Insulin-Like Growth Factor 2 (IGF2) Signaling in Colorectal Cancer—From Basic Research to Potential Clinical Applications
Abstract
1. Introduction
2. The IGF System in Carcinogenesis
IGF2 as an Enigmatic Component of IGF System—Role in Physiology
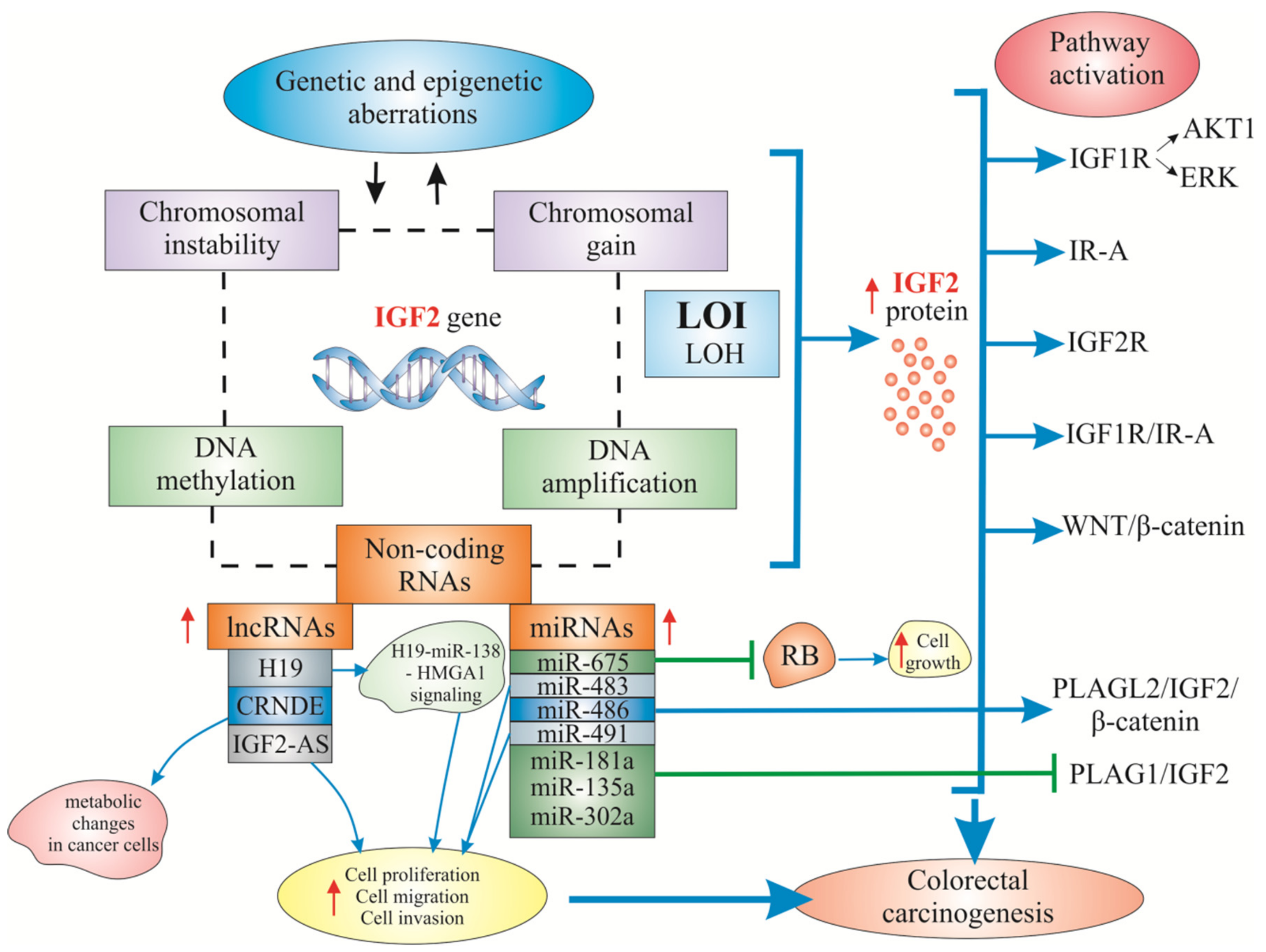
3. Known and Less Known Alterations of IGF2 in Colorectal Carcinogenesis
3.1. Autocrine/Paracrine IGF2 Secretion
3.2. Mechanisms of Increased IGF2 Gene/Protein Activities
3.3. IGF2 Actions Relevant to CRC Development
3.4. Epidemiological Evidence of Circulating IGF2 Association with CRC Risk
3.5. Tissue Expression of IGF2 in Primary and Metastatic CRC
3.6. Non-coding RNAs Regulated by IGF2 in CRC
4. New Targets in Anti-IGF2 Colorectal Cancer Therapy
4.1. Cell Line Research
4.2. CRC Xenograft Model Research
5. Concluding Remarks and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| A | Adenine |
| AKT1 | AKT serine/threonine kinase 1 |
| ALS | Acid-labile subunit |
| APC | Adenomatous polyposis coli |
| AS | Antisense |
| BRAF | Proto-oncogene B-Raf and v-Raf murine sarcoma viral oncogene homolog B |
| BRCA2 | Breast cancer 2 gene |
| C | Cytosine |
| CACNA1G | Calcium channel, voltage-dependent, T type alpha-1G subunit |
| CAFs | Cancer-associated fibroblasts |
| CA19-9 | Carbohydrate antigen 19-9 (cancer antigen 19-9) |
| CEA | Carcinoembryonic antigen |
| CDK1 | Cyclin-dependent kinase 1 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A |
| CEACAM6 | CEA related cell adhesion molecule 6 gene/protein |
| ceRNA | Competing endogenous RNA |
| CI | Confidence interval |
| CIMP | CpG island methylator phenotype |
| CIN | Chromosomal instability |
| CMS | Consensum molecular subtype |
| CRABP1 | Cellular retinoic acid-binding protein 1 |
| CRC | Colorectal cancer |
| CRIS | CRC intrinsic subtype |
| CRND | Colorectal neoplasia differentially expressed |
| CSCs | Cancer stem cells |
| CTCF | CCCTC-Binding Factor (Zinc Finger Protein) |
| DEGs | Differentially expressed genes |
| DMR | Differentially methylated region |
| ECM | Extracellular matrix |
| EGFR | Epidermal Growth Factor Receptor; HER1 in humans |
| eIF4A3 | Eukaryotic initiation factor 4A-III |
| ERBB2 | Erb-B2 Receptor Tyrosine Kinase |
| FSCN1 | Fascin Actin-Bundling Protein 1 |
| G | Guanine |
| GF | Growth factor |
| GH | Growth hormone |
| HMGA1 | High Mobility Group Protein A1 |
| hMLH1 | Human mutL homolog 1 |
| hsa-mir-483 | Stem-loop sequence-micro-RNA-483 |
| HyMiD | Hypermethylation of multiple iDMRs |
| iDMR | Imprinting-associated differentially methylated region |
| IGF1, -2 | Insulin growth factor 1, -2 |
| IGF1R, 2R | IGF Receptor type I, type II |
| IGF2-AS | IGF2 antisense |
| IGF2R/CI-M6PR | IGF2R/Cation-Independent Mannose-6-phosphate Receptor |
| IGFBPs | IGF binding proteins |
| IMPs | IGF2 mRNA-binding Proteins |
| INPP4B | Inositol polyphosphate-4-phosphatase, type II |
| INSR/InsR-A | Insulin receptor/IR isoform-A |
| IR | Insulin Receptor |
| IRS2 | Insulin receptor substrate 2 |
| KRAS | Proto-oncogene K-ras or Ki-ras; from Kirsten Rat Sarcoma Virus |
| lncRNAs | Long non-coding RNAs |
| LOH | Loss of heterozygosity |
| LOI | Loss of imprinting |
| MAPK | A mitogen-activated protein kinase |
| MEK1/2i | Mitogen-activated protein kinase (MAP2K, MEK, MAPKK) inhibitor |
| MLH1 | MutL homolog 1 |
| MMR | (DNA) mismatch repair |
| mRNA | Messenger RNA |
| miRNA | MicroRNA |
| MMP2 | Matrix Metallopeptidase 2, matrix metalloproteinase (MMP) gene family |
| MSI | Microsatellite instability |
| MSS | Microsatellite stable |
| mTORC1/2i | Mammalian Target of Rapamycin Complex 1/2 inhibitor |
| NEUROG1 | Neurogenin 1 |
| NICTH | Non-islet cell tumor hypoglycaemia |
| PCNA | Proliferating cell nuclear antigen |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| PLAGL2 | Pleomorphic adenoma gene-like 2 |
| PRC2 | Polycomb repressive complex 2 |
| PTEN | Phosphatase and tensin homolog deleted on chromosome ten |
| RB | Retinoblastoma |
| RUNX3 | Runt-related transcription factor 3 |
| SCNA | (DNA) Somatic copy number alteration |
| shRNA | Short hairpin RNA |
| sIGF2R | Soluble IGF Receptor type II |
| SOCS1 | Suppressor of cytokine signaling 1 |
| SSM | Stem/serrated/mesenchymal |
| TCGA | The Cancer Genome Atlas |
| TGF-α, β | Tumor Growth Factor α, β |
| T2DM | Type II Diabetes Mellitus |
| TNM | Tumor-Node-Metastasis |
| TP53 | Tumor gene/protein 53 |
| UTR | Untranslated region |
| VEGF-A | Vascular endothelial growth factor A |
| WNT1 | Wnt (gene wingless + integrated or int-1) Family Member 1 |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Nadal, C.; Maurel, J.; Gascon, P. Is there a genetic signature for liver metastasis in colorectal cancer? World J. Gastroenterol. 2007, 13, 5832–5844. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Chun, Y.S.; Kopetz, S.E.; Vauthey, J.N. Biomarkers in colorectal liver metastases. Br. J. Surg. 2018, 105, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Müller, M.F.; Ibrahim, A.E.; Arends, M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016, 469, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Molinari, C.; Marisi, G.; Passardi, A.; Matteucci, L.; De Maio, G.; Ulivi, P. Heterogeneity in Colorectal Cancer: A Challenge for Personalized Medicine? Int. J. Mol. Sci. 2018, 19, 3733. [Google Scholar] [CrossRef]
- Hühns, M.; Krohn, S.; Murua Escobar, H.; Prall, F. Genomic heterogeneity in primary colorectal carcinomas and their metastases: Born bad or brought up a villain? Hum. Pathol. 2018, 74, 54–63. [Google Scholar] [CrossRef]
- Puccini, A.; Berger, M.D.; Naseem, M.; Tokunaga, R.; Battaglin, F.; Cao, S.; Hanna, D.L.; McSkane, M.; Soni, S.; Zhang, W.; et al. Colorectal cancer: Epigenetic alterations and their clinical implications. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.R.; Chang, D.K. Colorectal cancer in inflammatory bowel disease: The risk, pathogenesis, prevention and diagnosis. World J. Gastroenterol. 2014, 7, 9872–9881. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Huo, T.; Canepa, R.; Sura, A.; Modave, F.; Gong, Y. Colorectal cancer stages transcriptome analysis. PLoS ONE 2017, 12, e0188697. [Google Scholar] [CrossRef]
- Zhang, Z.; Qian, W.; Wang, S.; Ji, D.; Wang, Q.; Li, J.; Peng, W.; Gu, J.; Hu, T.; Ji, B.; et al. Analysis of lncRNA-Associated ceRNA Network Reveals Potential lncRNA Biomarkers in Human Colon Adenocarcinoma. Cell Physiol. Biochem. 2018, 49, 1778–1791. [Google Scholar] [CrossRef] [PubMed]
- Isella, C.; Terrasi, A.; Bellomo, S.E.; Petti, C.; Galatola, G.; Muratore, A.; Mellano, A.; Senetta, R.; Cassenti, A.; Sonetto, C.; et al. Stromal contribution to the colorectal cancer transcriptome. Nat. Genet. 2015, 47, 312–329. [Google Scholar] [CrossRef]
- Isella, C.; Brundu, F.; Bellomo, S.E.; Galimi, F.; Zanella, E.; Porporato, R.; Petti, C.; Fiori, A.; Orzan, F.; Senetta, R.; et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat. Commun. 2017, 8, 15107. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Hagland, H.R.; Berg, M.; Jolma, I.W.; Carlsen, A.; Søreide, K. Molecular pathways and cellular metabolism in colorectal cancer. Dig. Surg. 2013, 30, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Lopez, G.; Boggio, F.; Ferrero, S.; Fusco, N.; Del Gobbo, A. Molecular and Immunohistochemical Markers with Prognostic and Predictive Significance in Liver Metastases from Colorectal Carcinoma. Int. J. Mol. Sci. 2018, 19, 3014. [Google Scholar] [CrossRef]
- Goldstein, N.S. Serrated pathway and APC (conventional)-type colorectal polyps: Molecular-morphologic correlations, genetic pathways, and implications for classification. Am. J. Clin. Pathol. 2006, 125, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Kalady, M.F. Sessile serrated polyps: An important route to colorectal cancer. J. Natl. Compr. Canc. Netw. 2013, 11, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Bao, Y.; Ma, M.; Yang, W. Identification of Key Candidate Genes and Pathways in Colorectal Cancer by Integrated Bioinformatical Analysis. Int. J. Mol. Sci. 2017, 18, 722. [Google Scholar] [CrossRef]
- Weischenfeldt, J.; Dubash, T.; Drainas, A.P.; Mardin, B.R.; Chen, Y.; Stütz, A.M.; Waszak, S.M.; Bosco, G.; Halvorsen, A.R.; Raeder, B.; et al. Pan-cancer analysis of somatic copy number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 2017, 49, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.A.; Yu, M.; Baker, K.; Redman, M.; Wu, C.; Heinzerling, T.J.; Wirtz, R.M.; Charalambous, E.; Pentheroudakis, G.; Kotoula, V.; et al. The CpG island methylator phenotype is concordant between primary colorectal carcinoma and matched distant metastases. Clin. Epigenetics 2017, 9, 46. [Google Scholar] [CrossRef]
- Han, B.; Feng, D.; Yu, X.; Zhang, Y.; Liu, Y.; Zhou, L. Identification and Interaction Analysis of Molecular Markers in Colorectal Cancer by Integrated Bioinformatics Analysis. Med. Sci. Monit. 2018, 24, 6059–6069. [Google Scholar] [CrossRef]
- Freitas, M.; Ferreira, F.; Carvalho, S.; Silva, F.; Lopes, P.; Antunes, L.; Salta, S.; Diniz, F.; Santos, L.L.; Videira, J.F.; et al. A novel DNA methylation panel accurately detects colorectal cancer independently of molecular pathway. J. Transl. Med. 2018, 16, 45. [Google Scholar] [CrossRef]
- Vanaja, K.G.; Timp, W.; Feinberg, A.P.; Levchenko, A. A Loss of Epigenetic Control Can Promote Cell Death through Reversing the Balance of Pathways in a Signaling Network. Mol. Cell 2018, 72, 60–70. [Google Scholar] [CrossRef]
- Menter, D.G.; Davis, J.S.; Broom, B.M.; Overman, M.J.; Morris, J.; Kopetz, S. Back to the Colorectal Cancer Consensus Molecular Subtype Future. Curr. Gastroenterol. Rep. 2019, 21, 5. [Google Scholar] [CrossRef]
- Tang, X.; Qiao, X.; Chen, C.; Liu, Y.; Zhu, J.; Liu, J. Regulation Mechanism of Long Noncoding RNAs in Colon Cancer Development and Progression. Yonsei Med. J. 2019, 60, 319–325. [Google Scholar] [CrossRef]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instability in colorectal cancers. Nature 1997, 386, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.M.; Zhou, W.; Goodman, S.N.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 2001, 61, 818–822. [Google Scholar] [PubMed]
- Zhu, H.; Yu, J.; Zhu, H.; Guo, Y.; Feng, S. Identification of key lncRNAs in colorectal cancer progression based on associated protein-protein interaction analysis. World J. Surg. Oncol. 2017, 15, 153. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, C.; Ma, M.H.; Dai, D.Q. Identification and prediction of novel non-coding and coding RNA-associated competing endogenous RNA networks in colorectal cancer. World J. Gastroenterol. 2018, 24, 5259–5270. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.; Raveh, E.; Ohana, P.; Lail, R.A.; Gershtain, E.; Gilon, M.; De Groot, N.; Czerniak, A.; Hochberg, A. The increasing complexity of the oncofetal h19 gene locus: Functional dissection and therapeutic intervention. Int. J. Mol. Sci. 2013, 14, 4298–4316. [Google Scholar] [CrossRef] [PubMed]
- Pizzini, S.; Bisognin, A.; Mandruzzato, S.; Biasiolo, M.; Facciolli, A.; Perilli, L.; Rossi, E.; Esposito, G.; Rugge, M.; Pilati, P.; et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics 2013, 14, 589. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.C.; Graham, L.D.; Molloy, P.L. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim. Biophys. Acta. 2014, 1843, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Rokavec, M.; Horst, D.; Hermeking, H. Cellular Model of Colon Cancer Progression Reveals Signatures of mRNAs, miRNA, lncRNAs, and Epigenetic Modifications Associated with Metastasis. Cancer Res. 2017, 77, 1854–1867. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, H.; Al-Ghafari, A.; Choudhry, H.; Al Doghaither, H. Roles of long non-coding RNAs in colorectal cancer tumorigenesis: A Review. Mol. Clin. Oncol. 2019, 11, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhao, Y.; Zhang, W.; Bai, Y. Whole transcriptome sequencing identifies crucial genes associated with colon cancer and elucidation of their possible mechanisms of action. Onco. Targets Ther. 2019, 12, 2737–2747. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Croce, C.M. Long noncoding RNAs: Undeciphered cellular codes encrypting keys of colorectal cancer pathogenesis. Cancer Lett. 2018, 417, 89–95. [Google Scholar] [CrossRef]
- Liu, H.; Ye, D.; Chen, A.; Tan, D.; Zhang, W.; Jiang, W.; Wang, M.; Zhang, X. A pilot study of new promising non-coding RNA diagnostic biomarkers for early-stage colorectal cancers. Clin. Chem. Lab. Med. 2019, 57, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Strubberg, A.M.; Madison, B.B. MicroRNAs in the etiology of colorectal cancer: Pathways and clinical implications. Dis. Model. Mech. 2017, 10, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Hayashi, N.; Kuroda, Y.; Ito, S.; Eguchi, H.; Mimori, K. MicroRNAs as Biomarkers in Colorectal Cancer. Cancers 2017, 9, 124. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhao, Z.Y.; Wu, R.; Zhang, Y.; Zhang, Z.Y. Prognostic value of microRNAs in colorectal cancer: A meta-analysis. Cancer Manag. Res. 2018, 10, 907–929. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Li, J.; Dong, Z.; Liu, Y.; Chen, Y.; Chen, N.; Cheng, L.; Fang, C.; Wang, H.; Ji, Y.; et al. Temporal expression and functional analysis of long non-coding RNAs in colorectal cancer initiation. J. Cell Mol. Med. 2019, 23, 4127–4138. [Google Scholar] [CrossRef]
- Weroha, S.J.; Haluska, P. The insulin-like growth factor system in cancer. Endocrinol. Metab. Clin. North. Am. 2012, 41, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Brouwer-Visser, J.; Huang, G.S. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 2015, 26, 371–377. [Google Scholar] [CrossRef]
- Bieghs, L.; Johnsen, H.E.; Maes, K.; Menu, E.; Van Valckenborgh, E.; Overgaard, M.T.; Nyegaard, M.; Conover, C.A.; Vanderkerken, K.; De Bruyne, E. The insulin-like growth factor system in multiple myeloma: Diagnostic and therapeutic potential. Oncotarget 2016, 7, 48732–48752. [Google Scholar] [CrossRef]
- Vishwamitra, D.; George, S.K.; Shi, P.; Kaseb, A.O.; Amin, H.M. Type I insulin-like growth factor receptor signaling in hematological malignancies. Oncotarget 2017, 8, 1814–1844. [Google Scholar] [CrossRef]
- Pavelic, K.; Buković, D.; Pavelić, J. The role of insulin-like growth factor 2 and its receptors in human tumors. Mol. Med. 2002, 8, 771–780. [Google Scholar] [CrossRef]
- Gallagher, E.J.; Fierz, Y.; Ferguson, R.D.; LeRoith, D. The pathway from diabetes and obesity to cancer, on the route to targeted therapy. Endocr. Pract. 2010, 16, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Muc-Wierzgoń, M.; Nowakowska-Zajdel, E.; Dzięgielewska-Gęsiak, S.; Kokot, T.; Klakla, K.; Fatyga, E.; Grochowska-Niedworok, E.; Waniczek, D.; Wierzgoń, J. Specific metabolic biomarkers as risk and prognostic factors in colorectal cancer. World J. Gastroenterol. 2014, 20, 9759–9774. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, J.Z. Obesity, the PI3K/Akt signal pathway and colon cancer. Obes. Rev. 2009, 10, 610–616. [Google Scholar] [CrossRef]
- Sax, A.T.; Jenkins, D.G.; Devin, J.L.; Hughes, G.I.; Bolam, K.A.; Skinner, T.L. The insulin-like growth factor axis: A biological mechanism linking physical activity to colorectal cancer survival. Cancer Epidemiol. 2014, 38, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.D.; Vidigal, V.M.; Felipe, A.V.; DE Lima, J.M.; Neto, R.A.; Saad, S.S.; Forones, N.M. DNA methylation as an epigenetic biomarker in colorectal cancer. Oncol. Lett. 2013, 6, 1687–1692. [Google Scholar] [CrossRef]
- Harrela, M.; Koistinen, H.; Kaprio, J.; Lehtovirta, M.; Tuomilehto, J.; Eriksson, J.; Toivanen, L.; Koskenvuo, M.; Leinonen, P.; Koistinen, R.; et al. Genetic and environmental components of interindividual variation in circulating levels of IGF-I, IGF-II, IGFBP-1, and IGFBP-3. J. Clin. Invest. 1996, 98, 2612–2615. [Google Scholar] [CrossRef]
- Livingstone, C. IGF2 and cancer. Endocr. Relat. Cancer. 2013, 20, R321–R339. [Google Scholar] [CrossRef] [PubMed]
- Hoyo, C.; Murphy, S.K.; Schildkraut, J.M.; Vidal, A.C.; Skaar, D.; Millikan, R.C.; Galanko, J.; Sandler, R.S.; Jirtle, R.; Keku, T. IGF2R genetic variants, circulating IGF2 concentrations and colon cancer risk in African Americans and Whites. Dis. Markers 2012, 32, 133–141. [Google Scholar] [CrossRef]
- Nosho, K.; Yamamoto, H.; Taniguchi, H.; Adachi, Y.; Yoshida, Y.; Arimura, Y.; Endo, T.; Hinoda, Y.; Imai, K. Interplay of insulin-like growth factor-II, insulin-like growth factor-I, insulin-like growth factor-I receptor, COX-2, and matrix metalloproteinase-7, play key roles in the early stage of colorectal carcinogenesis. Clin. Cancer Res. 2004, 10, 7950–7957. [Google Scholar] [CrossRef]
- Jirtle, R.L. IGF2 loss of imprinting: A potential heritable risk factor for colorectal cancer. Gastroenterology 2004, 126, 1190–1193. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.; D’Amore, P.A. IGF2: Epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev. 2008, 19, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Belharazem, D.; Magdeburg, J.; Berton, A.K.; Beissbarth, L.; Sauer, C.; Sticht, C.; Marx, A.; Hofheinz, R.; Post, S.; Kienle, P.; et al. Carcinoma of the colon and rectum with deregulation of insulin-like growth factor 2 signaling: Clinical and molecular implications. J. Gastroenterol. 2016, 51, 971–984. [Google Scholar] [CrossRef] [PubMed]
- DeChiara, T.M.; Robertson, E.J.; Efstratiadis, A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 1991, 64, 849–859. [Google Scholar] [CrossRef]
- Vu, T.H.; Hoffman, A.R. Promoter-specific imprinting of the human insulin-like growth factor-II gene. Nature 1994, 371, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, A.; Feinberg, A.P. Loss of imprinting of IGF2: A common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005, 65, 11236–11240. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Cruz-Correa, M.; Giardiello, F.M.; Hutcheon, D.F.; Kafonek, D.R.; Brandenburg, S.; Wu, Y.; He, X.; Powe, N.R.; Feinberg, A.P. Loss of IGF2 imprinting: A potential marker of colorectal cancer risk. Science 2003, 299, 1753–1755. [Google Scholar] [CrossRef]
- Cui, H. Loss of imprinting of IGF2 as an epigenetic marker for the risk of human cancer. Dis. Markers 2007, 23, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Tang, Z.; Song, G.; Pan, Y.; He, B.; Bao, Q.; Wang, S. Loss of imprinting of IGF2 correlates with hypomethylation of the H19 differentially methylated region in the tumor tissue of colorectal cancer patients. Mol. Med. Rep. 2012, 5, 1536–1540. [Google Scholar] [CrossRef]
- Takano, Y.; Shiota, G.; Kawasaki, H. Analysis of genomic imprinting of insulin-like growth factor 2 in colorectal cancer. Oncology 2000, 59, 210–216. [Google Scholar] [CrossRef]
- Nishihara, R.; Wang, M.; Qian, Z.R.; Baba, Y.; Yamauchi, M.; Mima, K.; Sukawa, Y.; Kim, S.A.; Inamura, K.; Zhang, X.; et al. Alcohol, one-carbon nutrient intake, and risk of colorectal cancer according to tumor methylation level of IGF2 differentially methylated region. Am. J. Clin. Nutr. 2014, 100, 1479–1488. [Google Scholar] [CrossRef] [PubMed]
- Daughaday, W.H.; Trivedi, B. Measurement of derivatives of proinsulin-like growth factor-II in serum by a radioimmunoassay directed against the E-domain in normal subjects and patients with nonislet cell tumor hypoglycemia. J. Clin. Endocrinol. Metab. 1992, 75, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Devanathan, N.; Kimble-Hill, A.C. Systematic Survey of the Role of IGF in the Link Between Diabetes and Cancer. Indiana Univ. J. Undergrad Res. 2018, 4, 17–26. [Google Scholar] [CrossRef] [PubMed]
- LeRoith, D.; Roberts, C.T., Jr. The insulin-like growth factor system and cancer. Cancer Lett. 2003, 195, 127–137. [Google Scholar] [CrossRef]
- Ding, J.; Li, C.; Tang, J.; Yi, C.; Liu, J.Y.; Qiu, M. Higher Expression of Proteins in IGF/IR Axes in Colorectal Cancer is Associated with Type 2 Diabetes Mellitus. Pathol. Oncol. Res. 2016, 22, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Alfares, M.N.; Perks, C.M.; Hamilton-Shield, J.P.; Holly, J.M.P. Insulin-like growth factor-II in adipocyte regulation: Depot-specific actions suggest a potential role limiting excess visceral adiposity. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E1098–E1107. [Google Scholar] [CrossRef] [PubMed]
- Matuschek, C.; Rudoy, M.; Peiper, M.; Gerber, P.A.; Hoff, N.P.; Buhren, B.A.; Flehmig, B.; Budach, W.; Knoefel, W.T.; Bojar, H.; et al. Do insulin-like growth factor associated proteins qualify as a tumor marker? Results of a prospective study in 163 cancer patients. Eur. J. Med. Res. 2011, 16, 451–456. [Google Scholar] [CrossRef]
- Kushlinskii, N.E.; Gershtein, E.S.; Nikolaev, A.A.; Delektorskaya, V.V.; Korotkova, E.A.; Dvorova, E.K.; Kostyleva, O.I. Insulin-like growth factors (IGF), IGF-binding proteins (IGFBP), and vascular endothelial growth factor (VEGF) in blood serum of patients with colorectal cancer. Bull. Exp. Biol. Med. 2014, 156, 684–688. [Google Scholar] [CrossRef]
- Zhao, R.; Berho, M.; Nogueras, J.; Sands, D.; Weiss, E.; Wexner, S.; Giardiello, F.M.; Cruz-Correa, M. Positive correlation of insulin-like growth factor-II with proliferating cell index in patients with colorectal neoplasia. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 1819–1822. [Google Scholar] [CrossRef]
- Liou, J.M.; Shun, C.T.; Liang, J.T.; Chiu, H.M.; Chen, M.J.; Chen, C.C.; Wang, H.P.; Wu, M.S.; Lin, J.T. Plasma insulin-like growth factor-binding protein-2 levels as diagnostic and prognostic biomarker of colorectal cancer. J. Clin. Endocrinol. Metab. 2010, 95, 1717–1725. [Google Scholar] [CrossRef]
- Chi, F.; Wu, R.; Zeng, Y.C.; Xing, R.; Liu, Y. Circulation insulin-like growth factor peptides and colorectal cancer risk: An updated systematic review and meta-analysis. Mol. Biol. Rep. 2013, 40, 3583–3590. [Google Scholar] [CrossRef] [PubMed]
- Tricoli, J.V.; Rall, L.B.; Karakousis, C.P.; Herrera, L.; Petrelli, N.J.; Bell, G.I.; Shows, T.B. Enhanced levels of insulin-like growth factor messenger RNA in human colon carcinomas and liposarcomas. Cancer Res. 1986, 46, 6169–6173. [Google Scholar] [PubMed]
- Lambert, S.; Vivario, J.; Boniver, J.; Gol-Winkler, R. Abnormal expression and structural modification of the insulin-like growth-factor-II gene in human colorectal tumors. Int. J. Cancer 1990, 46, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, K.; Onodera, H.; Kondo, S.; Kan, S.; Ikeuchi, D.; Maetani, S.; Imamura, M. Expression of insulin-like growth factor-2 can predict the prognosis of human colorectal cancer patients: Correlation with tumor progression, proliferative activity and survival. Oncology 1998, 55, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Freier, S.; Weiss, O.; Eran, M.; Flyvbjerg, A.; Dahan, R.; Nephesh, I.; Safra, T.; Shiloni, E.; Raz, I. Expression of the insulin-like growth factors and their receptors in adenocarcinoma of the colon. Gut 1999, 44, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.R.; He, X.B.; Yang, Y.H.; Xie, W. The expression and imprinting status of insulin-like growth factor 2 gene in colorectal cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2003, 20, 31–34. [Google Scholar]
- Li, S.R.; Ng, C.F.; Banerjea, A.; Ahmed, S.; Hands, R.; Powar, M.; Ogunkolade, W.; Dorudi, S.; Bustin, S.A. Differential expression patterns of the insulin-like growth factor 2 gene in human colorectal cancer. Tumour. Biol. 2004, 25, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.; Carlisi, A.; Collette, J.; Franchimont, P.; Gol-Winkler, R. Insulin-like growth factor II in two human colon-carcinoma cell lines: Gene structure and expression, and protein secretion. Int. J. Cancer 1992, 52, 404–408. [Google Scholar] [CrossRef]
- Zarrilli, R.; Pignata, S.; Romano, M.; Gravina, A.; Casola, S.; Bruni, C.B.; Acquaviva, A.M. Expression of insulin-like growth factor (IGF)-II and IGF-I receptor during proliferation and differentiation of CaCo-2 human colon carcinoma cells. Cell Growth Differ. 1994, 5, 1085–1091. [Google Scholar]
- Guo, Y.S.; Jin, G.F.; Townsend, C.M., Jr.; Zhang, T.; Sheng, H.M.; Beauchamp, R.D.; Thompson, J.C. Insulin-like growth factor-II expression in carcinoma in colon cell lines: Implications for autocrine actions. J. Am. Coll. Surg. 1995, 181, 145–154. [Google Scholar]
- Zhong, H.; Fazenbaker, C.; Chen, C.; Breen, S.; Huang, J.; Yao, X.; Ren, P.; Yao, Y.; Herbst, R.; Hollingsworth, R.E. Overproduction of IGF-2 drives a subset of colorectal cancer cells, which specifically respond to an anti-IGF therapeutic antibody and combination therapies. Oncogene 2017, 36, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, K.; Onodera, H.; Kan, S.; Kondo, S.; Imamura, M. Possible paracrine mechanism of insulin-like growth factor-2 in the development of liver metastases from colorectal carcinoma. Cancer 1999, 85, 18–25. [Google Scholar] [CrossRef]
- Barozzi, C.; Ravaioli, M.; D’Errico, A.; Grazi, G.L.; Poggioli, G.; Cavrini, G.; Mazziotti, A.; Grigioni, W.F. Relevance of biologic markers in colorectal carcinoma: A comparative study of a broad panel. Cancer 2002, 94, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Peters, G.; Gongoll, S.; Langner, C.; Mengel, M.; Piso, P.; Klempnauer, J.; Rüschoff, J.; Kreipe, H.; von Wasielewski, R. IGF-1R, IGF-1 and IGF-2 expression as potential prognostic and predictive markers in colorectal-cancer. Virchows Arch. 2003, 443, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Forbes, B.E. Two years in IGF research. Growth Horm. IGF Res. 2016, 30, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Kessler, S.M.; Haybaeck, J.; Kiemer, A.K. Insulin-Like Growth Factor 2-The Oncogene and its Accomplices. Curr. Pharm. Des. 2016, 22, 5948–5961. [Google Scholar] [CrossRef]
- Kasprzak, A.; Kwasniewski, W.; Adamek, A.; Gozdzicka-Jozefiak, A. Insulin-like growth factor (IGF) axis in cancerogenesis. Mutat. Res. Rev. Mutat. Res. 2017, 772, 78–104. [Google Scholar] [CrossRef]
- Diehl, D.; Oesterle, D.; Elmlinger, M.W.; Hoeflich, A.; Wolf, E.; Lahm, H. IGF-II transgenic mice display increased aberrant colon crypt multiplicity and tumor volume after 1,2-dimethylhydrazine treatment. J. Carcinog. 2006, 5, 24. [Google Scholar] [CrossRef]
- Witsch, E.; Sela, M.; Yarden, Y. Roles for growth factors in cancer progression. Physiology 2010, 25, 85–101. [Google Scholar] [CrossRef]
- Fenton, J.I.; Birmingham, J.M. Adipokine regulation of colon cancer: Adiponectin attenuates interleukin-6-induced colon carcinoma cell proliferation via STAT-3. Mol. Carcinog. 2010, 49, 700–709. [Google Scholar] [CrossRef]
- Liu, X.; Chen, X.; Zeng, K.; Xu, M.; He, B.; Pan, Y.; Sun, H.; Pan, B.; Xu, X.; Xu, T.; et al. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell Death Dis. 2018, 9, 1037. [Google Scholar] [CrossRef] [PubMed]
- Giovannucci, E. Nutrition, insulin, insulin-like growth factors and cancer. Horm. Metab. Res. 2003, 35, 694–704. [Google Scholar] [CrossRef]
- Vigneri, P.G.; Tirrò, E.; Pennisi, M.S.; Massimino, M.; Stella, S.; Romano, C.; Manzella, L. The Insulin/IGF System in Colorectal Cancer Development and Resistance to Therapy. Front. Oncol. 2015, 5, 230. [Google Scholar] [CrossRef] [PubMed]
- Bowers, L.W.; Rossi, E.L.; O’Flanagan, C.H.; deGraffenried, L.A.; Hursting, S.D. The Role of the Insulin/IGF System in Cancer: Lessons Learned from Clinical Trials and the Energy Balance-Cancer Link. Front. Endocrinol. 2015, 6, 77. [Google Scholar] [CrossRef]
- Mishra, L.; Bass, B.; Ooi, B.S.; Sidawy, A.; Korman, L. Role of insulin-like growth factor-I (IGF-I) receptor, IGF-I, and IGF binding protein-2 in human colorectal cancers. Growth Horm. IGF Res. 1998, 8, 473–479. [Google Scholar] [CrossRef]
- Cohen, P.; Clemmons, D.R.; Rosenfeld, R.G. Does the GH-IGF axis play a role in cancer pathogenesis? Growth Horm. IGF. Res. 2000, 10, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Giovannucci, E. Insulin, insulin-like growth factors and colon cancer: A review of the evidence. J. Nutr. 2001, 131, 3109S–3120S. [Google Scholar] [CrossRef]
- Humbel, R.E. Insulin-like growth factors I and II. Eur. J. Biochem. 1990, 190, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef]
- Rinderknecht, E.; Humbel, R.E. Polypeptides with nonsuppressible insulin-like and cell-growth promoting activities in human serum: Isolation, chemical characterization, and some biological properties of forms I and II. Proc. Natl. Acad. Sci. USA 1976, 73, 2365–2369. [Google Scholar] [CrossRef]
- Rinderknecht, E.; Humbel, R.E. Amino-terminal sequences of two polypeptides from human serum with nonsuppressible insulin-like and cell-growth-promoting activities: Evidence for structural homology with insulin B chain. Proc. Natl. Acad. Sci. USA 1976, 73, 4379–4381. [Google Scholar] [CrossRef]
- Rinderknecht, E.; Humbel, R.E. Primary structure of human insulin-like growth factor II. FEBS Lett. 1978, 89, 283–286. [Google Scholar] [CrossRef]
- De Pagter-Holthuizen, P.; van Schaik, F.M.; Verduijn, G.M.; van Ommen, G.J.; Bouma, B.N.; Jansen, M.; Sussenbach, J.S. Organization of the human genes for insulin-like growth factors I and II. FEBS Lett. 1986, 195, 179–184. [Google Scholar] [CrossRef]
- O’Dell, S.D.; Day, I.N. Insulin-like growth factor II (IGF-II). Int. J. Biochem. Cell Biol. 1998, 30, 767–771. [Google Scholar] [CrossRef]
- Butler, A.A.; LeRoith, D. Minireview: Tissue-specific versus generalized gene targeting of the igf1 and igf1r genes and their roles in insulin-like growth factor physiology. Endocrinology 2001, 142, 1685–1688. [Google Scholar] [CrossRef] [PubMed]
- Duguay, S.J.; Jin, Y.; Stein, J.; Duguay, A.N.; Gardner, P.; Steiner, D.F. Post-translational processing of the insulin-like growth factor-2 precursor. Analysis of O-glycosylation and endoproteolysis. J. Biol. Chem. 1998, 273, 18443–18451. [Google Scholar] [CrossRef]
- Pereira, G.T.A.; Utsunomiya, Y.T.; Milanesi, M.; Torrecilha, R.B.; Carmo, A.S.; Neves, H.H.; Carvalheiro, R.; Ajmone-Marsan, P.; Sonstegard, T.S.; Sölkner, J.; et al. Pleiotropic Genes Affecting Carcass Traits in Bos indicus (Nellore) Cattle Are Modulators of Growth. PLoS ONE 2016, 11, e0158165. [Google Scholar] [CrossRef] [PubMed]
- Baral, K.; Rotwein, P. The insulin-like growth factor 2 gene in mammals: Organizational complexity within a conserved locus. PLoS ONE 2019, 14, e0219155. [Google Scholar] [CrossRef] [PubMed]
- Firth, S.M.; Baxter, R.C. Cellular actions of the insulin-like growth factor binding proteins. Endocr. Rev. 2002, 23, 824–854. [Google Scholar] [CrossRef]
- Marks, A.G.; Carroll, J.M.; Purnell, J.Q.; Roberts, C.T., Jr. Plasma distribution and signaling activities of IGF-II precursors. Endocrinology 2011, 152, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Mistry, J.; Nicar, M.J.; Khosravi, M.J.; Diamandis, A.; van Doorn, J.; Juul, A. Insulin-like growth factors (IGF-I, free IGF-I and IGF-II) and insulin-like growth factor binding proteins (IGFBP-2, IGFBP-3, IGFBP-6, and ALS) in blood circulation. J. Clin. Lab. Anal. 1999, 13, 166–172. [Google Scholar] [CrossRef]
- Brissenden, J.E.; Ullrich, A.; Francke, U. Human chromosomal mapping of genes for insulin-like growth factors I and II and epidermal growth factor. Nature 1984, 310, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Tricoli, J.V.; Rall, L.B.; Scott, J.; Bell, G.I.; Shows, T.B. Localization of insulin-like growth factor genes to human chromosomes 11 and 12. Nature 1984, 5, 310–784. [Google Scholar] [CrossRef]
- Rotwein, P. Similarity and variation in the insulin-like growth factor 2-H19 locus in primates. Physiol. Genom. 2018, 50, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Holthuizen, P.; Van Dijk, M.A.; Rodenburg, R.J.; Koonen-Reemst, A.M.; Sussenbach, J.S. Transcriptional regulation of the major promoters of the human IGF-II gene. Mol. Reprod. Dev. 1993, 35, 391–393. [Google Scholar] [CrossRef]
- Daughaday, W.H.; Trivedi, B.; Baxter, R.C. Serum “big insulin-like growth factor II” from patients with tumor hypoglycemia lacks normal E-domain O-linked glycosylation, a possible determinant of normal propeptide processing. Proc. Natl. Acad. Sci. USA 1993, 90, 5823–5827. [Google Scholar] [CrossRef] [PubMed]
- Frasca, F.; Pandini, G.; Scalia, P.; Sciacca, L.; Mineo, R.; Costantino, A.; Goldfine, I.D.; Belfiore, A.; Vigneri, R. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol. Cell Biol. 1999, 19, 3278–3288. [Google Scholar] [CrossRef]
- Belfiore, A.; Frasca, F.; Pandini, G.; Sciacca, L.; Vigneri, R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 2009, 30, 586–623. [Google Scholar] [CrossRef]
- Denley, A.; Brierley, G.V.; Carroll, J.M.; Lindenberg, A.; Booker, G.W.; Cosgrove, L.J.; Wallace, J.C.; Forbes, B.E.; Roberts, C.T., Jr. Differential activation of insulin receptor isoforms by insulin-like growth factors is determined by the C domain. Endocrinology 2006, 147, 1029–1036. [Google Scholar] [CrossRef][Green Version]
- Henderson, S.T.; Brierley, G.V.; Surinya, K.H.; Priebe, I.K.; Catcheside, D.E.; Wallace, J.C.; Forbes, B.E.; Cosgrove, L.J. Delineation of the IGF-II C domain elements involved in binding and activation of the IR-A., IR-B and IGF-IR. Growth Horm. IGF Res. 2015, 25, 20–27. [Google Scholar] [CrossRef]
- Szebenyi, G.; Rotweien, P. The mouse insulin-like growth factor II/cation-independent mannose 6-phosphate (IGFII/MPR) receptor gene: Molecular cloning and genomic organization. Genomics 1994, 19, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Rezgui, D.; Prince, S.N.; Zaccheo, O.J.; Foulstone, E.J.; Forbes, B.E.; Norton, R.S.; Crosby, J.; Hassan, A.B.; Crump, M.P. Structural insights into the interaction of insulin-like growth factor 2 with IGF2R domain 11. Structure 2007, 15, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; MacDonald, R.G.; Thinakaran, G.; Kar, S. Insulin-Like Growth Factor-II/Cation-Independent Mannose 6-Phosphate Receptor in Neurodegenerative Diseases. Mol. Neurobiol. 2017, 54, 2636–2658. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Hernández, A.; Escribano, Ó.; Perdomo, L.; Otero, Y.F.; García-Gómez, G.; Fernández, S.; Beneit, N.; Benito, M. Implication of insulin receptor A isoform and IRA/IGF-IR hybrid receptors in the aortic vascular smooth muscle cell proliferation: Role of TNF-α and IGF-II. Endocrinology 2013, 154, 2352–2364. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.; Surakhy, M.; Can, S.; Ducker, M.; Davies, N.; Szele, F.; Bühnemann, C.; Carter, E.; Trikin, R.; Crump, M.P.; et al. Maternal transmission of an Igf2r domain 11: IGF2 binding mutant allele (Igf2rI1565A) results in partial lethality, overgrowth and intestinal adenoma progression. Sci. Rep. 2019, 9, 11388. [Google Scholar] [CrossRef]
- Unger, C.; Kramer, N.; Unterleuthner, D.; Scherzer, M.; Burian, A.; Rudisch, A.; Stadler, M.; Schlederer, M.; Lenhardt, D.; Riedl, A.; et al. Stromal-derived IGF2 promotes colon cancer progression via paracrine and autocrine mechanisms. Oncogene 2017, 36, 5341–5355. [Google Scholar] [CrossRef]
- Lamonerie, T.; Lavialle, C.; Haddada, H.; Brison, O. IGF-2 autocrine stimulation in tumorigenic clones of a human colon-carcinoma cell line. Int. J. Cancer 1995, 61, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Lamonerie, T.; Lavialle, C.; de Galle, B.; Binoux, M.; Brison, O. Constitutive or inducible overexpression of the IGF-2 gene in cells of a human colon carcinoma cell line. Exp. Cell Res. 1995, 216, 342–351. [Google Scholar] [CrossRef]
- Sanderson, M.P.; Hofmann, M.H.; Garin-Chesa, P.; Schweifer, N.; Wernitznig, A.; Fischer, S.; Jeschko, A.; Meyer, R.; Moll, J.; Pecina, T.; et al. The IGF1R/INSR Inhibitor BI 885578 Selectively Inhibits Growth of IGF2-Overexpressing Colorectal Cancer Tumors and Potentiates the Efficacy of Anti-VEGF Therapy. Mol. Cancer Ther. 2017, 16, 2223–2233. [Google Scholar] [CrossRef]
- Cheng, Y.W.; Idrees, K.; Shattock, R.; Khan, S.A.; Zeng, Z.; Brennan, C.W.; Paty, P.; Barany, F. Loss of imprinting and marked gene elevation are 2 forms of aberrant IGF2 expression in colorectal cancer. Int. J. Cancer 2010, 127, 568–577. [Google Scholar] [CrossRef]
- Stange, D.E.; Engel, F.; Longerich, T.; Koo, B.K.; Koch, M.; Delhomme, N.; Aigner, M.; Toedt, G.; Schirmacher, P.; Lichter, P.; et al. Expression of an ASCL2 related stem cell signature and IGF2 in colorectal cancer liver metastases with 11p15.5 gain. Gut 2010, 59, 1236–1244. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, X.; Wang, G.; Wen, X.; Zhang, X.; Hoffman, A.R.; Li, W.; Hu, J.F.; Cui, J. Loss of insulin-like growth factor II imprinting is a hallmark associated with enhanced chemo/radiotherapy resistance in cancer stem cells. Oncotarget 2016, 7, 51349–51364. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Liu, Y.; Jiang, J.; Liu, Y.; Yang, Z.; Wu, S.; Cao, W.; Cui, I.H.; Yu, C. IGF2-derived miR-483 mediated oncofunction by suppressing DLC-1 and associated with colorectal cancer. Oncotarget 2016, 7, 48456–48466. [Google Scholar] [CrossRef] [PubMed]
- Lahm, H.; Suardet, L.; Laurent, P.L.; Fischer, J.R.; Ceyhan, A.; Givel, J.C.; Odartchenko, N. Growth regulation and co-stimulation of human colorectal cancer cell lines by insulin-like growth factor I, II and transforming growth factor alpha. Br. J. Cancer 1992, 65, 341–346. [Google Scholar] [CrossRef]
- Lahm, H.; Amstad, P.; Wyniger, J.; Yilmaz, A.; Fischer, J.R.; Schreyer, M.; Givel, J.C. Blockade of the insulin-like growth-factor-I receptor inhibits growth of human colorectal cancer cells: Evidence of a functional IGF-II-mediated autocrine loop. Int. J. Cancer 1994, 58, 452–459. [Google Scholar] [CrossRef]
- Fu, P.; Thompson, J.A.; Leeding, K.S.; Bach, L.A. Insulin-like growth factors induce apoptosis as well as proliferation in LIM 1215 colon cancer cells. J. Cell Biochem. 2007, 100, 58–68. [Google Scholar] [CrossRef]
- Rogers, M.A.; Kalter, V.; Strowitzki, M.; Schneider, M.; Lichter, P. IGF2 knockdown in two colorectal cancer cell lines decreases survival, adhesion and modulates survival-associated genes. Tumour Biol. 2016, 37, 12485–12495. [Google Scholar] [CrossRef]
- Dupont, J.; Pierre, A.; Froment, P.; Moreau, C. The insulin-like growth factor axis in cell cycle progression. Horm. Metab. Res. 2003, 35, 740–750. [Google Scholar] [CrossRef]
- Singh, P.; Dai, B.; Yallampalli, U.; Lu, X.; Schroy, P.C. Proliferation and differentiation of a human colon cancer cell line (CaCo2) is associated with significant changes in the expression and secretion of insulin-like growth factor (IGF) IGF-II and IGF binding protein-4: Role of IGF-II. Endocrinology 1996, 137, 1764–1774. [Google Scholar] [CrossRef]
- Lee, H.; Kim, N.; Yoo, Y.J.; Kim, H.; Jeong, E.; Choi, S.; Moon, S.U.; Oh, S.H.; Mills, G.B.; Yoon, S.; et al. β-catenin/TCF activity regulates IGF-1R tyrosine kinase inhibitor sensitivity in colon cancer. Oncogene 2018, 37, 5466–5475. [Google Scholar] [CrossRef]
- Ma, J.; Pollak, M.N.; Giovannucci, E.; Chan, J.M.; Tao, Y.; Hennekens, C.H.; Stampfer, M.J. Prospective study of colorectal cancer risk in men and plasma levels of insulin-like growth factor (IGF)-I and IGF-binding protein-3. J. Natl. Cancer Inst. 1999, 91, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Manousos, O.; Souglakos, J.; Bosetti, C.; Tzonou, A.; Chatzidakis, V.; Trichopoulos, D.; Adami, H.O.; Mantzoros, C. IGF-I and IGF-II in relation to colorectal cancer. Int. J. Cancer 1999, 83, 15–17. [Google Scholar] [CrossRef]
- Probst-Hensch, N.M.; Yuan, J.M.; Stanczyk, F.Z.; Gao, Y.T.; Ross, R.K.; Yu, M.C. IGF-1, IGF-2 and IGFBP-3 in prediagnostic serum: Association with colorectal cancer in a cohort of Chinese men in Shanghai. Br. J. Cancer 2001, 85, 1695–1699. [Google Scholar] [CrossRef] [PubMed]
- Renehan, A.G.; Jones, J.; Potten, C.S.; Shalet, S.M.; O’Dwyer, S.T. Elevated serum insulin-like growth factor (IGF)-II and IGF binding protein-2 in patients with colorectal cancer. Br. J. Cancer 2000, 83, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Akaike, M.; Yoshihara, K.; Shiozawa, M.; Yamamoto, N.; Sato, T.; Yamada, R.; Fujii, S.; Rino, Y.; Kunisaki, C.; et al. Clinicopathological significance of the gene expression of matrix metalloproteinase-7, insulin-like growth factor-1, insulin-like growth factor-2 and insulin-like growth factor-1 receptor in patients with colorectal cancer: Insulin-like growth factor-1 receptor gene expression is a useful predictor of liver metastasis from colorectal cancer. Oncol. Rep. 2008, 20, 359–364. [Google Scholar] [PubMed]
- Yamamoto, N.; Oshima, T.; Yoshihara, K.; Aoyama, T.; Hayashi, T.; Yamada, T.; Sato, T.; Shiozawa, M.; Yoshikawa, T.; Morinaga, S.; et al. Clinicopathological significance and impact on outcomes of the gene expression levels of IGF-1, IGF-2 and IGF-1R, IGFBP-3 in patients with colorectal cancer: Overexpression of the IGFBP-3 gene is an effective predictor of outcomes in patients with colorectal cancer. Oncol. Lett. 2017, 13, 3958–3966. [Google Scholar] [CrossRef]
- Vrieling, A.; Voskuil, D.W.; Bosma, A.; Majoor, D.M.; van Doorn, J.; Cats, A.; Depla, A.C.; Timmer, R.; Witteman, B.J.; Wesseling, J.; et al. Expression of insulin-like growth factor system components in colorectal tissue and its relation with serum IGF levels. Growth Horm. IGF Res. 2009, 19, 126–135. [Google Scholar] [CrossRef]
- Hegde, P.; Qi, R.; Gaspard, R.; Abernathy, K.; Dharap, S.; Earle-Hughes, J.; Gay, C.; Nwokekeh, N.U.; Chen, T.; Saeed, A.I.; et al. Identification of tumor markers in models of human colorectal cancer using a 19,200-element complementary DNA microarray. Cancer Res. 2001, 61, 7792–7797. [Google Scholar]
- Yamasaki, M.; Takemasa, I.; Komori, T.; Watanabe, S.; Sekimoto, M.; Doki, Y.; Matsubara, K.; Monden, M. The gene expression profile represents the molecular nature of liver metastasis in colorectal cancer. Int. J. Oncol. 2007, 30, 129–138. [Google Scholar] [CrossRef][Green Version]
- Kang, K.J.; Min, B.H.; Ryu, K.J.; Kim, K.M.; Chang, D.K.; Kim, J.J.; Rhee, J.C.; Kim, Y.H. The role of the CpG island methylator phenotype on survival outcome in colon cancer. Gut Liver. 2015, 9, 202–207. [Google Scholar] [CrossRef][Green Version]
- Tapial, S.; Olmedillas-López, S.; Rueda, D.; Arriba, M.; García, J.L.; Vivas, A.; Pérez, J.; Pena-Couso, L.; Olivera, R.; Rodríguez, Y.; et al. Cimp-Positive Status is More Representative in Multiple Colorectal Cancers than in Unique Primary Colorectal Cancers. Sci. Rep. 2019, 9, 10516. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Nosho, K.; Shima, K.; Huttenhower, C.; Tanaka, N.; Hazra, A.; Giovannucci, E.L.; Fuchs, C.S.; Ogino, S. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology 2010, 139, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wang, Y.B.; Zeng, P.; Yan, G.Q.; Xin, L.; Hu, X.Y. Expression of long non-coding RNA (lncRNA) H19 in immunodeficient mice induced with human colon cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4880–4884. [Google Scholar] [PubMed]
- Yang, Y.; Zhao, L.; Lei, L.; Lau, W.B.; Lau, B.; Yang, Q.; Le, X.; Yang, H.; Wang, C.; Luo, Z.; et al. LncRNAs: The bridge linking RNA and colorectal cancer. Oncotarget 2017, 8, 12517–12532. [Google Scholar] [CrossRef][Green Version]
- Yang, Q.; Wang, X.; Tang, C.; Chen, X.; He, J. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int. J. Oncol. 2017, 50, 1801–1809. [Google Scholar] [CrossRef]
- He, T.Y.; Li, S.H.; Huang, J.; Gong, M.; Li, G. Prognostic value of long non-coding RNA CRNDE in gastrointestinal cancers: A meta-analysis. Cancer Manag. Res. 2019, 11, 5629–5642. [Google Scholar] [CrossRef]
- Hibner, G.; Kimsa-Furdzik, M.; Francuz, T. Relevance of MicroRNAs as Potential Diagnostic and Prognostic Markers in Colorectal Cancer. Int. J. Mol. Sci. 2018, 19, 2944. [Google Scholar] [CrossRef]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef]
- Steinhoff, C.; Paulsen, M.; Kielbasa, S.; Walter, J.; Vingron, M. Expression profile and transcription factor binding site exploration of imprinted genes in human and mouse. BMC Genomics. 2009, 10, 144. [Google Scholar] [CrossRef]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- Nakagawa, H.; Chadwick, R.B.; Peltomaki, P.; Plass, C.; Nakamura, Y.; de La Chapelle, A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc. Natl. Acad. Sci USA 2001, 98, 591–596. [Google Scholar] [CrossRef]
- Cui, H.; Onyango, P.; Brandenburg, S.; Wu, Y.; Hsieh, C.L.; Feinberg, A.P. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002, 62, 6442–6446. [Google Scholar] [PubMed]
- Zhang, H.; Niu, B.; Hu, J.F.; Ge, S.; Wang, H.; Li, T.; Ling, J.; Steelman, B.N.; Qian, G.; Hoffman, A.R. Interruption of intrachromosomal looping by CCCTC binding factor decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. J. Cell Biol. 2011, 193, 475–487. [Google Scholar] [CrossRef]
- Hidaka, H.; Higashimoto, K.; Aoki, S.; Mishima, H.; Hayashida, C.; Maeda, T.; Koga, Y.; Yatsuki, H.; Joh, K.; Noshiro, H. Comprehensive methylation analysis of imprinting-associated differentially methylated regions in colorectal cancer. Clin. Epigenetics. 2018, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Gao, X.; Wang, M.; Qiao, Y.; Xu, Y.; Yang, J.; Dong, N.; He, J.; Sun, Q.; Lv, G.; et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget 2016, 7, 22159–22173. [Google Scholar] [CrossRef]
- Tsang, W.P.; Ng, E.K.; Ng, S.S.; Jin, H.; Yu, J.; Sung, J.J.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010, 31, 350–358. [Google Scholar] [CrossRef]
- Lu, L.; Cai, M.; Peng, M.; Wang, F.; Zhai, X. miR-491-5p functions as a tumor suppressor by targeting IGF2 in colorectal cancer. Cancer Manag. Res. 2019, 11, 1805–1816. [Google Scholar] [CrossRef]
- Shi, L.; Li, X.; Wu, Z.; Li, X.; Nie, J.; Guo, M.; Mei, Q.; Han, W. DNA methylation-mediated repression of miR-181a/135a/302c expression promotes the microsatellite-unstable colorectal cancer development and 5-FU resistance via targeting PLAG1. J. Genet. Genomics. 2018, 45, 205–214. [Google Scholar] [CrossRef]
- Simpson, A.; Petnga, W.; Macaulay, V.M.; Weyer-Czernilofsky, U.; Bogenrieder, T. Insulin-Like Growth Factor (IGF) Pathway Targeting in Cancer: Role of the IGF Axis and Opportunities for Future Combination Studies. Target. Oncol. 2017, 12, 571–597. [Google Scholar] [CrossRef]
- Weber, D.; Gödde, D.; Postberg, J.; Zirngibl, H.; Prinz, C. IGF-2 growth factor expression in human rectal adenocarcinoma. J. Gastrointest. Dig. Syst. 2016, 6, 451. [Google Scholar] [CrossRef]
- Nie, Z.L.; Pan, Y.Q.; He, B.S.; Gu, L.; Chen, L.P.; Li, R.; Xu, Y.Q.; Gao, T.Y.; Song, G.Q.; Hoffman, A.R.; et al. Gene therapy for colorectal cancer by an oncolytic adenovirus that targets loss of the insulin-like growth factor 2 imprinting system. Mol. Cancer 2012, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; He, B.; Chen, J.; Sun, H.; Deng, Q.; Wang, F.; Ying, H.; Liu, X.; Lin, K.; Peng, H.; et al. Gene therapy for colorectal cancer by adenovirus-mediated siRNA targeting CD147 based on loss of the IGF2 imprinting system. Int, J. Oncol. 2015, 47, 1881–1889. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.Q.; He, B.S.; Zhu, C.; Qu, L.L.; Xu, X.F.; Wang, S.K. Effect of recombinant adenovirus Ad-DT-A in targeted therapy for malignant cancer cell lines with loss of IGF2 imprinting. Zhonghua Zhong Liu Za Zhi 2011, 33, 816–821. [Google Scholar] [PubMed]
- Sun, H.; Pan, Y.; He, B.; Deng, Q.; Li, R.; Xu, Y.; Chen, J.; Gao, T.; Ying, H.; Wang, F.; et al. Gene therapy for human colorectal cancer cell lines with recombinant adenovirus 5 based on loss of the insulin-like growth factor 2 imprinting. Int J. Oncol. 2015, 46, 1759–1767. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zanella, E.R.; Galimi, F.; Sassi, F.; Migliardi, G.; Cottino, F.; Leto, S.M.; Lupo, B.; Erriquez, J.; Isella, C.; Comoglio, P.M.; et al. IGF2 is an actionable target that identifies a distinct subpopulation of colorectal cancer patients with marginal response to anti-EGFR therapies. Sci. Transl. Med. 2015, 7, 272ra12. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.; Vrignaud, P.; Vacher, S.; Richon, S.; Lièvre, A.; Cacheux, W.; Weiswald, L.B.; Massonnet, G.; Chateau-Joubert, S.; Nicolas, A.; et al. Evaluating patient-derived colorectal cancer xenografts as preclinical models by comparison with patient clinical data. Cancer Res. 2015, 75, 1560–1566. [Google Scholar] [CrossRef]
- Harper, J.; Burns, J.L.; Foulstone, E.J.; Pignatelli, M.; Zaina, S.; Hassan, A. Soluble IGF2 receptor rescues Apc(Min/+) intestinal adenoma progression induced by Igf2 loss of imprinting. Cancer Res. 2006, 66, 1940–1948. [Google Scholar] [CrossRef]
- Gyparaki, M.T.; Basdra, E.K.; Papavassiliou, A.G. DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. J. Mol. Med. (Berl). 2013, 91, 1249–1256. [Google Scholar] [CrossRef]
- Mäki-Nevala, S.; Valo, S.; Ristimäki, A.; Sarhadi, V.; Knuutila, S.; Nyström, M.; Renkonen-Sinisalo, L.; Lepistö, A.; Mecklin, J.P.; Peltomäki, P. DNA methylation changes and somatic mutations as tumorigenic events in Lynch syndrome-associated adenomas retaining mismatch repair protein expression. EBioMedicine 2019, 39, 280–291. [Google Scholar] [CrossRef]

| Biomarker | Type of Change | Summary of the Findings | Potential Clinical Significance |
|---|---|---|---|
| IGF2 gene | LOI | The adjusted OR for IGF2 LOI in lymphocytes was 5.15 for patients with a positive family history, 3.46 for patients with adenomas, and 21.7 for patients with CRC [67]. | Diagnosis |
| IGF2 LOI occurs not only in colon cancer tissues but also in matched normal tissues and peripheral blood cells [68]. | Screening and diagnosis | ||
| IGF2 LOI were associated with increased risk of mortality in patients with stage IV disease; higher plasma IGF2 levels were associated with reduced risk of mortality [80]. | Diagnosis and prognosis | ||
| IGF2 LOI was in 63% of CRC and in 21.7% of the normal control group (p < 0.01) [69]. | Screening and diagnosis | ||
| IGF2 LOI with IGF2 overexpression [63] | Screening and diagnosis | ||
| IGF2 LOI was a common feature in CSCs; increased IGF2 was found in CSCs isolated from CRC cell lines (HT29, HRT18, and HCT116); higher rate of colony formation and greater resistance to chemotherapy and radiotherapy were found in vitro [142]. | Diagnosis, mechanisms, and therapeutics | ||
| IGF2 LOI leads to rebalancing of activities of canonical AKT and ERK pathways; altered signaling balance leads to rebalancing of pro- and antiapoptotic control [28]. | Diagnosis, mechanisms, and therapeutics | ||
| Amplification | Colorectal intrinsic D (CRIS-D) subtype was specifically enriched for amplification of chromosome 11p15.5 and IGF2 high-level overexpression in CRC [17]. | Diagnosis and mechanisms | |
| DNA methylation | Biallelic hypermethylation of a core of five CpG sites in the insulator region of IGF2/H19 correlated strongly with IGF2 LOI [171]. | Diagnosis and mechanisms | |
| Hypomethylation of the H19 differentially methylated region (DMR) and DMR upstream of exon 3 of IGF2 was found in CRC and normal colon mucosa; normal imprinting in the colon and LOI in CRC is specifically linked to the methylation status of a DMR within IGF2 and not H19 [172]. | Diagnosis and mechanisms | ||
| DMR IGF2/H19 hypomethylation correlated with IGF2 LOI; IGF2 overexpression did not correlate with IGF2/H19 hypomethylation but negatively correlated with MSI [140]. | Diagnosis and mechanisms | ||
| Lower levels of IGF2 DMR0 methylation were found in CRC than in control mucosa; IGF2 DMR0 hypomethylation associated with male sex, low tumor grade, microsatellite instability (MSI)-low/microsatellite stable (MSS), CpG island methylator phenotype (CIMP)-low/0, wild-type proto-oncogene B-Raf (wt BRAF), and proto-oncogene K-ras (KRAS) mutation and higher overall mortality [162]. | Diagnosis and prognosis | ||
| Hypomethylation of the six CTCF-binding sites in IGF2/H19 DMR were linked to IGF2 LOI (two forms of aberrant IGF2 expression) and promotes MSI and oncogenesis. [69] | Diagnosis and mechanisms | ||
| IGF2 was between the five genes with the highest average hypermethylated percentages (50.4%) of CRC patients [56]. | Diagnosis | ||
| Lower levels of IGF2 DMR0 methylation and ≥15 g/day alcohol consumption were associated with elevated risk of CRC [71]. | Diagnosis and mechanisms | ||
| IGF2 DMR0 hypomethylation and aberrant methylation of other iDMRs within the IGF2/H19 domain with no association with IGF2 LOI [174] | Diagnosis and mechanisms | ||
| The “insular” genomic aberrations in IGF2; mosaic distribution of methylation in 1 region of the primaries or the metastases [10] | Diagnosis and prognosis | ||
| Chromosomal gains | IGF2 overexpression in primary CRC and liver metastases was accompanied by chromosomal gains at 11p15.5 in a subset of CRC patients [6,141]. | Diagnosis and prognosis | |
| LOH | LOH of IGF2 was present in 13% of CRC patients with 33% with LOI of the IGF2 gene [70]. | Diagnosis | |
| Others | A tumor type-specific analysis uncovered that enhancer hijacking mediates gene dysregulation at the IGF2 locus in CRC [24]. | Mechanisms | |
| IGF2R gene | Genetic variants | Women homozygous for the IGF2R c.5002 G > A allele had higher mean levels of sIGF2; Whites homozygous for IGF2R c.901 C > G variant had a higher risk of CRC [59]. | Diagnosis and prognosis |
| lncRNAs | IGF2-AS | This key type of lncRNAs has correlation with certain clinical features (e.g., negative correlation between upregulation of IGF2-AS and distant metastasis) [15]. | Diagnosis and prognosis |
| This type of differentially expressed genes (DEG) was negatively correlated with overall survival (OS) [34]. | Prognosis | ||
| H19 | This type of lncRNA was increased significantly in immunodeficient mice induced with human colon cancer cells when compared with controls [163]. | Prognosis and therapeutics | |
| H19 upregulated a series of cell-cycle genes; eIF4A3 binds to H19. Higher expression of H19 was correlated with tumor differentiation and advanced Tumor-Node-Metastasis (TNM) stage [175]. | Prognosis, mechanisms, and therapeutics | ||
| H19 overexpressed in CRC tissues and cell lines; the interference of H19 by shRNA effectively decreased the migration and invasion of CRC cells. H19 shRNA strongly reduced the tumor growth and tumor volume in vivo [165]. | Mechanisms and therapeutics | ||
| Elevated expression of H19 lncRNA due to promoter demethylation was observed in cells isolated from metastases and was associated with poor survival of CRC patients [15]. | Diagnosis and prognosis | ||
| H19 was significantly upregulated in CRC tissues compared with normal control, which might regulate FSCN1 expression by competitively sponging miR-29b-3p [40]. | Diagnosis and mechanisms | ||
| H19 was upregulated in colon tumors and correlated with poor prognosis [46]. | Diagnosis, mechanisms, and prognosis | ||
| CRNDE | Regulates cellular metabolism, which may correlate with their upregulation in CRC; can promote the metabolic changes in cancer cells (switch to aerobic glycolysis) [37]. | Diagnosis and mechanisms | |
| miRNAs | miR-675 | H19-derived miRNA was upregulated in cell lines and primary CRC as compared to noncancerous tissues through downregulation of RB, increased tumor cells growth, and regulation of the CRC development [176]. | Mechanisms and therapeutics |
| miR-483 | miR-483 was a dominant driver oncogene at the IGF2 11p15.5 CRC amplicon, inducing dysplasia in vitro and tumorigenicity in vivo [109]. | Mechanisms and therapeutics | |
| miR-483-3p, miR-483-5p | The levels of IGF2, miR-483-3p, and -5p were synchronously increased in CRC tissues; IGF2 correlated with both types of miRNAs; and higher smiR-483-5p levels were found compared to controls [143]. | Diagnosis, mechanisms, and therapeutics | |
| miR-486-5p | Plasma miR-486-5p expression was upregulated in CRC; decreased levels were associated with TNM stage, larger tumor size, lymphatic metastasis, and poor prognosis [101]. | Diagnosis, mechanisms, prognosis, and therapeutics | |
| miR-491-5p | The overexpression of IGF2 rescued the miR-491-5p-induced suppression of proliferation in CRC cells; decreased plasma miR-491-5p expression in CRC was found compared to controls [177]. | Mechanisms, prognosis, and therapeutics | |
| miR-181a/135a/302c | DNA methylation mediated repression via repressing PLAG1/IGF2 signaling; promotes the microsatellite-unstable CRC development and 5-FU resistance [178]. | Mechanisms, prognosis, and therapeutics |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasprzak, A.; Adamek, A. Insulin-Like Growth Factor 2 (IGF2) Signaling in Colorectal Cancer—From Basic Research to Potential Clinical Applications. Int. J. Mol. Sci. 2019, 20, 4915. https://doi.org/10.3390/ijms20194915
Kasprzak A, Adamek A. Insulin-Like Growth Factor 2 (IGF2) Signaling in Colorectal Cancer—From Basic Research to Potential Clinical Applications. International Journal of Molecular Sciences. 2019; 20(19):4915. https://doi.org/10.3390/ijms20194915
Chicago/Turabian StyleKasprzak, Aldona, and Agnieszka Adamek. 2019. "Insulin-Like Growth Factor 2 (IGF2) Signaling in Colorectal Cancer—From Basic Research to Potential Clinical Applications" International Journal of Molecular Sciences 20, no. 19: 4915. https://doi.org/10.3390/ijms20194915
APA StyleKasprzak, A., & Adamek, A. (2019). Insulin-Like Growth Factor 2 (IGF2) Signaling in Colorectal Cancer—From Basic Research to Potential Clinical Applications. International Journal of Molecular Sciences, 20(19), 4915. https://doi.org/10.3390/ijms20194915





