Chromosome Conformation Capture Reveals Two Elements That Interact with the PTBP3 (ROD1) Transcription Start Site
Abstract
1. Introduction
2. Results
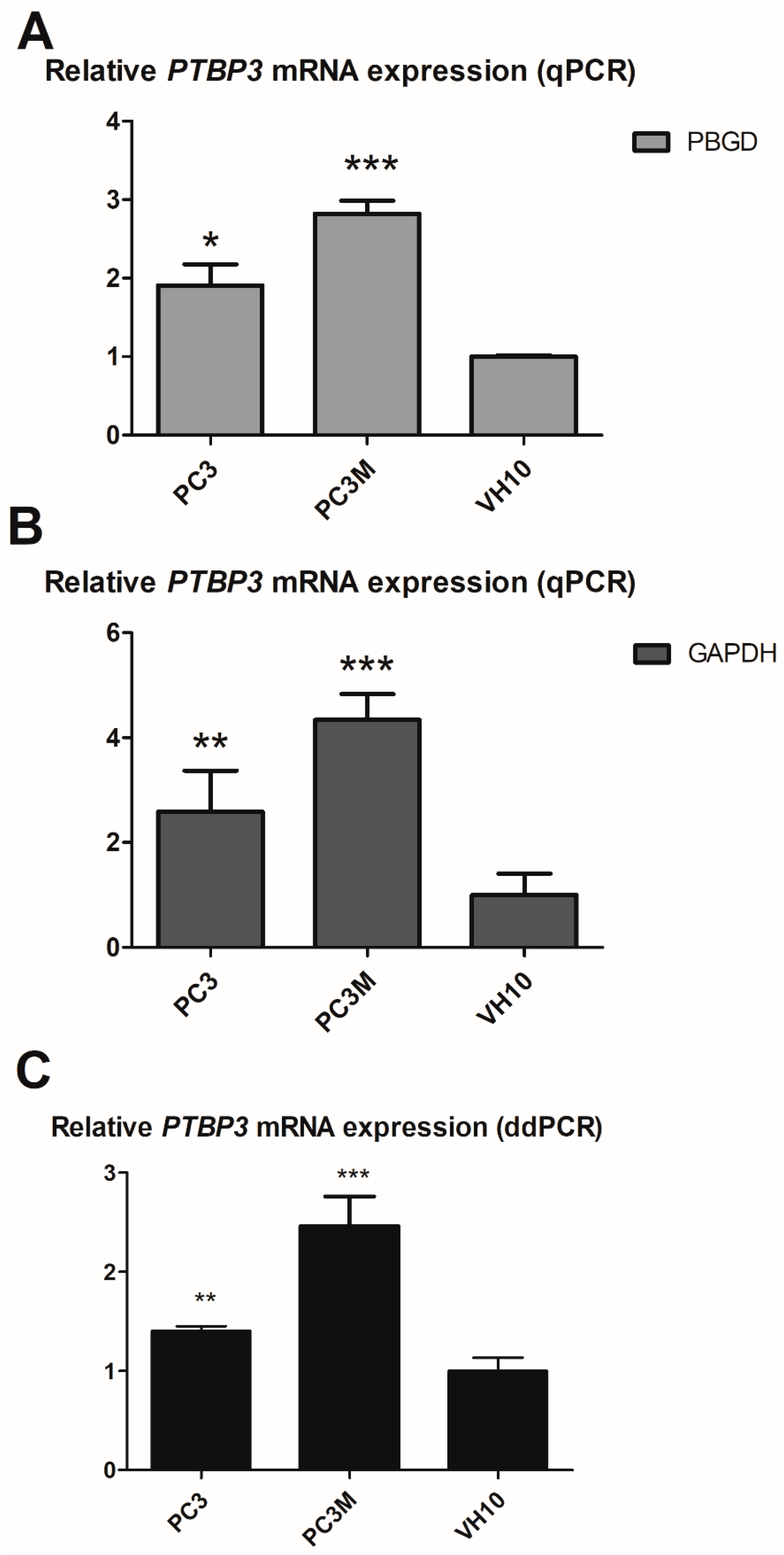
2.1. Levels of PTBP3 mRNA in Prostate Cancer Cell Lines
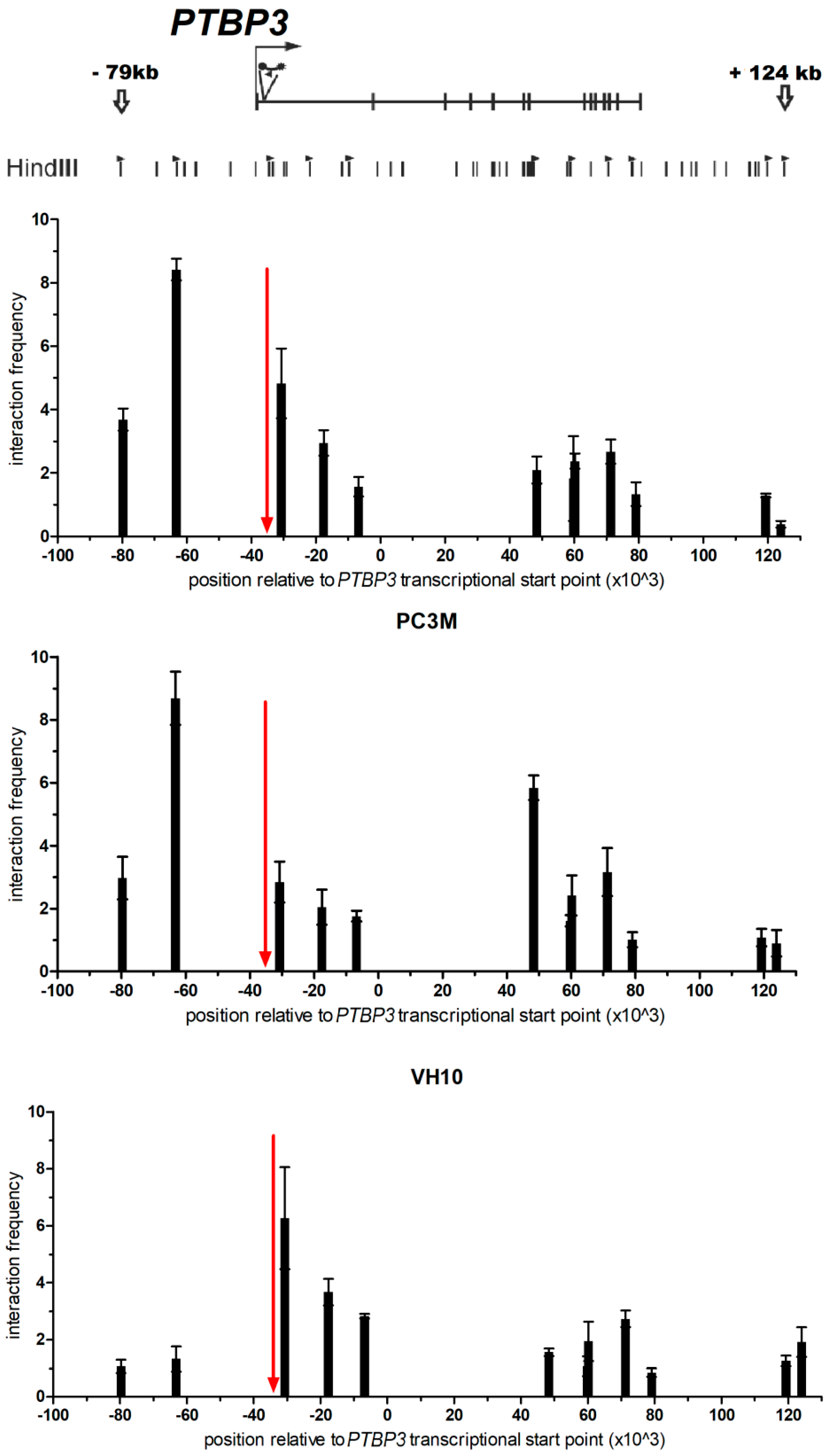
2.2. Looping Organization of the PTBP3 Locus in Prostate Cancer Cell Lines
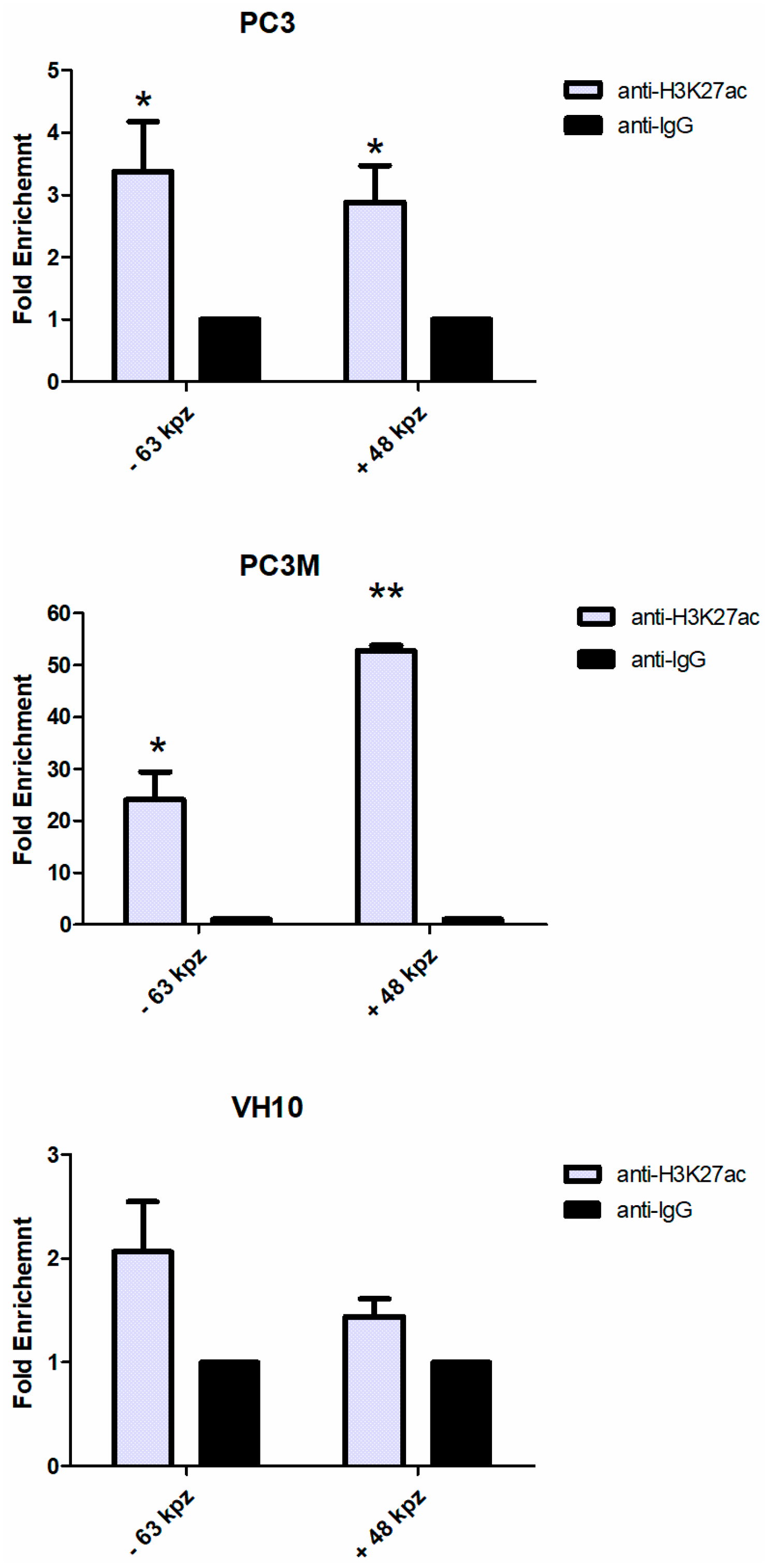
2.3. The −63 kbp and +48 kbp Regions of the PTBP3 Promoter are Open Chromatin Regions
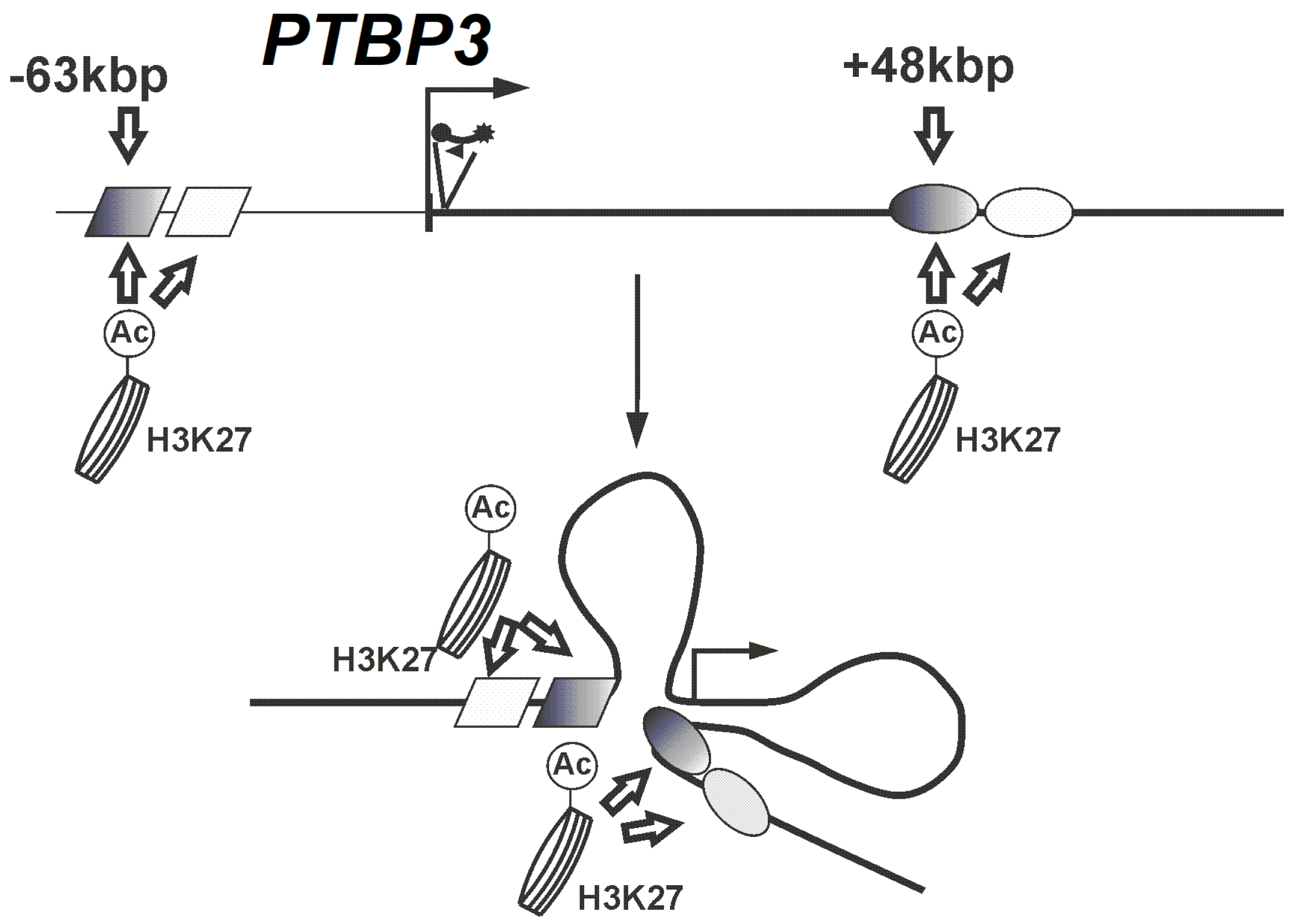
3. Discussion
4. Materials and Methods
4.1. Cell Line Cultures
4.2. RNA Isolation and cDNA Synthesis
4.3. Real-time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
4.4. Digital Droplet Polymerase Chain Reaction (ddPCR)
4.5. Gene Expression Databases
4.6. Chromosome Conformation Capture (3C)
4.7. Chromatin Immunoprecipitation (ChIP)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Narlikar, G.J.; Fan, H.-Y.; Kingston, R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell 2002, 108, 475–487. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Ceriani, M.C.; Capaccioli, S.; Nicolin, A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 2003, 195, 356–372. [Google Scholar] [CrossRef] [PubMed]
- Stoneley, M.; Willis, A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene 2004, 23, 3200–3207. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Campbell, M.J. The Interactions of microRNA and Epigenetic Modifications in Prostate Cancer. Cancers 2013, 5, 998–1019. [Google Scholar] [CrossRef] [PubMed]
- Ballestar, E.; Esteller, M. The impact of chromatin in human cancer: Linking DNA methylation to gene silencing. Carcinogenesis 2002, 23, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Dekker, J. Gene regulation in the third dimension. Science 2008, 319, 1793–1794. [Google Scholar] [CrossRef]
- Simonis, M.; Klous, P.; Splinter, E.; Moshkin, Y.; Willemsen, R.; de Wit, E.; van Steensel, B.; de Laat, W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 2006, 38, 1348–1354. [Google Scholar] [CrossRef]
- Zhao, Z.; Tavoosidana, G.; Sjölinder, M.; Göndör, A.; Mariano, P.; Wang, S.; Kanduri, C.; Lezcano, M.; Sandhu, K.S.; Singh, U.; et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006, 38, 1341–1347. [Google Scholar] [CrossRef]
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Kolovos, P.; van de Werken, H.J.; Kepper, N.; Zuin, J.; Brouwer, R.W.; Kockx, C.E.; Wendt, K.S.; van IJcken, W.F.; Grosveld, F.; Knoch, T.A. Targeted Chromatin Capture (T2C): A novel high resolution high throughput method to detect genomic interactions and regulatory elements. Epigenet. Chromatin 2014, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.R.; Roberts, N.; McGowan, S.; Hay, D.; Giannoulatou, E.; Lynch, M.; De Gobbi, M.; Taylor, S.; Gibbons, R.; Higgs, D.R. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 2014, 46, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Kubiak, M.; Lewandowska, M.A. Can chromatin conformation technologies bring light into human molecular pathology? Acta Biochim. Pol. 2015, 62, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Spellman, R.; Rideau, A.; Matlin, A.; Gooding, C.; Robinson, F.; McGlincy, N.; Grellscheid, S.N.; Southby, J.; Wollerton, M.; Smith, C.W.J. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005, 33, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Sawicka, K.; Bushell, M.; Spriggs, K.A.; Willis, A.E. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008, 36, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Wollerton, M.C.; Gooding, C.; Wagner, E.J.; Garcia-Blanco, M.A.; Smith, C.W.J. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 2004, 13, 91–100. [Google Scholar] [CrossRef]
- Suckale, J.; Wendling, O.; Masjkur, J.; Jäger, M.; Münster, C.; Anastassiadis, K.; Stewart, A.F.; Solimena, M. PTBP1 Is Required for Embryonic Development before Gastrulation. PLoS ONE 2011, 6, e16992. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Tsukahara, K.; Kanaoka, Y.; Jinno, S.; Okayama, H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 1999, 19, 3829–3841. [Google Scholar] [CrossRef]
- Brazão, T.F.; Demmers, J.; van IJcken, W.; Strouboulis, J.; Fornerod, M.; Romão, L.; Grosveld, F.G. A new function of ROD1 in nonsense-mediated mRNA decay. FEBS Lett. 2012, 586, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Spellman, R.; Llorian, M.; Smith, C.W.J. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell 2007, 27, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Ott, C.J.; Blackledge, N.P.; Leir, S.-H.; Harris, A. Novel regulatory mechanisms for the CFTR gene. Biochem. Soc. Trans. 2009, 37, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ott, C.J.; Lewandowska, M.A.; Leir, S.-H.; Harris, A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell. Mol. Med. 2012, 16, 1321–1330. [Google Scholar] [CrossRef] [PubMed]
- Gheldof, N.; Smith, E.M.; Tabuchi, T.M.; Koch, C.M.; Dunham, I.; Stamatoyannopoulos, J.A.; Dekker, J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010, 38, 4325–4336. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wen, F.; Wiley, G.B.; Kinter, M.T.; Gaffney, P.M. An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression. PLoS Genet. 2013, 9, e1003750. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, S.E.; Wu, Q.; Barutcu, A.R.; Xiao, H.; Ohkawa, Y.; Imbalzano, A.N. The PPARγ locus makes long-range chromatin interactions with selected tissue-specific gene loci during adipocyte differentiation in a protein kinase A dependent manner. PLoS ONE 2014, 9, e86140. [Google Scholar] [CrossRef] [PubMed]
- Ooi, A.T.; Gower, A.C.; Zhang, K.X.; Vick, J.L.; Hong, L.; Nagao, B.; Wallace, W.D.; Elashoff, D.A.; Walser, T.C.; Dubinett, S.M.; et al. Molecular profiling of premalignant lesions in lung squamous cell carcinomas identifies mechanisms involved in stepwise carcinogenesis. Cancer Prev. Res. 2014, 7, 487–495. [Google Scholar] [CrossRef]
- Liang, X.; Shi, H.; Yang, L.; Qiu, C.; Lin, S.; Qi, Y.; Li, J.; Zhao, A.; Liu, J. Inhibition of polypyrimidine tract-binding protein 3 induces apoptosis and cell cycle arrest, and enhances the cytotoxicity of 5- fluorouracil in gastric cancer cells. Br. J. Cancer 2017, 116, 903–911. [Google Scholar] [CrossRef]
- Ernst, J.; Kheradpour, P.; Mikkelsen, T.S.; Shoresh, N.; Ward, L.D.; Epstein, C.B.; Zhang, X.; Wang, L.; Issner, R.; Coyne, M.; et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011, 473, 43–49. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, W. Enhancers, enhancers—From their discovery to today’s universe of transcription enhancers. Biol. Chem. 2015, 396, 311–327. [Google Scholar] [CrossRef] [PubMed]
- Ashoor, H.; Kleftogiannis, D.; Radovanovic, A.; Bajic, V.B. DENdb: Database of integrated human enhancers. Database 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Kellis, M. ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 2012, 9, 215–216. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.M.; Buske, O.J.; Wang, J.; Weng, Z.; Bilmes, J.A.; Noble, W.S. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods 2012, 9, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, N.; Xie, W.; Li, Y.; Wagner, U.; Wang, W.; Stamatoyannopoulos, J.; Ernst, J.; Kellis, M.; Ren, B. RFECS: A random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput. Biol. 2013, 9, e1002968. [Google Scholar] [CrossRef]
- Firpi, H.A.; Ucar, D.; Tan, K. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics 2010, 26, 1579–1586. [Google Scholar] [CrossRef]
- Landt, S.G.; Marinov, G.K.; Kundaje, A.; Kheradpour, P.; Pauli, F.; Batzoglou, S.; Bernstein, B.E.; Bickel, P.; Brown, J.B.; Cayting, P.; et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012, 22, 1813–1831. [Google Scholar] [CrossRef]
- Miele, A.; Dekker, J. Mapping cis- and trans-chromatin interaction networks using chromosome conformation capture (3C). Methods Mol. Biol. 2009, 464, 105–121. [Google Scholar]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef]
- Jia, R.; Chai, P.; Zhang, H.; Fan, X. Novel insights into chromosomal conformations in cancer. Mol. Cancer 2017, 16, 173. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhang, C.; Shen, Y.; Nephew, K.P.; Wang, Q. Androgen receptor-driven chromatin looping in prostate cancer. Trends Endocrinol. Metab. 2011, 22, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Carroll, J.S.; Brown, M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 2005, 19, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Kaneshiro, K.; Tsutsumi, S.; Horie-Inoue, K.; Ikeda, K.; Urano, T.; Ijichi, N.; Ouchi, Y.; Shirahige, K.; Aburatani, H.; et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene 2007, 26, 4453–4463. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, W.; Liu, X.S.; Carroll, J.S.; Jänne, O.A.; Keeton, E.K.; Chinnaiyan, A.M.; Pienta, K.J.; Brown, M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell 2007, 27, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, C.; Wu, D.; Chen, H.; Rorick, A.; Zhang, X.; Wang, Q. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011, 30, 2405–2419. [Google Scholar] [CrossRef] [PubMed]
- Home-GEO-NCBI. Available online: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 23 July 2018).
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Hagège, H.; Klous, P.; Braem, C.; Splinter, E.; Dekker, J.; Cathala, G.; de Laat, W.; Forné, T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2007, 2, 1722–1733. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubiak, M.; Jurek, A.; Kamińska, K.; Kowalewski, J.; Huang, S.; Lewandowska, M.A. Chromosome Conformation Capture Reveals Two Elements That Interact with the PTBP3 (ROD1) Transcription Start Site. Int. J. Mol. Sci. 2019, 20, 242. https://doi.org/10.3390/ijms20020242
Kubiak M, Jurek A, Kamińska K, Kowalewski J, Huang S, Lewandowska MA. Chromosome Conformation Capture Reveals Two Elements That Interact with the PTBP3 (ROD1) Transcription Start Site. International Journal of Molecular Sciences. 2019; 20(2):242. https://doi.org/10.3390/ijms20020242
Chicago/Turabian StyleKubiak, Marta, Anna Jurek, Katarzyna Kamińska, Janusz Kowalewski, Sui Huang, and Marzena Anna Lewandowska. 2019. "Chromosome Conformation Capture Reveals Two Elements That Interact with the PTBP3 (ROD1) Transcription Start Site" International Journal of Molecular Sciences 20, no. 2: 242. https://doi.org/10.3390/ijms20020242
APA StyleKubiak, M., Jurek, A., Kamińska, K., Kowalewski, J., Huang, S., & Lewandowska, M. A. (2019). Chromosome Conformation Capture Reveals Two Elements That Interact with the PTBP3 (ROD1) Transcription Start Site. International Journal of Molecular Sciences, 20(2), 242. https://doi.org/10.3390/ijms20020242



