enzyme in the Oxidative Catabolism of Abscisic Acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef]
- Hartung, W.; Sauter, A.; Hose, E. Abscisic acid in the xylem: Where does it come from, where does it go to? J. Exp. Bot. 2002, 53, 27–32. [Google Scholar] [CrossRef]
- Klein, M.; Perfus-Barbeoch, L.; Frelet, A.; Gaedeke, N.; Reinhardt, D.; Mueller-Roeber, B.; Martinoia, E.; Forestier, C. The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. Plant J. 2003, 33, 119–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Zhou, L.; Liu, X.; Liu, H.; Li, D.; Cao, M.; Chen, H.; Xu, L.; Zhu, J.-K.; Zhao, Y. A Novel Chemical Inhibitor of ABA Signaling Targets All ABA Receptors. Plant Physiol. 2017, 173, 2356–2369. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hwang, J.-U.; Lee, M.; Kim, Y.-Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuromori, T.; Sugimoto, E.; Shinozaki, K. Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 2011, 67, 885–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-f.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Xing, L.; Liu, X.; Hou, Y.-J.; Chinnusamy, V.; Wang, P.; Duan, C.; Zhu, J.-K. The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 2013, 23, 1380. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.-J.; Gao, J.; Wang, P.; Duan, C.-G.; Zhu, X.; Zhu, J.-K. The ABA Receptor PYL8 Promotes Lateral Root Growth by Enhancing MYB77-Dependent Transcription of Auxin-Responsive Genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.C.; Hong, S.W.; Whang, S.S.; Lim, P.O.; Nam, H.G.; Koo, J.C. Age-Dependent Action of an ABA-Inducible Receptor Kinase, RPK1, as a Positive Regulator of Senescence in Arabidopsis Leaves. Plant Cell Physiol. 2011, 52, 651–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Xiang, F.; Zhang, G.; Miao, Y.; Miao, C.; Song, C.-P. Abscisic Acid as an Internal Integrator of Multiple Physiological Processes Modulates Leaf Senescence Onset in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 181. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Zhang, Y.; Sinumporn, S.; Yu, N.; Zhan, X.; Shen, X.; Chen, D.; Yu, P.; Wu, W.; Liu, Q.; et al. Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. 2018, 95, 877–891. [Google Scholar] [CrossRef] [PubMed]
- Jibran, R.; Sullivan, K.L.; Crowhurst, R.; Erridge, Z.A.; Chagné, D.; McLachlan, A.R.G.; Brummell, D.A.; Dijkwel, P.P.; Hunter, D.A. Staying green postharvest: How three mutations in the Arabidopsis chlorophyll b reductase gene NYC1 delay degreening by distinct mechanisms. J. Exp. Bot. 2015, 66, 6849–6862. [Google Scholar] [CrossRef]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice NON-YELLOW COLORING1 Is Involved in Light-Harvesting Complex II and Grana Degradation during Leaf Senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Schelbert, S.; Park, S.-Y.; Han, S.-H.; Lee, B.-D.; Andrès, C.B.; Kessler, F.; Hörtensteiner, S.; Paek, N.-C. STAY-GREEN and Chlorophyll Catabolic Enzymes Interact at Light-Harvesting Complex II for Chlorophyll Detoxification during Leaf Senescence in Arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 Promote ABA-Mediated Chlorophyll Degradation and Leaf Senescence by Transcriptional Activation of Chlorophyll Catabolic Genes and Senescence-Associated Genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in Arabidopsis. Plos Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Horie, Y.; Ito, H.; Kusaba, M.; Tanaka, R.; Tanaka, A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 2009, 284, 17449–17456. [Google Scholar] [CrossRef] [PubMed]
- Distelfeld, A.; Avni, R.; Fischer, A.M. Senescence, nutrient remobilization, and yield in wheat and barley. J. Exp. Bot. 2014, 65, 3783–3798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.; Tao, F.; Li, W. Lipid Profiling Demonstrates That Suppressing Arabidopsis Phospholipase Dδ Retards ABA-Promoted Leaf Senescence by Attenuating Lipid Degradation. PLoS ONE 2013, 8, e65687. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.G.; Deng, K.J.; Hu, X.Y.; Li, L.Q.; Li, X.; Chen, J.B.; Zhang, H.Y.; Tang, Z.X.; Zhang, Y.; Sun, Q.X.; et al. Chloroplast ultrastructure regeneration with protection of photosystem II is responsible for the functional ‘stay-green’ trait in wheat. Plantcell Environ. 2013, 36, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, B.; Ziolkowska, A.; Bagard, M.; Keech, O.; Gardeström, P. The impact of light intensity on shade-induced leaf senescence. Plantcell Environ. 2012, 35, 1084–1098. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Amasino, R.M. Making Sense of Senescence (Molecular Genetic Regulation and Manipulation of Leaf Senescence). Plant Physiol. 1997, 113, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, I.N.; Caputo, C.; Criado, M.V.; Funk, C. Senescence-associated proteases in plants. Physiol. Plant. 2012, 145, 130–139. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.F.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.G.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Involvement of plasma-membrane NADPH oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 2002, 215, 1022–1030. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, H.; Sun, L.; Jiao, Y.; Zhang, G.; Miao, C.; Hao, F. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 2012, 63, 305–317. [Google Scholar] [CrossRef]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.G.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dα1 and Phosphatidic Acid Regulate NADPH Oxidase Activity and Production of Reactive Oxygen Species in ABA-Mediated Stomatal Closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, Y.; Wen, F.; Yao, D.; Wang, L.; Guo, J.; Ni, L.; Zhang, A.; Tan, M.; Jiang, M. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 2014, 65, 5795–5809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, Z.-M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L.; Jones, J.D.G. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef]
- Lin, F.; Ding, H.; Wang, J.; Zhang, H.; Zhang, A.; Zhang, Y.; Tan, M.; Dong, W.; Jiang, M. Positive feedback regulation of maize NADPH oxidase by mitogen-activated protein kinase cascade in abscisic acid signalling. J. Exp. Bot. 2009, 60, 3221–3238. [Google Scholar] [CrossRef] [Green Version]
- Hung, K.T.; Kao, C.H. Nitric oxide counteracts the senescence of rice leaves induced by abscisic acid. J. Plant Physiol. 2003, 160, 871–879. [Google Scholar] [CrossRef]
- Orozco-Cárdenas, M.L.; Narváez-Vásquez, J.; Ryan, C.A. Hydrogen Peroxide Acts as a Second Messenger for the Induction of Defense Genes in Tomato Plants in Response to Wounding, Systemin, and Methyl Jasmonate. Plant Cell 2001, 13, 179–191. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Li, L.; Zhang, Z.; Wu, J.-L. Identification and Comparative Analysis of Premature Senescence Leaf Mutants in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2018, 19, 140. [Google Scholar] [CrossRef]
- Price, A.M.; Aros Orellana, D.F.; Salleh, F.M.; Stevens, R.; Acock, R.; Buchanan-Wollaston, V.; Stead, A.D.; Rogers, H.J. A Comparison of Leaf and Petal Senescence in Wallflower Reveals Common and Distinct Patterns of Gene Expression and Physiology. Plant Physiol. 2008, 147, 1898–1912. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.E.; Legge, R.L.; Barber, R.F. The role of free radicals in senescence and wounding. New Phytol. 1987, 105, 317–344. [Google Scholar] [CrossRef]
- Saini, P.; Gani, M.; Kaur, J.J.; Godara, L.C.; Singh, C.; Chauhan, S.S.; Francies, R.M.; Bhardwaj, A.; Bharat Kumar, N.; Ghosh, M.K. Reactive Oxygen Species (ROS): A Way to Stress Survival in Plants. In Abiotic Stress-Mediated Sensing and Signaling in Plants: An Omics Perspective; Zargar, S.M., Zargar, M.Y., Eds.; Springer: Singapore, 2018; pp. 127–153. [Google Scholar]
- Dwivedi, S.K.; Arora, A.; Singh, V.P. Effects of exogenously applied plant growth regulators on the physiology and anti-oxidant activity of wheat under water deficit condition. Indian J. Plant Physiol. 2018. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, K.; Wang, Z.; Zhang, H.; Gu, J.; Liu, L.; Yang, J.; Zhang, J. Brassinosteroids function in spikelet differentiation and degeneration in rice. J. Integr. Plant Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hung, K.T.; Kao, C.H. Hydrogen peroxide is necessary for abscisic acid-induced senescence of rice leaves. J. Plant Physiol. 2004, 161, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Bueno, P.; Piqueras, A.; Kurepa, J.; Savouré, A.; Verbruggen, N.; Van Montagu, M.; Inzé, D. Expression of antioxidant enzymes in response to abscisic acid and high osmoticum in tobacco BY-2 cell cultures. Plant Sci. 1998, 138, 27–34. [Google Scholar] [CrossRef]
- Qin, P.; Fan, S.; Deng, L.; Zhong, G.; Zhang, S.; Li, M.; Chen, W.; Wang, G.; Tu, B.; Wang, Y.; et al. LML1, Encoding a Conserved Eukaryotic Release Factor 1 Protein, Regulates Cell Death and Pathogen Resistance by Forming a Conserved Complex with SPL33 in Rice. Plant Cell Physiol. 2018, 59, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [Green Version]
- Hao, Q.; Yin, P.; Li, W.; Wang, L.; Yan, C.; Lin, Z.; Wu, J.Z.; Wang, J.; Yan, S.F.; Yan, N. The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins. Mol. Cell 2011, 42, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Melcher, K.; Ng, L.-M.; Zhou, X.E.; Soon, F.-F.; Xu, Y.; Suino-Powell, K.M.; Park, S.-Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009, 462, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Hitomi, K.; Arvai, A.S.; Rambo, R.P.; Hitomi, C.; Cutler, S.R.; Schroeder, J.I.; Getzoff, E.D. Structural Mechanism of Abscisic Acid Binding and Signaling by Dimeric PYR1. Science 2009, 326, 1373–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiago, J.; Dupeux, F.; Round, A.; Antoni, R.; Park, S.-Y.; Jamin, M.; Cutler, S.R.; Rodriguez, P.L.; Márquez, J.A. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 2009, 462, 665–668. [Google Scholar] [CrossRef]
- Yin, P.; Fan, H.; Hao, Q.; Yuan, X.; Wu, D.; Pang, Y.; Yan, C.; Li, W.; Wang, J.; Yan, N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 2009, 16, 1230–1236. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR Receptors Play a Major Role in Quantitative Regulation of Stomatal Aperture and Transcriptional Response to Abscisic Acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hwang, H.; Hong, J.-W.; Lee, Y.-N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.-D.; Lee, S.; Lee, S.C.; Kim, B.-G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Hao, Q.; Li, W.; Yan, C.; Yan, N.; Yin, P. Identification and Characterization of ABA Receptors in Oryza sativa. PLoS ONE 2014, 9, e95246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gan, S.-S. An Abscisic Acid-AtNAP Transcription Factor-SAG113 Protein Phosphatase 2C Regulatory Chain for Controlling Dehydration in Senescing Arabidopsis Leaves. Plant Physiol. 2012, 158, 961–969. [Google Scholar] [CrossRef]
- Zhang, K.; Xia, X.; Zhang, Y.; Gan, S.-S. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 2012, 69, 667–678. [Google Scholar] [CrossRef]
- Ma, W.; Berkowitz, G.A. Cyclic nucleotide gated channel and Ca2+-mediated signal transduction during plant senescence signaling. Plant Signal. Behav. 2011, 6, 413–415. [Google Scholar] [CrossRef]
- Fujiki, Y.; Nakagawa, Y.; Furumoto, T.; Yoshida, S.; Biswal, B.; Ito, M.; Watanabe, A.; Nishida, I. Response to Darkness of Late-Responsive Dark-Inducible Genes is Positively Regulated by Leaf Age and Negatively Regulated by Calmodulin-Antagonist-Sensitive Signalling in Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 1741–1746. [Google Scholar] [CrossRef] [Green Version]
- McCormack, E.; Tsai, Y.-C.; Braam, J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005, 10, 383–389. [Google Scholar] [CrossRef]
- Osakabe, Y.; Maruyama, K.; Seki, M.; Satou, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Leucine-Rich Repeat Receptor-Like Kinase1 Is a Key Membrane-Bound Regulator of Abscisic Acid Early Signaling in Arabidopsis. Plant Cell 2005, 17, 1105–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Sensing the environment: Key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 2013, 64, 445–458. [Google Scholar] [CrossRef]
- Nodine, M.D.; Tax, F.E. Two receptor-like kinases required together for the establishment of Arabidopsis cotyledon primordia. Dev. Biol. 2008, 314, 161–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denness, L.; McKenna, J.F.; Segonzac, C.; Wormit, A.; Madhou, P.; Bennett, M.; Mansfield, J.; Zipfel, C.; Hamann, T. Cell Wall Damage-Induced Lignin Biosynthesis Is Regulated by a Reactive Oxygen Species- and Jasmonic Acid-Dependent Process in Arabidopsis. Plant Physiol. 2011, 156, 1364–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, J.C.; Lee, I.C.; Dai, C.; Lee, Y.; Cho, H.K.; Kim, Y.; Phee, B.-K.; Kim, H.; Lee, I.H.; Choi, S.H.; et al. The Protein Trio RPK1–CaM4–RbohF Mediates Transient Superoxide Production to Trigger Age-Dependent Cell Death in Arabidopsis. Cell Rep. 2017, 21, 3373–3380. [Google Scholar] [CrossRef]
- Dai, C.; Lee, Y.; Lee, I.C.; Nam, H.G.; Kwak, J.M. Calmodulin 1 Regulates Senescence and ABA Response in Arabidopsis. Front. Plant Sci. 2018, 9, 803. [Google Scholar] [CrossRef]
- Balazadeh, S.; Riaño-Pachón, D.M.; Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 2008, 10, 63–75. [Google Scholar] [CrossRef]
- Ren, T.; Wang, J.; Zhao, M.; Gong, X.; Wang, S.; Wang, G.; Zhou, C. Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta 2018, 247, 53–68. [Google Scholar] [CrossRef]
- Koyama, T. The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate Feed-Forward Regulation of Age-Dependent Cell Death Involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.-P.; Mueller-Roeber, B. ORS1, an H2O2-Responsive NAC Transcription Factor, Controls Senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC Gene Regulating Senescence Improves Grain Protein, Zinc, and Iron Content in Wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Derkx, A.P.; Liu, D.-C.; Buchner, P.; Hawkesford, M.J. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 2015, 17, 904–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Huang, W.; Liu, L.; Chen, T.; Zhou, F.; Lin, Y. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 2013, 13, 132. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [Green Version]
- Tweneboah, S.; Oh, S.-K. Biological roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in solanaceous crops. J. Plant Biotechnol. 2017, 44, 1–11. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.-I.; Köhler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Y.; Turečková, V.; Xue, G.-P.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. The NAC Transcription Factor SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiol. 2018, 177, 1286–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Worley, E.; Udvardi, M. A NAP-AAO3 Regulatory Module Promotes Chlorophyll Degradation via ABA Biosynthesis in Arabidopsis Leaves. Plant Cell 2014, 26, 4862–4874. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Watkins, C.B.; Gan, S.-S. Arabidopsis AtNAP regulates fruit senescence. J. Exp. Bot. 2012, 63, 6139–6147. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Bibi, N.; Gan, S.; Li, F.; Yuan, S.; Ni, M.; Wang, M.; Shen, H.; Wang, X. A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J. Exp. Bot. 2015, 66, 4669–4682. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Jeong, J.; Kang, M.-Y.; Kim, J.; Paek, N.-C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [Green Version]
- Zong, W.; Tang, N.; Yang, J.; Peng, L.; Ma, S.; Xu, Y.; Li, G.; Xiong, L. Feedback Regulation of ABA Signaling and Biosynthesis by a bZIP Transcription Factor Targets Drought-Resistance-Related Genes. Plant Physiol. 2016, 171, 2810–2825. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a Key Player of the Basic Leucine Zipper Transcription Factor Family for Conferring Abscisic Acid Sensitivity and Salinity and Drought Tolerance in Rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef] [Green Version]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive Activation of Transcription Factor OsbZIP46 Improves Drought Tolerance in Rice. Plant Physiol. 2012, 158, 1755–1768. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R. Abscisic Acid synthesis and response. Arab. Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, Q.; Guo, Y.; Yang, J.; Wang, M.; Duan, X.; Niu, J.; Liu, S.; Zhang, J.; Lu, Y.; et al. Arabidopsis subtilase SASP is involved in the regulation of ABA signaling and drought tolerance by interacting with OPEN STOMATA 1. J. Exp. Bot. 2018, 69, 4403–4417. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.E.; Borniego, M.L.; Battchikova, N.; Aro, E.-M.; Tyystjärvi, E.; Guiamét, J.J. SASP, a Senescence-Associated Subtilisin Protease, is involved in reproductive development and determination of silique number in Arabidopsis. J. Exp. Bot. 2015, 66, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Raab, S.; Drechsel, G.; Zarepour, M.; Hartung, W.; Koshiba, T.; Bittner, F.; Hoth, S. Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J. 2009, 59, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef] [Green Version]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR Signaling and Nutrient Sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef]
- Schepetilnikov, M.; Makarian, J.; Srour, O.; Geldreich, A.; Yang, Z.; Chicher, J.; Hammann, P.; Ryabova, L.A. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J. 2017, 36, 886–903. [Google Scholar] [CrossRef] [Green Version]
- Schepetilnikov, M.; Dimitrova, M.; Mancera-Martínez, E.; Geldreich, A.; Keller, M.; Ryabova, L.A. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013, 32, 1087–1102. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhu, J.-Y.; Roh, J.; Marchive, C.; Kim, S.-K.; Meyer, C.; Sun, Y.; Wang, W.; Wang, Z.-Y. TOR Signaling Promotes Accumulation of BZR1 to Balance Growth with Carbon Availability in Arabidopsis. Curr. Biol. 2016, 26, 1854–1860. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.-C.; Liu, X.; Fu, L.; Hou, Y.-J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2018, 69, 100–112.e6. [Google Scholar] [CrossRef]
- Cheng, W.-H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.-C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. A Unique Short-Chain Dehydrogenase/Reductase in Arabidopsis Glucose Signaling and Abscisic Acid Biosynthesis and Functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, M.R.; Feurtado, J.A.; Huang, D.; Lu, Y.; Cutler, A.J. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013, 13, 192. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cheng, J.; Zhuang, Y.; Ye, L.; Li, Z.; Wang, Y.; Qi, M.; Xu, L.; Zhang, Y. Polycomb repressive complex 2 attenuates ABA-induced senescence in Arabidopsis. Plant J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xu, M.; Koo, Y.; He, J.; Poethig, R.S. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2013, 2, e00260. [Google Scholar] [CrossRef] [PubMed]
- Wingler, A.; Purdy, S.; MacLean, J.A.; Pourtau, N. The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 2006, 57, 391–399. [Google Scholar] [CrossRef]
- Wingler, A.; Delatte, T.L.; O’Hara, L.E.; Primavesi, L.F.; Jhurreea, D.; Paul, M.J.; Schluepmann, H. Trehalose 6-Phosphate Is Required for the Onset of Leaf Senescence Associated with High Carbon Availability. Plant Physiol. 2012, 158, 1241–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balibrea Lara, M.E.; Gonzalez Garcia, M.-C.; Fatima, T.; Ehneß, R.; Lee, T.K.; Proels, R.; Tanner, W.; Roitsch, T. Extracellular Invertase Is an Essential Component of Cytokinin-Mediated Delay of Senescence. Plant Cell 2004, 16, 1276–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Ni, D.-A.; Ruan, Y.-L. Posttranslational Elevation of Cell Wall Invertase Activity by Silencing Its Inhibitor in Tomato Delays Leaf Senescence and Increases Seed Weight and Fruit Hexose Level. Plant Cell 2009, 21, 2072–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, G.; David-Schwartz, R.; Sade, N.; Moshelion, M.; Levi, A.; Alchanatis, V.; Granot, D. The Pitfalls of Transgenic Selection and New Roles of AtHXK1: A High Level of AtHXK1 Expression Uncouples Hexokinase1-Dependent Sugar Signaling from Exogenous Sugar. Plant Physiol. 2012, 159, 47–51. [Google Scholar] [CrossRef]
- Tsai, A.Y.-L.; Gazzarrini, S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012, 69, 809–821. [Google Scholar] [CrossRef]
- Wingler, A. Transitioning to the Next Phase: The Role of Sugar Signaling throughout the Plant Life Cycle. Plant Physiol. 2018, 176, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Sasaki, Y.; Li, X.; Mori, I.C.; Matsuura, T.; Hirayama, T.; Sato, T.; Yamaguchi, J. ABI1 regulates carbon/nitrogen-nutrient signal transduction independent of ABA biosynthesis and canonical ABA signalling pathways in Arabidopsis. J. Exp. Bot. 2015, 66, 2763–2771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Maekawa, S.; Yasuda, S.; Domeki, Y.; Sueyoshi, K.; Fujiwara, M.; Fukao, Y.; Goto, D.B.; Yamaguchi, J. Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J. 2011, 68, 137–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
 indicate induction and
indicate induction and  indicates suppression). Abiotic stresses induce ABA production, which is sensed by ABA signaling receptors (ABF, PYL). ABA signal induce expression of NAC, bZIP and NYC TFs. The activated TFs induce expression of SAGs which are translated in RBs (ribosomes) to synthesize (NOL, NADPH oxidase, Chl-b reductase). NDPH oxidase increase ROS production and Chl b reductase reduce PSII (photosystem II) efficiency and induce the degradation of D1 protein.
indicates suppression). Abiotic stresses induce ABA production, which is sensed by ABA signaling receptors (ABF, PYL). ABA signal induce expression of NAC, bZIP and NYC TFs. The activated TFs induce expression of SAGs which are translated in RBs (ribosomes) to synthesize (NOL, NADPH oxidase, Chl-b reductase). NDPH oxidase increase ROS production and Chl b reductase reduce PSII (photosystem II) efficiency and induce the degradation of D1 protein.
 indicate induction and
indicate induction and  indicates suppression). Abiotic stresses induce ABA production, which is sensed by ABA signaling receptors (ABF, PYL). ABA signal induce expression of NAC, bZIP and NYC TFs. The activated TFs induce expression of SAGs which are translated in RBs (ribosomes) to synthesize (NOL, NADPH oxidase, Chl-b reductase). NDPH oxidase increase ROS production and Chl b reductase reduce PSII (photosystem II) efficiency and induce the degradation of D1 protein.
indicates suppression). Abiotic stresses induce ABA production, which is sensed by ABA signaling receptors (ABF, PYL). ABA signal induce expression of NAC, bZIP and NYC TFs. The activated TFs induce expression of SAGs which are translated in RBs (ribosomes) to synthesize (NOL, NADPH oxidase, Chl-b reductase). NDPH oxidase increase ROS production and Chl b reductase reduce PSII (photosystem II) efficiency and induce the degradation of D1 protein.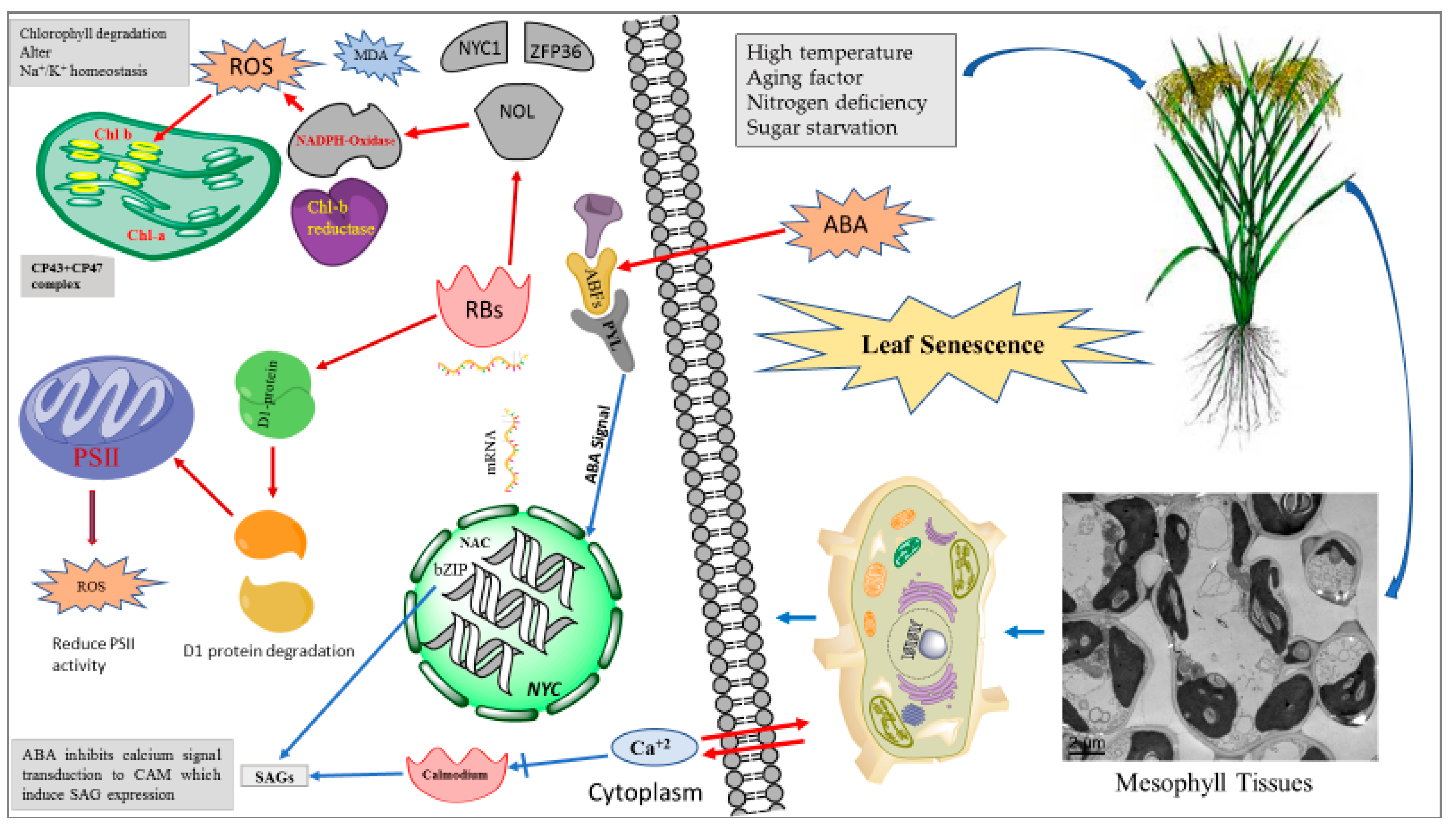
| S. No | Genotype | Mutation | Treatment | Transgene | Effect | Reference |
|---|---|---|---|---|---|---|
| 1 | Arabidopsis | − | Cold 4 °C, 400 mM NaCl, 500 mM mannitol, 10 mM H2O2 in 7-day-old plants | eas1-1 | Accelerated leaf senescence by reducing Ca2+ concentration | [54] |
| 2 | Rice | EMS | Darkness and 200 µM exogenous ABA for 5 days | psl15, psl50, psl85, psl89, psl117 and psl270 | Induced early and premature leaf senescence, increased malondialdehyde content | [80] |
| 3 | Rice | + | nyc1 | [57] | ||
| 4 | Rice | + | 5 d dark, exogenous ABA in detached leaves (4 µM) | OsNAC2-OX | Induce leaf senescence (4-week-old + grain filling) | [12] |
| 5 | Rice | − | 5-d dark treatment, exogenous ABA in detached leaves (4 µM) | OsNAC2-RNAi18 | Delayed senescence (4-week-old + grain filling) | [12] |
| 6 | Rice | + | ps1-D | Promoted premature leaf senescence | [119] | |
| 7 | Rice | − | OsNAP | Delayed leaf senescence | [119] | |
| 8 | Rice | EMS | 10 days dark treatment in detached leaves, exogenous ABA to detached leaves after 10 day of flowering | bml | Promoted leaf senescence by reducing chlorophyll contents | [22] |
| 9 | Arabidopsis | + | Drought/exogenous 100 µM ABA | pRD29A:PYL9 | Induced resistance to drought and accelerated ABA-induced leaf senescence | [52] |
| 10 | Arabidopsis | + | Exogenous 100 µM ABA in 4-week-old detached leaves | abf2abf3abf4 (triple mutant | Delayed leaf senescence by blocking signal for chlorophyll degradation | [60] |
| 11 | Arabidopsis | + | Exogenous 100 µM ABA in 4-week-old detached leaves | snrk2.2/2.3/2.6 (Triple mutant) | Inhibited chlorophyll degradation with stay-green phenotype | [60] |
| 12 | Arabidopsis | + | Exogenous 100 µM ABA in 4-week-old detached leaves | acd1-20, nyc1-1 | Inhibited signaling in Chl and LHC (light harvesting complex) degradation pathways | [60] |
| 13 | Arabidopsis | + | Exogenous 50 µM ABA in 3-week-old detached leaves | CaM1 | Triggered the accumulation of ROS and SAG12 expression | [108] |
| 14 | Arabidopsis | − | Exogenous 50 µM ABA in 3-week-old detached leaves | amiRNA-CaM1 | Delayed leaf senescence | [108] |
| 15 | Arabidopsis | − | 100 µM ABA of 20 DAG leaves for 20 h | SAG113 | Exhibited delayed leaf senescence | [99] |
| 16 | Arabidopsis | + | Drought stress at 18 DAG, dark-induced treatment at 30 DAG for 7 days | OxMYBR1 | Delayed leaf senescence with strong holding capacity | [144] |
| 17 | Arabidopsis | − | Drought stress at 18 DAG, dark-induced treatment at 30 DAG for 7 days | mybr1 | Reduced water loss, more rapid chlorophyll loss, and induced leaf senescence | [144] |
| 18 | Rice | − | 3 μM exogenous ABA on 4-week-old detached leaves | osrl3 | Showed ABA insensitivity and stay-green phenotype | [1] |
| 19 | Arabidopsis | − | 50 μM exogenous ABA for 3 days | Sasp | Enhanced leaf senescence by increasing SAG12 expression and ROS production | [136] |
| 20 | Arabidopsis | − | 50 μm exogenous ABA for 8 days after 10 DAG | clf-50 swn-1 | Induced leaf senescence | [145] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asad, M.A.U.; Zakari, S.A.; Zhao, Q.; Zhou, L.; Ye, Y.; Cheng, F. Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants. Int. J. Mol. Sci. 2019, 20, 256. https://doi.org/10.3390/ijms20020256
Asad MAU, Zakari SA, Zhao Q, Zhou L, Ye Y, Cheng F. Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants. International Journal of Molecular Sciences. 2019; 20(2):256. https://doi.org/10.3390/ijms20020256
Chicago/Turabian StyleAsad, Muhammad Asad Ullah, Shamsu Ado Zakari, Qian Zhao, Lujian Zhou, Yu Ye, and Fangmin Cheng. 2019. "Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants" International Journal of Molecular Sciences 20, no. 2: 256. https://doi.org/10.3390/ijms20020256
APA StyleAsad, M. A. U., Zakari, S. A., Zhao, Q., Zhou, L., Ye, Y., & Cheng, F. (2019). Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants. International Journal of Molecular Sciences, 20(2), 256. https://doi.org/10.3390/ijms20020256




