piggyBac Transposon-Based Immortalization of Human Deciduous Tooth Dental Pulp Cells with Multipotency and Non-Tumorigenic Potential
Abstract
1. Introduction
2. Results
2.1. Strategy for Generating Immortalized HDDPCs
2.2. Successful Establishment of Immortalized HDDPCs
2.3. Characterization of Established MT_E7 and MT_hTERT Lines
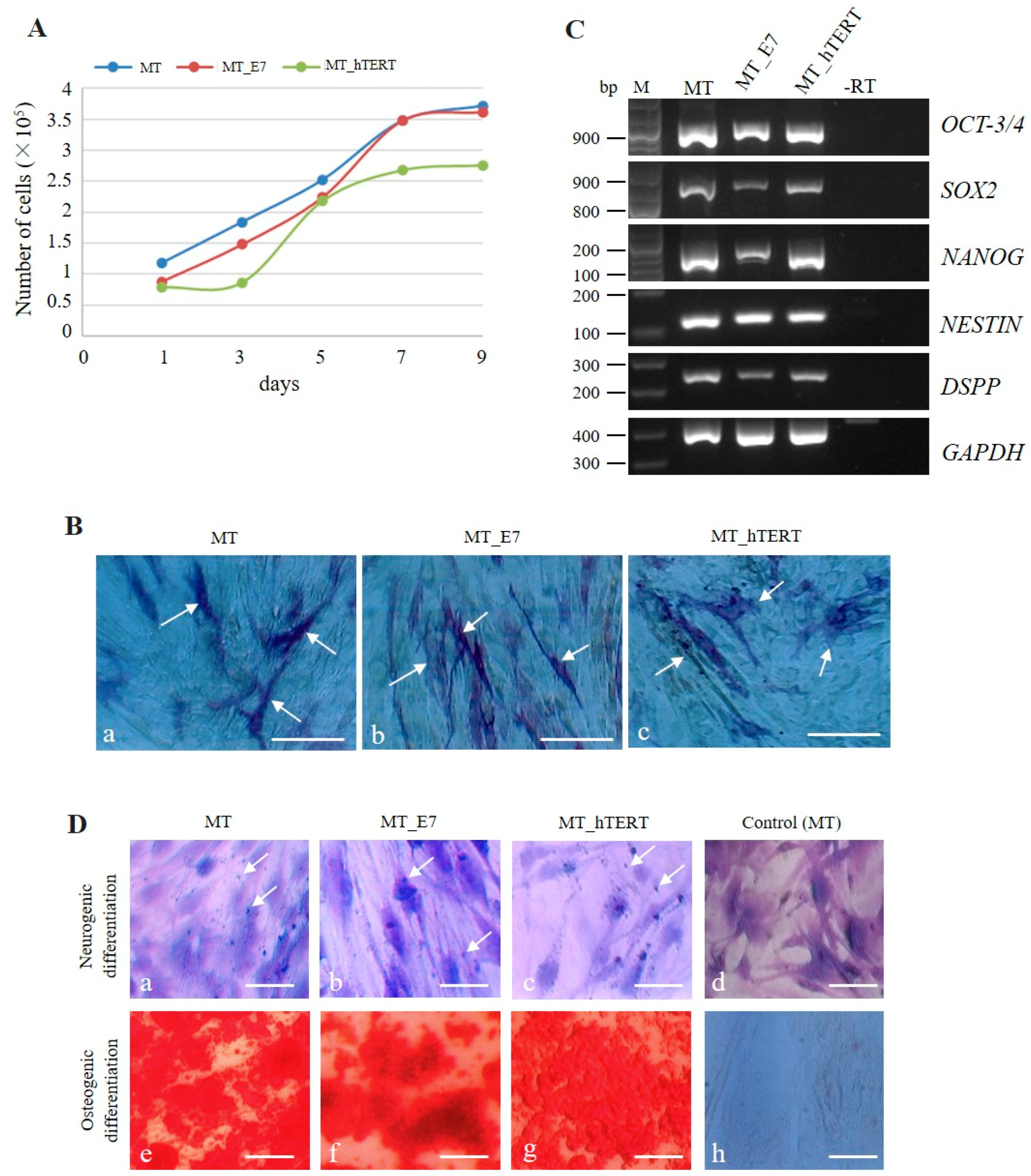
2.3.1. Transgenic Gene Expression and Cell Proliferation
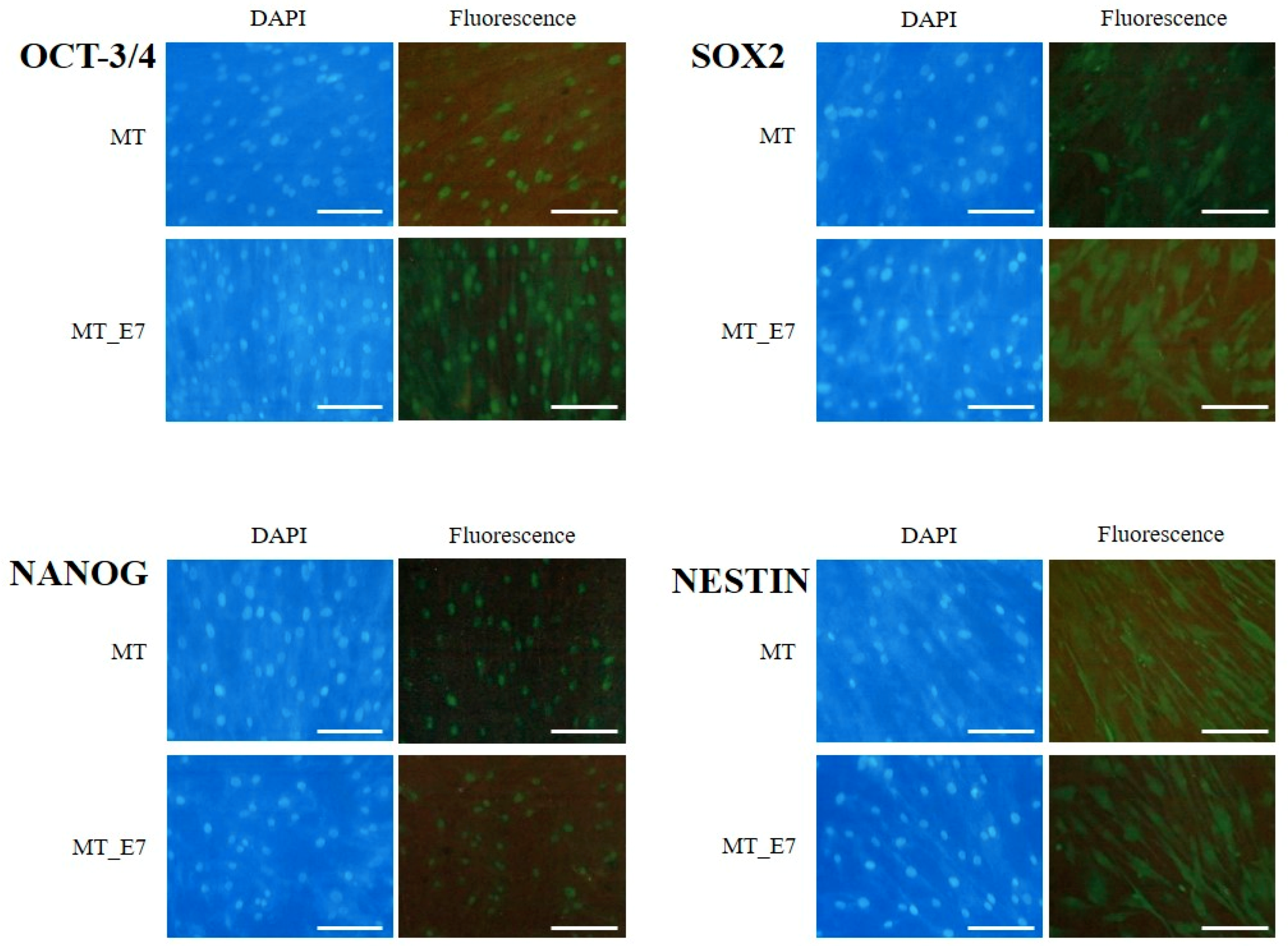
2.3.2. Stemness
2.3.3. Multipotency
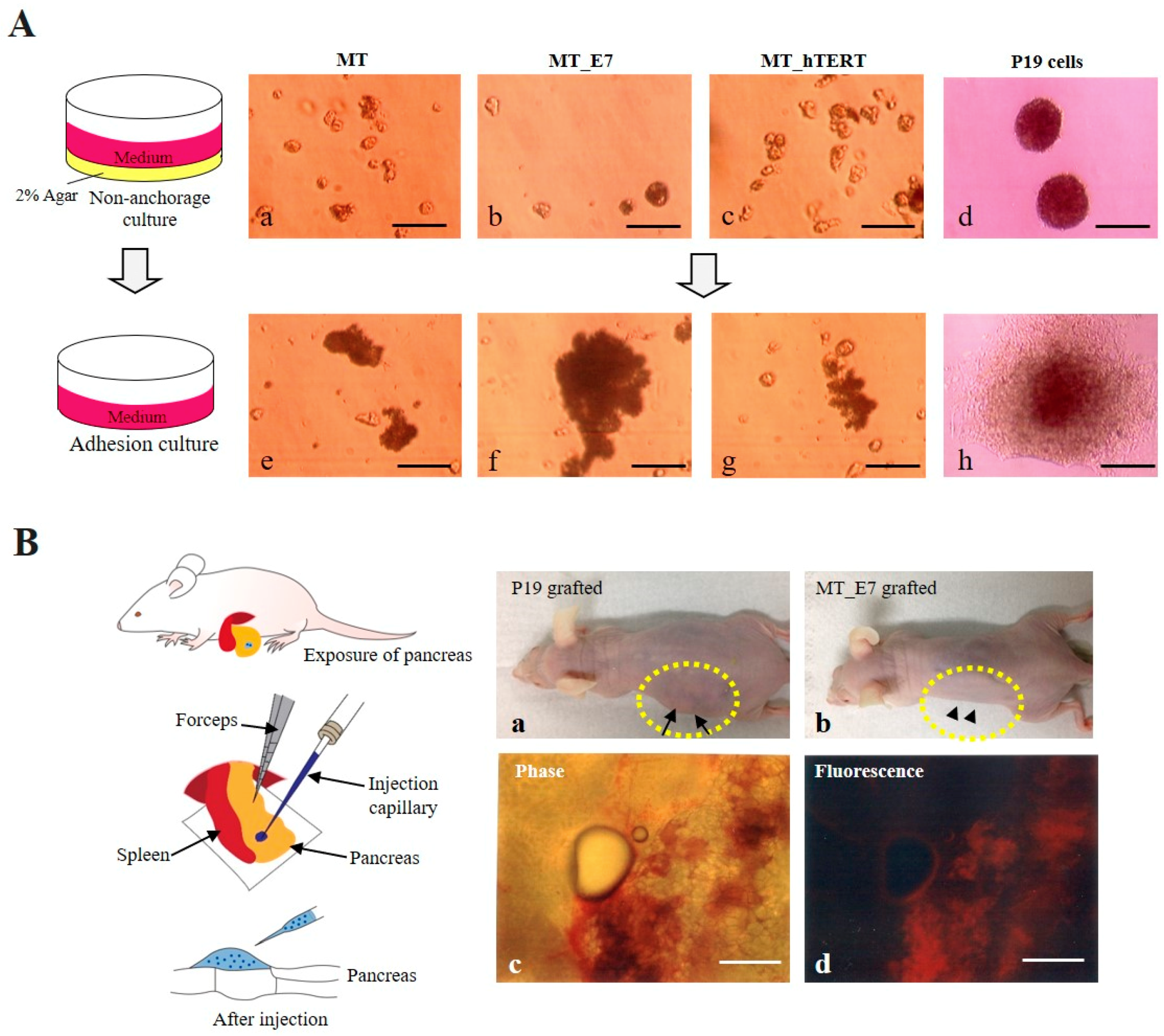
2.3.4. Immortalization
2.3.5. Tumorigenic potential
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Cells
4.3. Construction of PB Transposon Vectors
4.4. Generation of Immortalized HDDPC Lines
4.5. Fluorescence Observation
4.6. Cell Growth Assay
4.7. ALP Assay
4.8. PCR Analysis
4.9. RT-PCR Analysis
4.10. Immunocytochemical Staining
4.11. Induction of In Vitro Differentiation
4.12. Anchorage-Dependent Growth Assay
4.13. In Vivo Tumorigenic Assay
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HD | Human deciduous teeth |
| DPCs | Dental pulp cells |
| HDDPCs | Human deciduous teeth-derived dental pulp cells |
| SV40 | Simian virus 40 |
| HPV16 | Human papilloma virus 16 |
| SV40 TAg | SV40 large T antigen |
| HPV16-E7 | E7 oncoprotein of HPV16 |
| hTR | Human telomerase RNA |
| hTERT | Human telomerase reverse transcriptase |
| hTEP1 | Human telomerase-associated protein 1 |
| DPSCs | Dental pulp stem cells |
| PB | piggyBac |
| CAG | Chicken β-actin gene-based promoter |
| pac | Puromycin acetyltransferase gene |
| PGK | Phoshoglycerate kinase |
| tdTomato | Tandem dimer Tomato |
| EGFP | Enhanced green fluorescent protein |
| GEU | Gene expression unit |
| ALP | Alkaline phosphatase |
| PBS | Dulbecco’s modified Ca2+, Mg2+-free phosphate-buffered saline |
| iPS | Induced pluripotent stem |
| OCT-3/4 | Octamer-binding transcription factor-3/4 |
| SOX2 | Sex determining region Y-box 2 |
| DSPP | Dentin sialophosphoprotein |
| IP | Intraperitoneal |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | Fetal bovine serum |
| PFA | Paraformaldehyde |
| RT | Room temperature |
| PBS | Dulbecco’s modified Ca2+, Mg2+-free phosphate-buffered saline |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| NGS | Normal goat serum |
| DAPI | 4′,6-diamidino-2-phenylindole |
References
- Miura, M.; Gronthos, S.; Zhao, M.; Lu, B.; Fisher, L.W.; Robey, P.G.; Shi, S. SHED: Stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. USA 2003, 100, 5807–5812. [Google Scholar] [CrossRef] [PubMed]
- Martinez Saez, D.; Sasaki, R.T.; Neves, A.D.; da Silva, M.C. Stem cells from human exfoliated deciduous teeth: A growing literature. Cells Tissues Organs 2016, 202, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Sang, Y.; Zhang, F.; Liu, Z.; Qi, N.; Chen, Y. Comparative analysis of human mesenchymal stem cells from umbilical cord, dental pulp, and menstrual blood as sources for cell therapy. Stem Cells Int. 2016, 2016, 3516574. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Chang, P.L.; Gunby, J.L.; Tomkins, D.J.; Mak, I.; Rosa, N.E.; Mak, S. Transformation of human cultured fibroblasts with plasmids carrying dominant selection markers and immortalizing potential. Exp. Cell Res. 1986, 167, 407–416. [Google Scholar] [CrossRef]
- Pirisi, L.; Yasumoto, S.; Feller, M.; Doniger, J.; DiPaolo, J.A. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. J. Virol. 1987, 61, 1061–1066. [Google Scholar] [PubMed]
- Lechner, M.S.; Laimins, L.A. Human epithelial cells immortalized by SV40 retain differentiation capabilities in an in vitro raft system and maintain viral DNA extrachromosomally. Virology 1991, 185, 563–571. [Google Scholar] [CrossRef]
- Shay, J.W.; Pereira-Smith, O.M.; Wright, W.E. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 1991, 196, 33–39. [Google Scholar] [CrossRef]
- Stewart, N.; Bacchetti, S. Expression of SV40 large T antigen, but not small t antigen, is required for the induction of chromosomal aberrations in transformed human cells. Virology 1991, 180, 49–57. [Google Scholar] [CrossRef]
- Halbert, C.L.; Demers, G.W.; Galloway, D.A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 1991, 65, 473–478. [Google Scholar]
- Zwerschke, W.; Jansen-Durr, P. Cell transformation by the E7 oncoprotein of human papillomavirus type 16: Interactions with nuclear and cytoplasmic target proteins. Adv. Cancer Res. 2000, 78, 1–29. [Google Scholar] [PubMed]
- Huang, S.M.; McCance, D.J. Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J. Virol. 2002, 76, 8710–8721. [Google Scholar] [CrossRef] [PubMed]
- Darimont, C.; Mace, K. Immortalization of human preadipocytes. Biochimie 2003, 85, 1231–1233. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W. Telomere length regulation. Annu. Rev. Biochem. 1996, 65, 337–365. [Google Scholar] [CrossRef]
- Watson, J.D. Origin of concatemeric T7 DNA. Nat.: New Biol. 1972, 239, 197–201. [Google Scholar]
- Lustig, A.J. Crisis intervention: The role of telomerase. Proc. Natl. Acad. Sci. USA 1999, 96, 3339–3341. [Google Scholar] [CrossRef]
- Harrington, L.; McPhail, T.; Mar, V.; Zhou, W.; Oulton, R.; Bass, M.B.; Arruda, I.; Robinson, M.O. A mammalian telomerase-associated protein. Science 1997, 275, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Avilion, A.A.; Piatyszek, M.A.; Gupta, J.; Shay, J.W.; Bacchetti, S.; Greider, C.W. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996, 56, 645–650. [Google Scholar]
- Harrington, L.; Zhou, W.; McPhail, T.; Oulton, R.; Yeung, D.S.; Mar, V.; Bass, M.B.; Robinson, M.O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997, 11, 3109–3115. [Google Scholar] [CrossRef]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of life-span by introduction of telomerase into normal human cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef]
- Cong, Y.S.; Wright, W.E.; Shay, J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002, 66, 407–425. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, K.; Qu, X.; Wang, H.; Zhu, H.; Xu, X.; Zhang, M.; Tang, Y.; Yang, X. hTERT gene immortalized human adipose-derived stem cells and its multiple differentiations: A preliminary investigation. Appl. Biochem. Biotechnol. 2013, 169, 1546–1556. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, O.A.; Kaplunov, J.; Herbig, U.; Detoledo, S.; Azzam, E.I.; Santos, J.H. Expression of (NES-)hTERT in cancer cells delays cell cycle progression and increases sensitivity to genotoxic stress. PLoS ONE 2010, 5, e10812. [Google Scholar] [CrossRef] [PubMed]
- Potdar, P.D.; Jethmalani, Y.D. Human dental pulp stem cells: Applications in future regenerative medicine. World J. Stem Cells 2015, 7, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Kamata, N.; Fujimoto, R.; Tomonari, M.; Taki, M.; Nagayama, M.; Yasumoto, S. Immortalization of human dental papilla, dental pulp, periodontal ligament cells and gingival fibroblasts by telomerase reverse transcriptase. J. Oral Pathol. Med. 2004, 33, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Ueda, H.; Iizuka, S.; Sakamoto, K.; Oka, H.; Kudo, Y.; Ogawa, I.; Miyauchi, M.; Tahara, H.; Takata, T. Immortalization and characterization of human dental pulp cells with odontoblastic differentiation. Arch. Oral Biol. 2007, 52, 727–731. [Google Scholar] [CrossRef]
- Ikbale, el-A.; Goorha, S.; Reiter, L.T.; Miranda-Carboni, G.A. Effects of hTERT immortalization on osteogenic and adipogenic differentiation of dental pulp stem cells. Data Brief 2016, 6, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Maeda, H.; Wada, N.; Kano, Y.; Akamine, A. Establishing and characterizing human periodontal ligament fibroblasts immortalized by SV40T-antigen and hTERT gene transfer. Cell Tissue Res. 2006, 324, 117–125. [Google Scholar] [CrossRef]
- Inada, E.; Saitoh, I.; Kubota, N.; Soda, M.; Matsueda, K.; Murakami, T.; Sawami, T.; Kagoshima, A.; Yamasaki, Y.; Sato, M. Alkaline phosphatase and OCT-3/4 as useful markers for predicting susceptibility of human deciduous teeth-derived dental pulp cells to reprogramming factor-induced iPS cells. J. Investig. Clin. Dent. 2017, 8. [Google Scholar] [CrossRef]
- Wilson, M.H.; Coates, C.J.; George, A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. J. Am. Soc. Gene Ther. 2007, 15, 139–145. [Google Scholar] [CrossRef]
- Kahlig, K.M.; Saridey, S.K.; Kaja, A.; Daniels, M.A.; George, A.L., Jr.; Wilson, M.H. Multiplexed transposon-mediated stable gene transfer in human cells. Proc. Natl. Acad. Sci. USA 2010, 107, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhuang, Y.; Han, M.; Xu, T.; Wu, X. piggyBac as a high-capacity transgenesis and gene-therapy vector in human cells and mice. Dis. Models Mech. 2013, 6, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Inada, E.; Saitoh, I.; Watanabe, S.; Aoki, R.; Miura, H.; Ohtsuka, M.; Murakami, T.; Sawami, T.; Yamasaki, Y.; Sato, M. PiggyBac transposon-mediated gene delivery efficiently generates stable transfectants derived from cultured primary human deciduous tooth dental pulp cells (HDDPCs) and HDDPC-derived iPS cells. Int. J. Oral Sci. 2015, 7, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.P.; Yang, M.M.; Chen, Y.L. PiggyBac transposon-mediated gene transfer in Cashmere goat fetal fibroblast cells. Biosci. Biotechnol. Biochem. 2012, 76, 933–937. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Maeda, K.; Koriyama, M.; Inada, E.; Saitoh, I.; Miura, H.; Ohtsuka, M.; Nakamura, S.; Sakurai, T.; Watanabe, S.; et al. The piggyBac-based gene delivery system can confer successful production of cloned porcine blastocysts with multigene constructs. Int. J. Mol. Sci. 2016, 17, 1424. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Wu, X.; Li, G.; Han, M.; Zhuang, Y.; Xu, T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 2005, 122, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Inada, E.; Saitoh, I.; Matsumoto, Y.; Ohtsuka, M.; Miura, H.; Nakamura, S.; Sakurai, T.; Watanabe, S. A combination of targeted toxin technology and the piggyBac-mediated gene transfer system enables efficient isolation of stable transfectants in nonhuman mammalian cells. Biotechnol. J. 2015, 10, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Yamamura, K.; Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991, 108, 193–199. [Google Scholar] [PubMed]
- Fraser, M.J.; Ciszczon, T.; Elick, T.; Bauser, C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996, 5, 141–151. [Google Scholar] [CrossRef]
- Bauser, C.A.; Elick, T.A.; Fraser, M.J. Proteins from nuclear extracts of two lepidopteran cell lines recognize the ends of TTAA-specific transposons piggyBac and tagalong. Insect Mol. Biol. 1999, 8, 223–230. [Google Scholar] [CrossRef]
- Ginsburg, M.; Snow, M.H.; McLaren, A. Primordial germ cells in the mouse embryo during gastrulation. Development 1990, 110, 521–528. [Google Scholar] [PubMed]
- Berrill, A.; Tan, H.L.; Wuang, S.C.; Fong, W.J.; Choo, A.B.; Oh, S.K. Assessment of stem cell markers during long-term culture of mouse embryonic stem cells. Cytotechnology 2004, 44, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Morin, G.B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 1989, 59, 521–529. [Google Scholar] [CrossRef]
- Takubo, K.; Nakamura, K.; Izumiyama, N.; Mafune, K.; Tanaka, Y.; Miyashita, M.; Sasajima, K.; Kato, M.; Oshimura, M. Telomerase activity in esophageal carcinoma. J. Surg. Oncol. 1997, 66, 88–92. [Google Scholar] [CrossRef]
- DeRita, R.M.; Sayeed, A.; Garcia, V.; Krishn, S.R.; Shields, C.D.; Sarker, S.; Friedman, A.; McCue, P.; Molugu, S.K.; Rodeck, U.; et al. Tumor-Derived Extracellular Vesicles Require beta1 Integrins to Promote Anchorage-Independent Growth. iScience 2019, 14, 199–209. [Google Scholar] [CrossRef] [PubMed]
- McBurney, M.W. P19 embryonal carcinoma cells. Int. J. Dev. Biol. 1993, 37, 135–140. [Google Scholar] [PubMed]
- Gronthos, S.; Brahim, J.; Li, W.; Fisher, L.W.; Cherman, N.; Boyde, A.; DenBesten, P.; Robey, P.G.; Shi, S. Stem cell properties of human dental pulp stem cells. J. Dent. Res. 2002, 81, 531–535. [Google Scholar] [CrossRef]
- Yamazaki, H.; Tsuneto, M.; Yoshino, M.; Yamamura, K.; Hayashi, S. Potential of dental mesenchymal cells in developing teeth. Stem Cells 2007, 25, 78–87. [Google Scholar] [CrossRef]
- Wlodarski, K.H.; Reddi, A.H. Alkaline phosphatase as a marker of osteoinductive cells. Calcif. Tissue Int. 1986, 39, 382–385. [Google Scholar] [CrossRef]
- Soda, M.; Saitoh, I.; Murakami, T.; Inada, E.; Iwase, Y.; Noguchi, H.; Shibasaki, S.; Kurosawa, M.; Sawami, T.; Terunuma, M.; et al. Repeated human deciduous tooth-derived dental pulp cell reprogramming factor transfection yields multipotent intermediate cells with enhanced iPS cell formation capability. Sci. Rep. 2019, 9, 1490. [Google Scholar] [CrossRef]
- Cristofalo, V.J.; Parris, N.; Kritchevsky, D. Enzyme activity during the growth and aging of human cells in vitro. J. Cell. Physiol. 1967, 69, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Rikitake, Y. [Immortalization of human dental pulp cells with transfecting of the plasmid, pMT1-neo]. Kokubyo Gakkai Zasshi. J. Stomatol. Soc. Jpn. 1989, 56, 540–561. [Google Scholar] [CrossRef][Green Version]
- Galler, K.M.; Schweikl, H.; Thonemann, B.; D’Souza, R.N.; Schmalz, G. Human pulp-derived cells immortalized with Simian Virus 40 T-antigen. Eur. J. Oral Sci. 2006, 114, 138–146. [Google Scholar] [CrossRef]
- Kim, C.W.; Go, R.E.; Lee, G.A.; Kim, C.D.; Chun, Y.J.; Choi, K.C. Immortalization of human corneal epithelial cells using simian virus 40 large T antigen and cell characterization. J. Pharmacol. Toxicol. Methods 2016, 78, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Perez-Campo, F.M.; May, T.; Zauers, J.; Sanudo, C.; Delgado-Calle, J.; Arozamena, J.; Berciano, M.T.; Lafarga, M.; Riancho, J.A. Generation and characterization of two immortalized human osteoblastic cell lines useful for epigenetic studies. J. Bone Miner. Metab. 2017, 35, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.S.; Gomi, K.; Ashbridge, B.; Moore, M.A.; Arbelaez, V.; Heldrich, J.; Ding, B.S.; Rafii, S.; Staudt, M.R.; Crystal, R.G. Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir. Res. 2013, 14, 135. [Google Scholar] [CrossRef]
- Li, H.; Zhou, J.; Miki, J.; Furusato, B.; Gu, Y.; Srivastava, S.; McLeod, D.G.; Vogel, J.C.; Rhim, J.S. Telomerase-immortalized non-malignant human prostate epithelial cells retain the properties of multipotent stem cells. Exp. Cell Res. 2008, 314, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Hosomichi, J.; Ge, C.; Xu, J.; Franceschi, R.; Kapila, S. Immortalization and characterization of mouse temporomandibular joint disc cell clones with capacity for multi-lineage differentiation. Osteoarthr. Cartil. 2015, 23, 1532–1542. [Google Scholar] [CrossRef]
- Kocki, J.; Kolano, J.; Cioch, M.; Dmoszynska, A.; Wojcierowski, J. The activity of human telomerase in the cells of acute leukaemias. Folia Morphol. 2004, 63, 127–128. [Google Scholar]
- Boldrini, L.; Faviana, P.; Gisfredi, S.; Donati, V.; Zucconi, Y.; Ursino, S.; Simi, P.; Baldinotti, F.; Berti, P.; Galleri, D.; et al. Regulation of telomerase and its hTERT messenger in colorectal cancer. Oncol. Rep. 2004, 11, 395–400. [Google Scholar] [CrossRef]
- Duensing, S.; Duensing, A.; Flores, E.R.; Do, A.; Lambert, P.F.; Munger, K. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinocytes. J. Virol. 2001, 75, 7712–7716. [Google Scholar] [CrossRef] [PubMed]
- Perez-Reyes, N.; Halbert, C.L.; Smith, P.P.; Benditt, E.P.; McDougall, J.K. Immortalization of primary human smooth muscle cells. Proc. Natl. Acad. Sci. USA 1992, 89, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.Y.; Cen, S.; Xu, L.Y.; Cai, W.J.; Chen, M.H.; Shen, J.; Zeng, Y. E6/E7 genes of human papilloma virus type 18 induced immortalization of human fetal esophageal epithelium. Oncol. Rep. 2003, 10, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Kyo, S.; Nakamura, M.; Kiyono, T.; Maida, Y.; Kanaya, T.; Tanaka, M.; Yatabe, N.; Inoue, M. Successful immortalization of endometrial glandular cells with normal structural and functional characteristics. Am. J. Pathol. 2003, 163, 2259–2269. [Google Scholar] [CrossRef]
- He, Y.L.; Wu, Y.H.; He, X.N.; Liu, F.J.; He, X.Y.; Zhang, Y. An immortalized goat mammary epithelial cell line induced with human telomerase reverse transcriptase (hTERT) gene transfer. Theriogenology 2009, 71, 1417–1424. [Google Scholar] [CrossRef]
- Yin, Z.; Wang, Q.; Li, Y.; Wei, H.; Shi, J.; Li, A. A novel method for banking stem cells from human exfoliated deciduous teeth: Lentiviral TERT immortalization and phenotypical analysis. Stem Cell Res. Ther. 2016, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Feng, G.; Song, J.; Zhang, Y.; Yu, Y.; Huang, L.; Zheng, L.; Deng, F. TrAmplification of Human Dental Follicle Cells by piggyBac Transposon - Mediated Reversible Immortalization System. PLoS ONE 2015, 10, e0130937. [Google Scholar] [CrossRef]
- Telles, P.D.; Machado, M.A.; Sakai, V.T.; Nor, J.E. Pulp tissue from primary teeth: New source of stem cells. J. Appl. Oral Sci. Rev. Fob 2011, 19, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Campanella, V. Dental Stem Cells: Current research and future applications. Eur. J. Paediatr. Dent. 2018, 19, 257. [Google Scholar]
- Badal, S.; Badal, V.; Calleja-Macias, I.E.; Kalantari, M.; Chuang, L.S.; Li, B.F.; Bernard, H.U. The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation. Virology 2004, 324, 483–492. [Google Scholar] [CrossRef]
- Sato, M.I.E.; Saitoh, I.; Matsumoto, Y. Microbial and enzyme technology: An efficient and convenient method for MiniPrep analysis of recombinant plasmids. J. Biomed. Sci. Eng. 2014, 4, 105–107. [Google Scholar] [CrossRef][Green Version]
- Sato, M.; Ishikawa, A.; Kimura, M. Direct injection of foreign DNA into mouse testis as a possible in vivo gene transfer system via epididymal spermatozoa. Mol. Reprod. Dev. 2002, 61, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Saitoh, I.; Murakami, T.; Kubota, N.; Nakamura, S.; Watanabe, S.; Inada, E. Intrapancreatic parenchymal injection of cells as a useful tool for allowing a small number of proliferative cells to grow in vivo. Int. J. Mol. Sci. 2017, 18, 1678. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inada, E.; Saitoh, I.; Kubota, N.; Iwase, Y.; Kiyokawa, Y.; Shibasaki, S.; Noguchi, H.; Yamasaki, Y.; Sato, M. piggyBac Transposon-Based Immortalization of Human Deciduous Tooth Dental Pulp Cells with Multipotency and Non-Tumorigenic Potential. Int. J. Mol. Sci. 2019, 20, 4904. https://doi.org/10.3390/ijms20194904
Inada E, Saitoh I, Kubota N, Iwase Y, Kiyokawa Y, Shibasaki S, Noguchi H, Yamasaki Y, Sato M. piggyBac Transposon-Based Immortalization of Human Deciduous Tooth Dental Pulp Cells with Multipotency and Non-Tumorigenic Potential. International Journal of Molecular Sciences. 2019; 20(19):4904. https://doi.org/10.3390/ijms20194904
Chicago/Turabian StyleInada, Emi, Issei Saitoh, Naoko Kubota, Yoko Iwase, Yuki Kiyokawa, Shinji Shibasaki, Hirofumi Noguchi, Youichi Yamasaki, and Masahiro Sato. 2019. "piggyBac Transposon-Based Immortalization of Human Deciduous Tooth Dental Pulp Cells with Multipotency and Non-Tumorigenic Potential" International Journal of Molecular Sciences 20, no. 19: 4904. https://doi.org/10.3390/ijms20194904
APA StyleInada, E., Saitoh, I., Kubota, N., Iwase, Y., Kiyokawa, Y., Shibasaki, S., Noguchi, H., Yamasaki, Y., & Sato, M. (2019). piggyBac Transposon-Based Immortalization of Human Deciduous Tooth Dental Pulp Cells with Multipotency and Non-Tumorigenic Potential. International Journal of Molecular Sciences, 20(19), 4904. https://doi.org/10.3390/ijms20194904





