Comparative Genome Analyses Reveal the Genomic Traits and Host Plant Adaptations of Flavobacterium akiainvivens IK-1T
Abstract
1. Introduction
2. Results
2.1. Genome Features of IK-1T
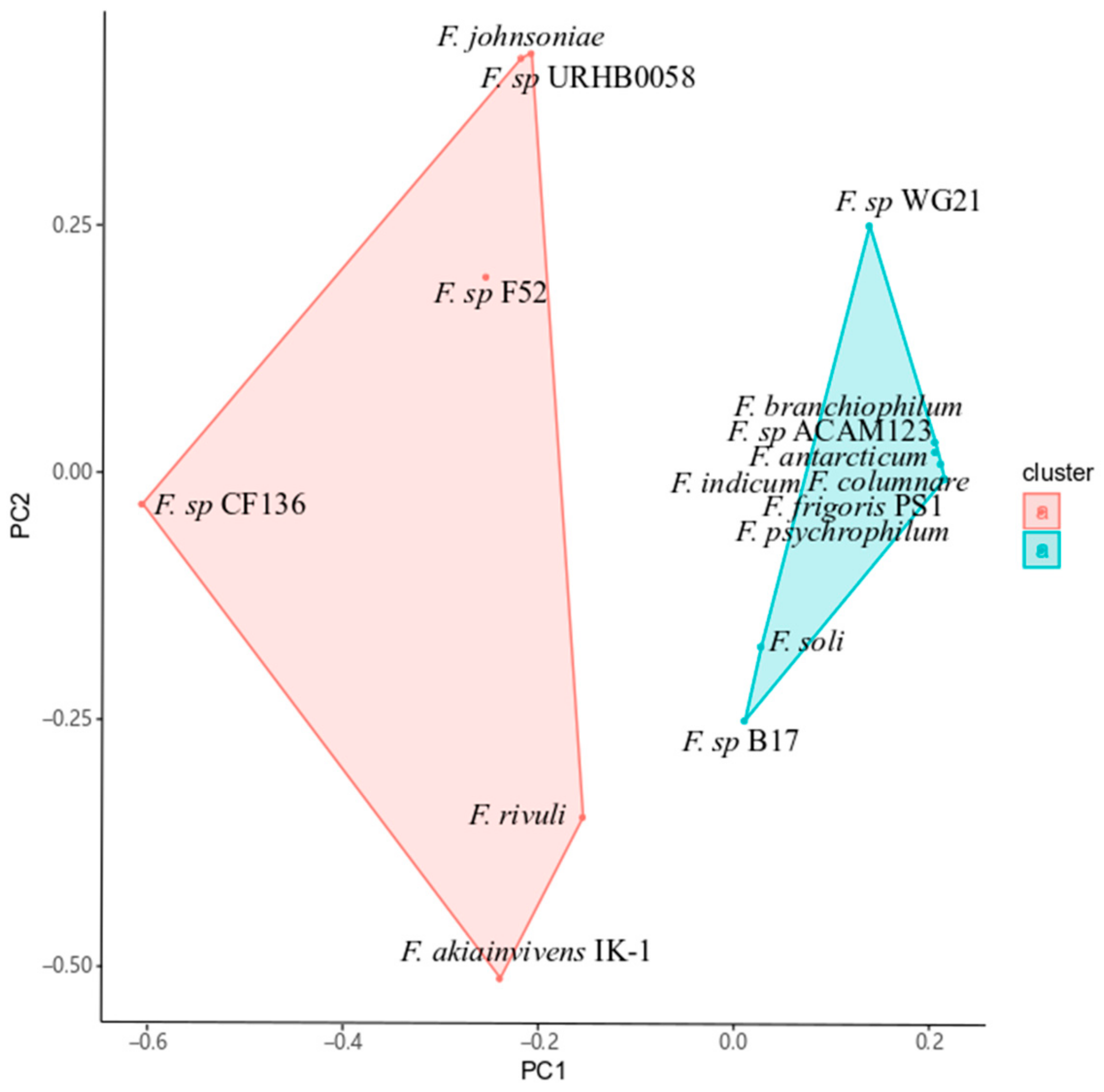
2.2. IK-1T is a Member of Terrestrial Clade-Affiliated Flavobacteria
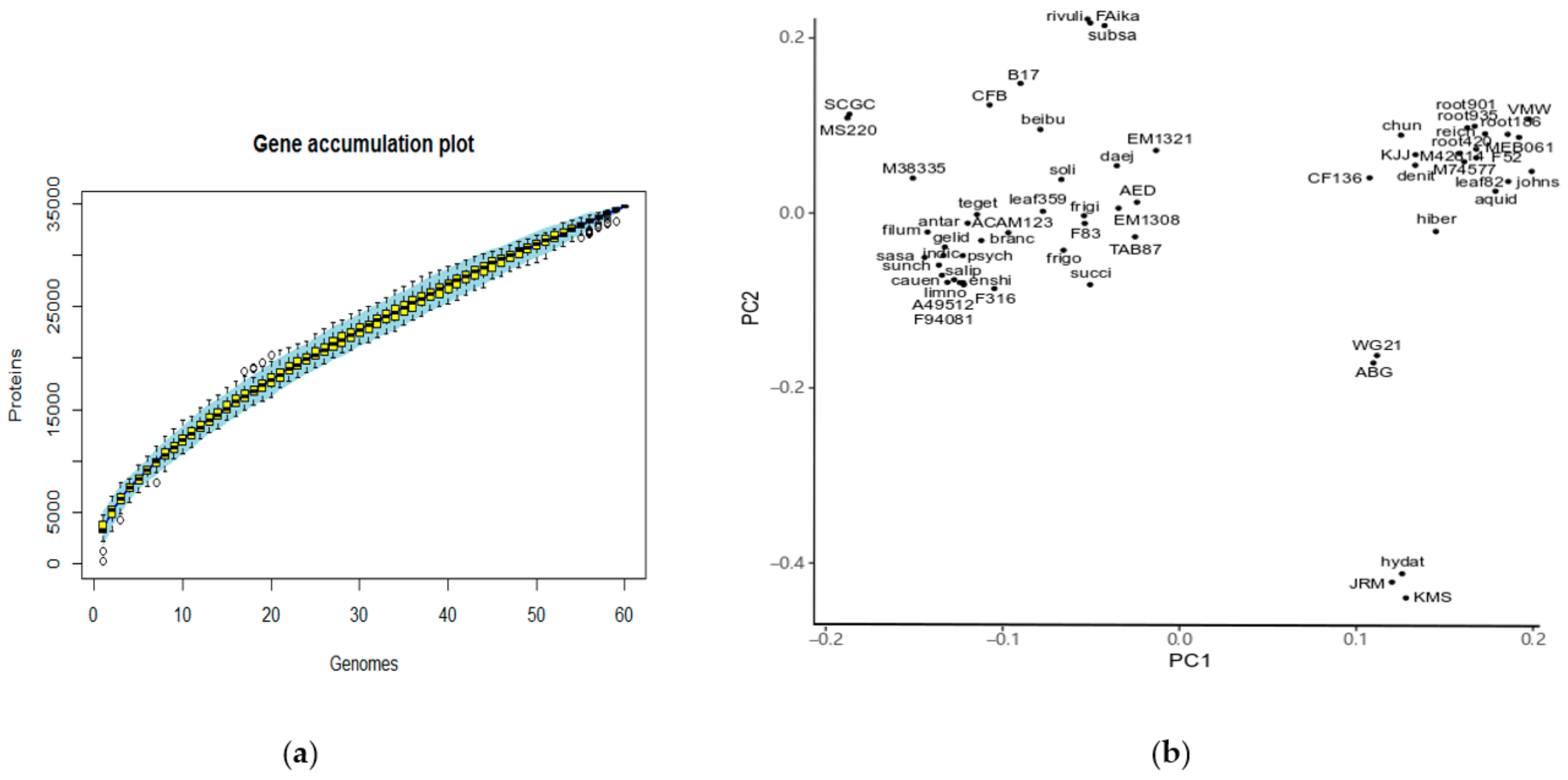
2.3. An Open Pan-Genome of Flavobacteria
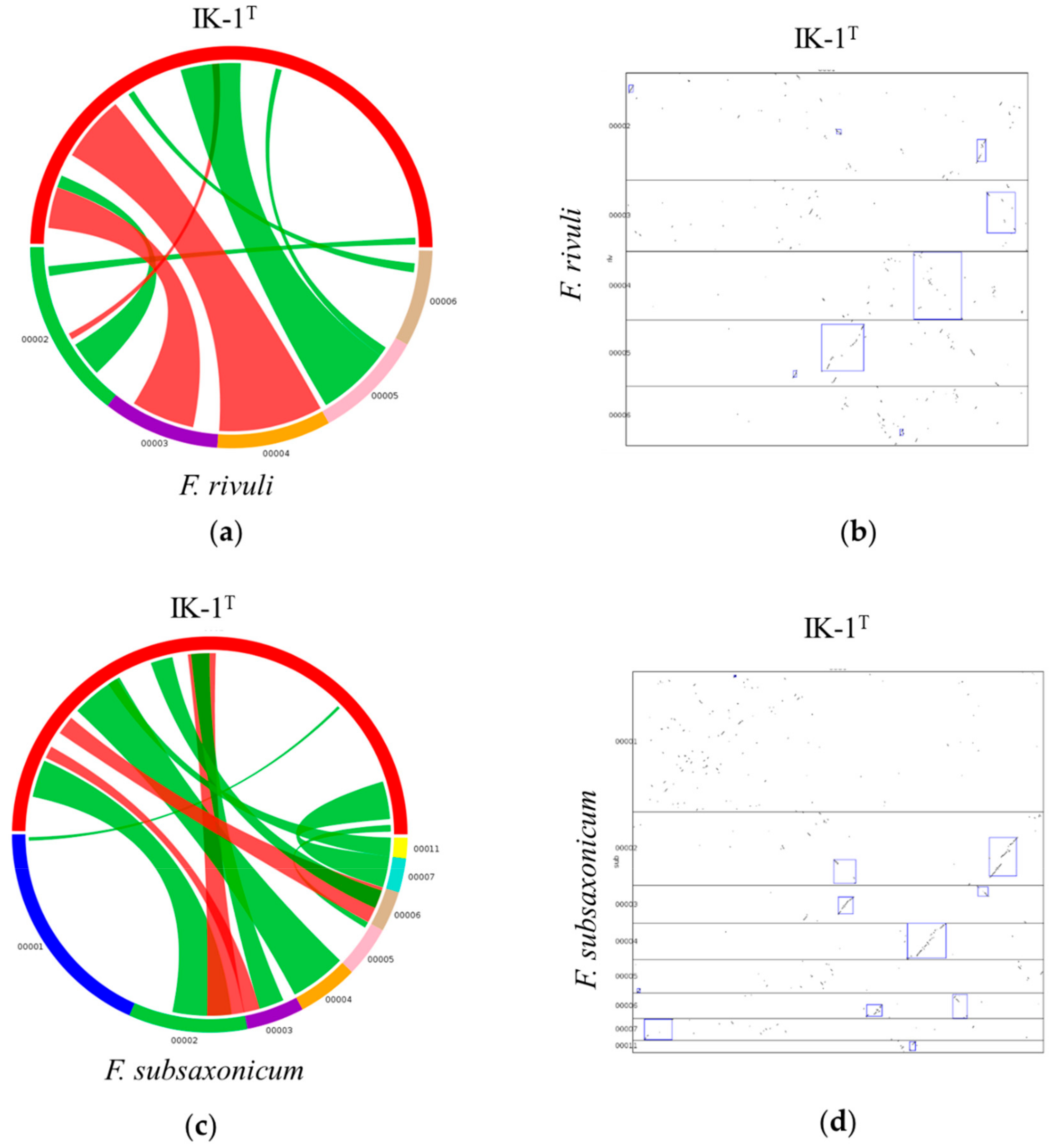
2.4. Synteny Between IK-1T and F. rivuli/F. subsaxonicum
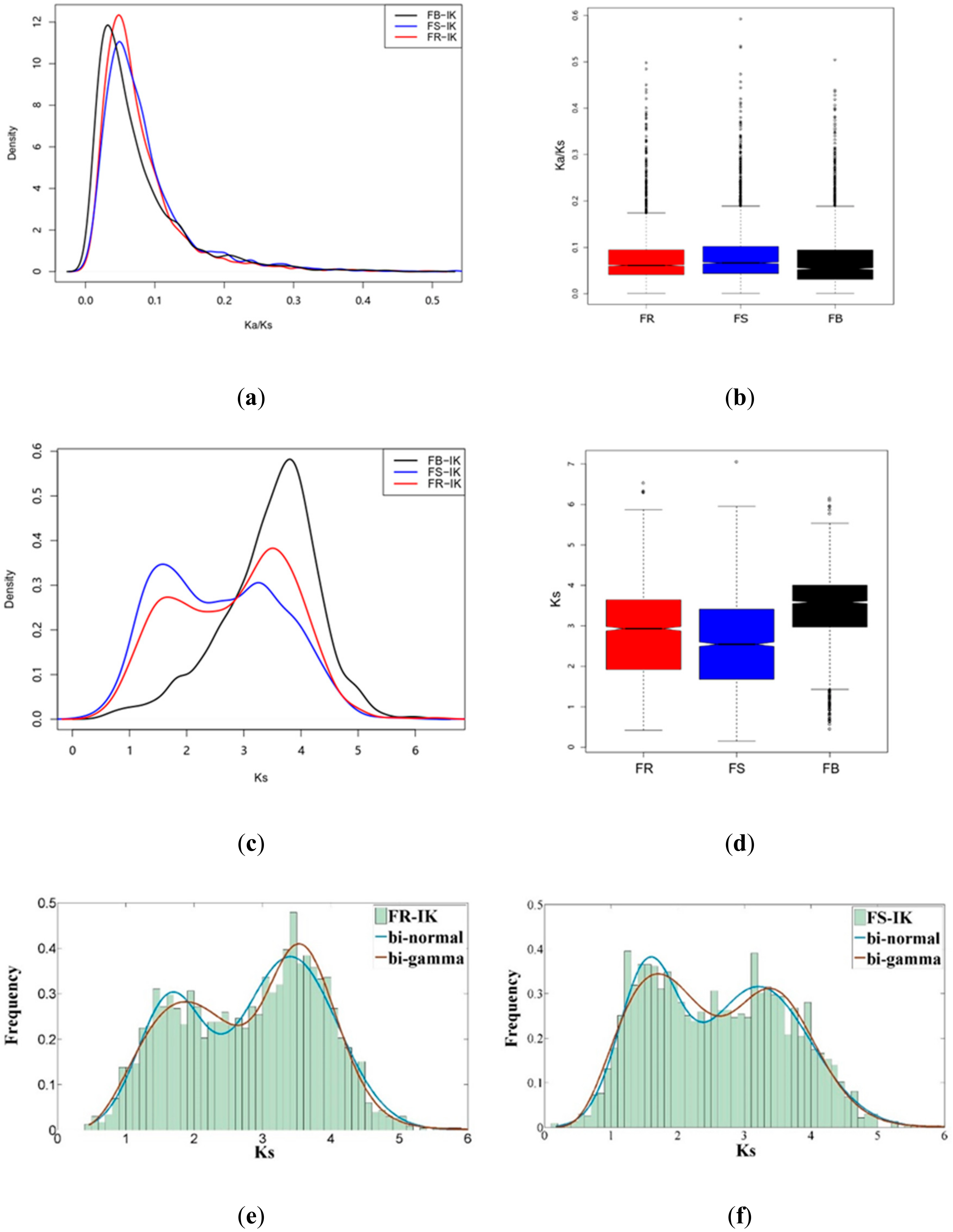
2.5. Evolution Direction in the IK-1T Genome
2.6. Gliding Motility and Type 9 Secretion System in IK-1T
2.7. Type 2 Secretion System in IK-1T
2.8. Eukaryotic-Like Domain Containing Proteins in IK-1T
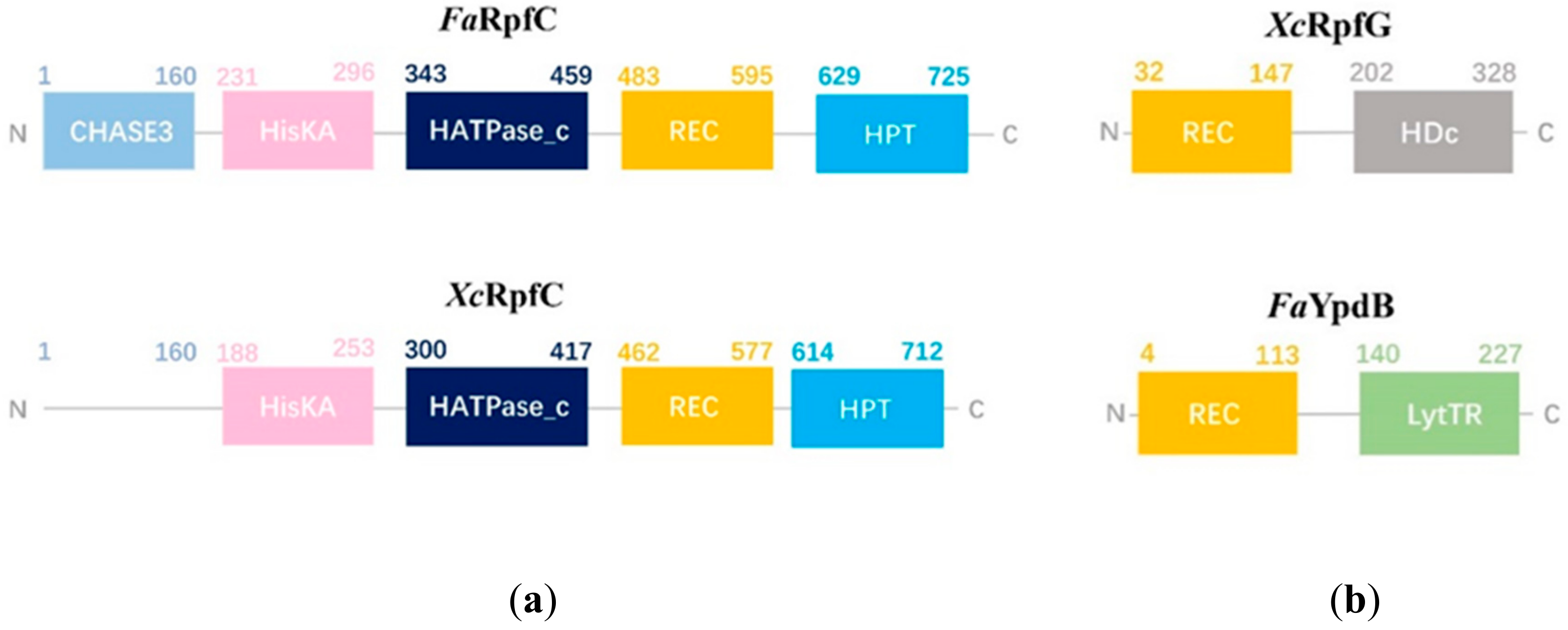
2.9. Quorum Sensing Mechanism
3. Discussion
4. Materials and Methods
4.1. Genome Functional Annotation
4.2. Annotation of Carbohydrate-Active Enzymes and Peptidases
4.3. Clustering of Orthologous Groups
4.4. Analysis of Synonymous/Nonsynonymous Substitution Rates
4.5. Analysis of Flavobacterium Akiainvivens IK-1T Secretion System
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| IK-1T | Flavobacterium akiainvivens IK-1T |
| LUCA | last universal common ancestor |
| MRCA | most recent common ancestor |
| HGT | horizontal gene transfer |
| ELD | eukaryotic-like domains |
| ANK | ankyrin repeat |
| vWFA | von Willebrand factor type A domain |
| MRJP | major royal jelly proteins |
References
- Bergey, D.H.; Harrison, F.C.; Breed, R.S.; Hammer, B.W.; Huntoon, F.M. Bergey’s Manual of Determinative Bacteriology, 1st ed.; The Williams and Wilkins Co.: Baltimore, MD, USA, 1923; pp. 1–442. [Google Scholar]
- Kirchman, D.L. The ecology of Cytophaga-Flavobacteria in aquatic environments. Fems Microbiol. Ecol. 2002, 39, 91–100. [Google Scholar] [CrossRef]
- Fernández-Gómez, B.; Richter, M.; Schüler, M.; Pinhassi, J.; Acinas, S.G.; González, J.M.; Pedrós-Alió, C. Ecology of marine Bacteroidetes: A comparative genomics approach. ISME J. 2013, 7, 1026–1037. [Google Scholar] [CrossRef] [PubMed]
- Teeling, H.; Fuchs, B.M.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.; Mann, A.J.; Waldmann, J.; et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 2012, 336, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Kolton, M.; Sela, N.; Elad, Y.; Cytryn, E. Comparative Genomic Analysis Indicates that Niche Adaptation of Terrestrial Flavobacteria Is Strongly Linked to Plant Glycan Metabolism. PLoS ONE 2013, 8, e76704. [Google Scholar] [CrossRef] [PubMed]
- Soltani, A.A.; Khavazi, K.; Asadi-Rahmani, H.; Omidvari, M.; Dahaji, P.A.; Mirhoseyni, H. Plant growth promoting characteristics in some Flavobacterium spp. isolated from soils of Iran. J. Agric. Sci. 2010, 2, 106–115. [Google Scholar] [CrossRef]
- Bodour, A.A.; Guerrero-barajas, C.; Jiorle, B.V.; Malcomson, M.E.; Paull, A.K.; Somogyi, A.; Trinh, L.N.; Bates, R.B.; Maier, R.M. Structure and characterization of Flavolipids, a novel class of biosurfactants produced by Flavobacterium sp. strain MTN11. Appl. Environ. Microbiol. 2004, 70, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Xun, L. Purification and characterization of 2,6-di-chloro-p-hydroquinone chlorohydrolase from Flavobacterium sp. strain ATCC 39723. J. Bacteriol. 1997, 179, 1521–1524. [Google Scholar]
- Lo, K.V.; Zhu, C.M.; Cheuk, W. Biodegradation of pentachloro-phenol by Flavobacterium species in batch and immobilized continuous reactors. Env. Technol. 1998, 19, 91–96. [Google Scholar] [CrossRef]
- McAllister, K.A.; Lee, H.; Trevors, J.T. Microbial degradation of pentachlorophenol. Biodegradation. 1996, 7, 1–40. [Google Scholar] [CrossRef]
- Negoro, S. Biodegradation of nylon oligomers. Appl. Microbiol. Biotechnol. 2000, 54, 461–466. [Google Scholar] [CrossRef]
- Wang, S.Y.; Vipulanandan, C. 2001 Biodegradation of naphthalene-contaminated soils in slurry bioreactors. J. Env. Eng. Asce. 2000, 127, 748–754. [Google Scholar] [CrossRef]
- Bodour, A.A.; Drees, K.P.; Maier, R.M. Distribution of Biosurfactant-Producing Bacteria in Undisturbed and Contaminated Arid Southwestern Soils. Appl. Env. Microbiol. 2003, 69, 3280–3287. [Google Scholar] [CrossRef] [PubMed]
- Kuo, I.; Saw, J.; Kapan, D.D.; Christensen, S.; Kaneshiro, K.Y.; Donachie, S.P. Flavobacterium akiainvivens sp. nov., from decaying wood of Wikstroemia oahuensis, Hawai‘i, and emended description of the genus Flavobacterium. Int. J. Syst. Evol. Microbiol. 2013, 63, 3280–3286. [Google Scholar] [CrossRef] [PubMed]
- Hahnke, R.L.; Stackebrandt, E.; Meier-Kolthoff, J.P.; Tindall, B.J.; Huang, S.; Rohde, M.; Lapidus, A.; Han, J.; Trong, S.; Hayneset, M.; et al. High quality draft genome sequence of Flavobacterium rivuli type strain WB 3. 3-2 T (DSM 21788T), a valuable source of polysaccharide decomposing enzymes. Stand. Genom. Sci. 2015, 10. [Google Scholar] [CrossRef]
- Ali, Z.; Cousin, S.; Frühling, A.; Brambilla, E.; Schumann, P.; Yang, Y.; Stackebrandt, E. Flavobacterium rivuli sp. nov., Flavobacterium subsaxonicum sp. nov., Flavobacterium swingsii sp. nov. and Flavobacterium reichenbachii sp. nov., isolated from a hard water rivulet. Int. J. Syst. Evol. Microbiol. 2009, 59, 2610–2617. [Google Scholar] [CrossRef] [PubMed]
- Cousin, S. Flavobacterial community structure in a hardwater rivulet and adjacent forest soil, Harz mountain, Germany. Curr. Microbiol. 2009, 58, 409–415. [Google Scholar] [CrossRef]
- Wan, X.; Hou, S.; Saito, J.A.; Kaneshiro, K.Y.; Donachie, S.P. Genome Sequence of Flavobacterium akiainvivens IK-1T, Isolated from Decaying Wikstroemia oahuensis, an Endemic Hawaiian Shrub. Genome Announc. 2015, 3. [Google Scholar] [CrossRef]
- Heaps, H.S. Information Retrieval: Computational and Theoretical Aspects; Academic Press Inc.: Orlando, FL, USA, 1978. [Google Scholar]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, N.; Tai, P.C. SecE-depleted Membranes of Escherichia coli Are Active. J. Biol. Chem. 1997, 272, 13660–13665. [Google Scholar] [CrossRef]
- Beckwith, J. The Sec-dependent pathway. Res. Microbiol. 2014, 164, 497–504. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Aravind, L.; Koonin, E.V. Eukaryotic Signalling Domain Homologues in Archaea and Bacteria Ancient. J. Mol. Biol. 1999, 289, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, V.; Nussbaumer, T.; Platzer, A.; Jehl, M.A.; Arnold, R.; Rattei, T. EffectiveDB-updates and novel features for a better annotation of bacterial secreted proteins and Type III, IV, VI secretion systems. Nucleic Acids Res. 2016, 44, D669–D674. [Google Scholar] [CrossRef] [PubMed]
- Al-Khodor, S.; Price, C.T.; Kalia, A.; Abu Kwaik, Y. Ankyrin-repeat containing proteins of microbes: A conserved structure with functional diversity. Trends in Microbiol. 2010, 18, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, E.T.; Sarver, R.W.; Bryant, G.L., Jr.; Curry, K.A.; Fairbanks, M.B.; Finzel, B.C.; Garlick, R.L.; Heinrikson, R.L.; Horton, N.C.; Kelley, L.C.; et al. Cation binding to the integrin CD11b domain and activation model assessment. Structures 1998, 6, 923–935. [Google Scholar] [CrossRef]
- Drapeau, M.D.; Albert, S.; Kucharski, R.; Prusko, C.; Maleszka, R. Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees. Genome Res. 2006, 16, 1385–1404. [Google Scholar] [CrossRef]
- Da Silva, F.R.; Vettore, A.L.; Kemper, E.L.; Leite, A.; Arruda, P. Fastidian gum: The exopolysaccharide possibly involved in bacterial pathogenicity. FEMS Microbiol. Lett. 2001, 203, 165–171. [Google Scholar] [CrossRef]
- Locey, K.J.; Lennon, J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 5970–5975. [Google Scholar] [CrossRef]
- Mcbride, M.J.; Zhu, Y. Gliding Motility and Por Secretion System Genes Are Widespread among Members of the Phylum Bacteroidetes. J. Bacteriol. 2013, 195, 270–278. [Google Scholar] [CrossRef]
- Sato, K.; Naito, M.; Yukitake, H.; Hirakawa, H.; Shoji, M.; McBride, M.J.; Rhodes, R.G.; Nakayama, K. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 276–281. [Google Scholar] [CrossRef]
- Abby, S.S.; Rocha, E.P.C. Identification of protein secretion systems in bacterial genomes using MacSyFinder. Methods Mol. Biol. 2017, 1615, 1–21. [Google Scholar] [PubMed]
- Song, C.; Kumar, A.; Saleh, M. Bioinformatic Comparison of Bacterial Secretomes. Genom. Proteom. Bioinform. 2009, 7, 37–46. [Google Scholar] [CrossRef]
- Zhulin, I.B.; Nikolskaya, A.N.; Galperin, M.Y. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in Bacteria and Archaea. J. Bacteriol. 2003, 185, 285–294. [Google Scholar] [CrossRef]
- Mascher, T.; Helmann, J.D.; Unden, G. Stimulus Perception in Bacterial Signal-Transducing Histidine Kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 910–938. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarczyk, A.; Hochstrasser, R.; Vorholt, J.A.; Francez-Charlot, A. Two-Tiered Histidine Kinase Pathway Involved in Heat Shock and Salt Sensing in the General Stress Response of Sphingomonas melonis Fr1. J. Bacteriol. 2015, 197, 1466–1477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformation 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013–2015. Available online: http://repeatmasker.org/.
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, 116–120. [Google Scholar] [CrossRef]
- Burge, S.W.; Daub, J.; Eberhardt, R.; Tate, J.; Barquist, L.; Nawrocki, E.P.; Eddy, S.R.; Gardner, P.P.; Bateman, A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2012, 41, 226–232. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Soderlund, C.; Bomhoff, M.; Nelson, W. SyMAP v3.4: A turnkey synteny system with application to plant genomes. Nucleic Acids Res. 2011, 39, 68. [Google Scholar] [CrossRef]
- Soderlund, C.; Nelson, W.; Shoemaker, A.; Paterson, A. SyMAP: A system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006, 16, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2006, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. R package version 2.5–5. 2019. Available online: https://github.com/vegandevs/vegan.
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified Interface to Visualize Statistical Result of Popular R Packages. R. J. 2016, 8, 478–489. [Google Scholar] [CrossRef]
- Horikoshi, M.; Tang, Y. ggfortify: Data Visualization Tools for Statistical Analysis Results. 2016. Available online: https://CRAN.R-project.org/package=ggfortify (accessed on 30 August 2019).
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting secretory proteins with SignalP. Protein Funct. Predict. 2017, 1611, 59–73. [Google Scholar]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, X. Comparative Genome Analyses Reveal the Genomic Traits and Host Plant Adaptations of Flavobacterium akiainvivens IK-1T. Int. J. Mol. Sci. 2019, 20, 4910. https://doi.org/10.3390/ijms20194910
Wan X. Comparative Genome Analyses Reveal the Genomic Traits and Host Plant Adaptations of Flavobacterium akiainvivens IK-1T. International Journal of Molecular Sciences. 2019; 20(19):4910. https://doi.org/10.3390/ijms20194910
Chicago/Turabian StyleWan, Xuehua. 2019. "Comparative Genome Analyses Reveal the Genomic Traits and Host Plant Adaptations of Flavobacterium akiainvivens IK-1T" International Journal of Molecular Sciences 20, no. 19: 4910. https://doi.org/10.3390/ijms20194910
APA StyleWan, X. (2019). Comparative Genome Analyses Reveal the Genomic Traits and Host Plant Adaptations of Flavobacterium akiainvivens IK-1T. International Journal of Molecular Sciences, 20(19), 4910. https://doi.org/10.3390/ijms20194910




